Photosystem II Extrinsic Proteins and Their Putative Role in Abiotic Stress Tolerance in Higher Plants
Abstract
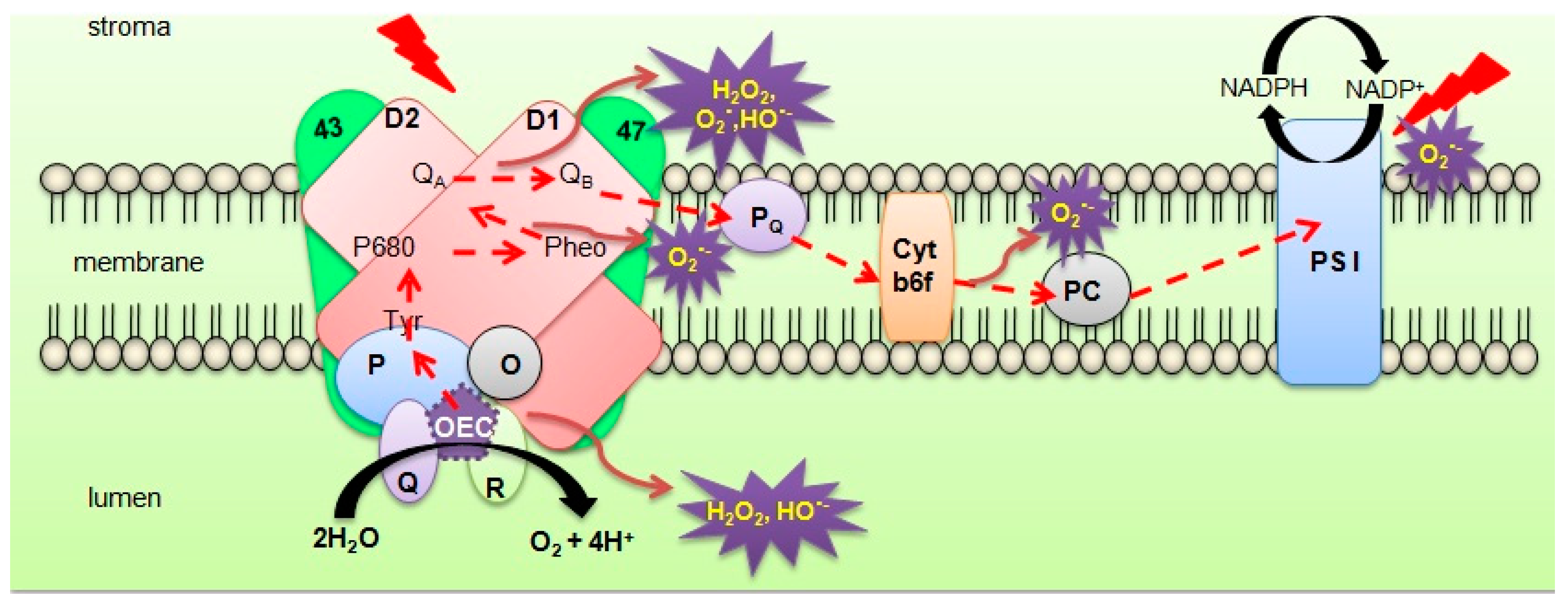
:1. Introduction
2. PsbO
3. PsbP
4. PsbQ
5. PsbR
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CP43 | chloroplast protein 43 |
| LHCII | light-harvesting complex II |
| NDH | NAPDH dehydrogenase |
| PQL-1 and 2 | PsbQ-like protein 1 and 2 |
| Psb | Photosystem b or Photosystem II |
| PSI | Photosystem I |
| PSII | Photosystem II |
| QA | bound plastoquinone |
| ROS | reactive oxygen species |
References
- Gururani, M.A.; Venkatesh, J.; Tran, L.S.P. Regulation of photosynthesis during abiotic stress-induced photoinhibition. Mol. Plant 2015, 8, 1304–1320. [Google Scholar] [CrossRef] [PubMed]
- Lundin, B.; Hansson, M.; Schoefs, B.; Vener, A.V.; Spetea, C. The Arabidopsis PsbO2 protein regulates dephosphorylation and turnover of the photosystem II reaction centre D1 protein. Plant J. 2007, 49, 528–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rochaix, J.-D. Regulation and dynamics of the light-harvesting system. Annu. Rev. Plant Biol. 2014, 65, 287–309. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Bueno, M.L.; Barón, M.; García-Luque, I. PsbO, PsbP, and PsbQ of photosystem II are encoded by gene families in Nicotiana benthamiana. Structure and functionality of their isoforms. Photosynthetica 2011, 49, 573–580. [Google Scholar] [CrossRef]
- Nath, K.; Jajoo, A.; Poudyal, R.S.; Timilsina, R.; Park, Y.S.; Aro, E.-M.; Nam, H.G.; Lee, C.-H. Towards a critical understanding of the photosystem II repair mechanism and its regulation during stress conditions. FEBS Lett. 2013, 587, 3372–3381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Q.; Bricker, T.M. Structural Organization of Proteins on the Oxidizing Side of photosystem II. Two molecules of the 33-kDa manganese-stabilizing proteins per reaction center. J. Biol. Chem. 1992, 267, 25816–25821. [Google Scholar] [PubMed]
- Murata, N.; Takahashi, S.; Nishiyama, Y.; Allakhverdiev, S.I. Photoinhibition of photosystem II under environmental stress. Biochim. Biophys. Acta 2007, 1767, 414–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seibert, M.; Toon, S.; Govindjee; O’Neil, M.P.; Wasielewski, M.R. Primary charge separation in isolated photosystem II reaction centers. Res. Photosynth. 1992, II, 41–44. [Google Scholar]
- Yano, J.; Yachandra, V. Mn4Ca cluster in photosynthesis: Where and how water is oxidized to dioxygen. Chem. Rev. 2014, 114, 4175–4205. [Google Scholar] [CrossRef] [PubMed]
- Roose, J.L.; Frankel, L.K.; Mummadisetti, M.P.; Bricker, T.M. The extrinsic proteins of photosystem II: Update. Planta 2016, 243, 889–908. [Google Scholar] [CrossRef] [PubMed]
- Ifuku, K.; Noguchi, T. Structural Coupling of Extrinsic Proteins with the Oxygen-Evolving Center in Photosystem II. Front Plant Sci. 2016, 7, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, J.; Thakur, J.K. Photosynthesis and Abiotic Stress in Plants. In Biotic and Abiotic Stress Tolerance in Plants; Springer: Singapore, 2018; pp. 27–46. ISBN 9789811090295. [Google Scholar]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017, 856–867. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Koussevitzky, S.; Mittler, R.; Miller, G. ROS and redox signalling in the response of plants to abiotic stress. Plant. Cell Environ. 2012, 35, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Roach, T.; Krieger-Liszkay, A. Regulation of photosynthetic electron transport and photoinhibition. Curr. Protein Pept. Sci. 2014, 15, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell 2005, 17, 1866–1875. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, Y.; Yamamoto, H.; Allakhverdiev, S.I.; Inaba, M.; Yokota, A.; Murata, N. Oxidative stress inhibits the repair of photodamage to the photosynthetic machinery. EMBO J. 2001, 20, 5587–5594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gururani, M.A.; Upadhyaya, C.P.; Strasser, R.J.; Woong, Y.J.; Park, S.W. Physiological and biochemical responses of transgenic potato plants with altered expression of PSII manganese stabilizing protein. Plant Physiol. Biochem. 2012, 58, 182–194. [Google Scholar] [CrossRef] [PubMed]
- Tikkanen, M.; Grieco, M.; Kangasjärvi, S.; Aro, E.-M. Thylakoid protein phosphorylation in higher plant chloroplasts optimizes electron transfer under fluctuating light. Plant Physiol. 2010, 152, 723–735. [Google Scholar] [CrossRef] [PubMed]
- Cazzaniga, S.; Dall’Osto, L.; Kong, S.G.; Wada, M.; Bassi, R. Interaction between avoidance of photon absorption, excess energy dissipation and zeaxanthin synthesis against photooxidative stress in Arabidopsis. Plant J. 2013, 76, 568–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gururani, M.A.; Upadhyaya, C.P.; Strasser, R.J.; Yu, J.W.; Park, S.W. Evaluation of abiotic stress tolerance in transgenic potato plants with reduced expression of PSII manganese stabilizing protein. Plant Sci. 2013, 198, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Herbstova, M.; Tietz, S.; Kinzel, C.; Turkina, M.V.; Kirchhoff, H. Architectural switch in plant photosynthetic membranes induced by light stress. Proc. Natl. Acad. Sci. USA 2012, 109, 20130–20135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toth, S.Z.; Nagy, V.; Puthur, J.T.; Kovacs, L.; Garab, G. The Physiological Role of Ascorbate as Photosystem II Electron Donor: Protection against Photoinactivation in Heat-Stressed Leaves. Plant Physiol. 2011, 156, 382–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, N.R. Chlorophyll Fluorescence: A Probe of Photosynthesis In Vivo. Annu. Rev. Plant Biol. 2008, 59, 89–113. [Google Scholar] [CrossRef] [PubMed]
- Pospíšil, P.; Yamamoto, Y. Damage to photosystem II by lipid peroxidation products. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Aro, E.-M.; Millar, A.H. Mechanisms of Photodamage and Protein Turnover in Photoinhibition. Trends Plant Sci. 2018, 23, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Bricker, T.; Burnap, R. Photosystem II Advances in Photosynthesis and Respiration; Wydrzynski, T., Satoh, K., Freeman, J., Eds.; Springer: Dordrecht, The Netherlands, 2005; pp. 95–120. ISBN 9781402042492. [Google Scholar]
- Kirchhoff, H. Diffusion of molecules and macromolecules in thylakoid membranes. Biochim. Biophys. Acta Bioenerg. 2014, 1837, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Kirchhoff, H. Molecular crowding and order in photosynthetic membranes. Trends Plant Sci. 2008, 13, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Fristedt, R.; Willig, A.; Granath, P.; Crèvecoeur, M.; Rochaix, J.-D.; Vener, A.V. Phosphorylation of photosystem II controls functional macroscopic folding of photosynthetic membranes in Arabidopsis. Plant Cell 2009, 21, 3950–3964. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Aminaka, R.; Yoshioka, M.; Khatoon, M.; Komayama, K.; Takenaka, D.; Yamashita, A.; Nijo, N.; Inagawa, K.; Morita, N.; et al. Quality control of photosystem II: Impact of light and heat stresses. Photosynth. Res. 2008, 98, 589–608. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.; Zhang, M.; Zhao, Y.; He, X.; Ding, C.; Wang, S.; Feng, Y.; Song, X.; Li, P.; Wang, B. Identification of Salt Tolerance-related microRNAs and Their Targets in Maize (Zea mays L.) Using High-throughput Sequencing and Degradome Analysis. Front. Plant Sci. 2017, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, S.; Tuteja, N. Cold, salinity and drought stresses: An overview. Arch. Biochem. Biophys. 2005, 444, 139–158. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.; Zhang, J.; Ma, Y.; Pan, X.; Dong, C.; Pang, S.; He, S.; Deng, L.; Yi, S.; Zheng, Y.; et al. Combined analysis of mRNA and miRNA identifies dehydration and salinity responsive key molecular players in citrus roots. Sci. Rep. 2017, 7, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Choi, J.; An, G.; Kim, S.-R. Ectopic Expression of OsSta2 Enhances Salt Stress Tolerance in Rice. Front. Plant Sci. 2017, 8, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, M.A.; Kumar, D.; Rajwanshi, R.; Strasser, R.J.; Tsimilli-Michael, M.; Govindjee; Sarin, N.B. Overexpression of gamma-tocopherol methyl transferase gene in transgenic Brassica juncea plants alleviates abiotic stress: Physiological and chlorophyll a fluorescence measurements. Biochim. Biophys. Acta 2010, 1797, 1428–1438. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.S.; Kim, Y.S.; Redillas, M.C.F.R.; Jang, G.; Jung, H.; Bang, S.W.; Choi, Y.D.; Ha, S.H.; Reuzeau, C.; Kim, J.K. OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol. J. 2013, 11, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Kuwabara, T.; Murata, N. Purification and Characterization of the 33 kDa protein of spinach chloroplasts. Biochim. Biophys. Acta 1979, 581, 228–236. [Google Scholar] [CrossRef]
- De Las Rivas, J.; Barber, J. Analysis of the structure of the PsbO protein and its implications. Photosynth. Res. 2004, 81, 329–343. [Google Scholar] [CrossRef] [PubMed]
- Duchoslav, M.; Fischer, L. Parallel subfunctionalisation of PsbO protein isoforms in angiosperms revealed by phylogenetic analysis and mapping of sequence variability onto protein structure. BMC Plant Biol. 2015, 15, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Murakami, R.; Ifuku, K.; Takabayashi, A.; Shikanai, T.; Endo, T.; Sato, F. Functional dissection of two Arabidopsis PsbO proteins PsbO1 and PsbO2. FEBS J. 2005, 272, 2165–2175. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.W.; Barber, J. Identification of a calcium-binding site in the PsbO protein of photosystem II. Biochemistry 2006, 45, 4128–4130. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y. Quality Control of Photosystem II: The Mechanisms for Avoidance and Tolerance of Light and Heat Stresses are Closely Linked to Membrane Fluidity of the Thylakoids. Front. Plant Sci. 2016, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Murakami, R.; Ifuku, K.; Takabayashi, A.; Shikanai, T.; Endo, T.; Sato, F. Characterization of an Arabidopsis thaliana mutant with impaired psbO, one of two genes encoding extrinsic 33-kDa proteins in photosystem II. FEBS Lett. 2002, 523, 138–142. [Google Scholar] [CrossRef]
- Yi, X.; McChargue, M.; Laborde, S.; Frankel, L.K.; Bricker, T.M. The manganese-stabilizing protein is required for photosystem II assembly/stability and photoautotrophy in higher plants. J. Biol. Chem. 2005, 280, 16170–16174. [Google Scholar] [CrossRef] [PubMed]
- Bricker, T.M.; Frankel, L.K. The psbo1 mutant of Arabidopsis cannot efficiently use calcium in support of oxygen evolution by photosystem II. J. Biol. Chem. 2008, 283, 29022–29027. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, S.; Chow, W.; Yamori, W.; Evans, J.; Kaines, S.; Badger, M.; Caemmerer, S. Antisense reductions in the PsbO protein of photosystem II leads to decreased quantum yield but similar maximal photosynthetic rates. J. Exp. Bot. 2012, 63, 4781–4795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ifuku, K.; Ishihara, S.; Sato, F. Molecular Functions of Oxygen-Evolving Complex Family Proteins in Photosynthetic Electron Flow. J. Integr. Plant Biol. 2010, 52, 723–734. [Google Scholar] [CrossRef] [PubMed]
- Umena, Y.; Kawakami, K.; Shen, J.R.; Kamiya, N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9Å. Nature 2011, 473, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Caffarri, S.; Kouřil, R.; Kereïche, S.; Boekema, E.J.; Croce, R. Functional architecture of higher plant photosystem II supercomplexes. EMBO J. 2009, 28, 3052–3063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slowik, D.; Rossmann, M.; Konarev, P.V.; Irrgang, K.D.; Saenger, W. Structural investigation of PsbO from plant and cyanobacterial photosystem II. J. Mol. Biol. 2011, 407, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Rapacz, M.; Ga̧sior, D.; Kościelniak, J.; Kosmala, A.; Zwierzykowski, Z.; Humphreys, M.W. The role of the photosynthetic apparatus in cold acclimation of Lolium multiflorum. Characteristics of novel genotypes low-sensitive to PSII over-reduction. Acta Physiol. Plant. 2007, 29, 309–316. [Google Scholar] [CrossRef]
- Ji, S.H.; Gururani, M.A.; Chun, S.-C. Isolation and characterization of plant growth promoting endophytic diazotrophic bacteria from Korean rice cultivars. Microbiol. Res. 2014, 169. [Google Scholar] [CrossRef] [PubMed]
- Oukarroum, A.; El Madidi, S.; Schansker, G.; Strasser, R.J. Probing the responses of barley cultivars (Hordeum vulgare L.) by chlorophyll a fluorescence OLKJIP under drought stress and re-watering. Environ. Exp. Bot. 2007, 60, 438–446. [Google Scholar] [CrossRef]
- Gururani, M.A.; Ganesan, M.; Song, I.-J.; Han, Y.; Kim, J.-I.; Lee, H.-Y.; Song, P.-S. Transgenic Turfgrasses Expressing Hyperactive Ser599Ala Phytochrome A Mutant Exhibit Abiotic Stress Tolerance. J. Plant Growth Regul. 2016, 35. [Google Scholar] [CrossRef]
- Gururani, M.A.; Venkatesh, J.; Ghosh, R.; Strasser, R.J.; Ponpandian, L.N.; Bae, H. Chlorophyll-a fluorescence evaluation of PEG-induced osmotic stress on PSII activity in Arabidopsis plants expressing SIP1. Plant Biosyst. Int. J. Deal. All Asp. Plant Biol. 2017, 3504, 1–8. [Google Scholar] [CrossRef]
- Henmi, T.; Miyao, M.; Yamamoto, Y. Release and Reactive-Oxygen-Mediated Damage of the Oxygen-Evolving Complex Subunits of PSII during Photoinhibition. Plant Cell Physiol. 2004, 45, 243–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pawłowicz, I.; Kosmala, A.; Rapacz, M. Expression pattern of the psbO gene and its involvement in acclimation of the photosynthetic apparatus during abiotic stresses in Festuca arundinacea and F. pratensis. Acta Physiol. Plant. 2012, 34, 1915–1924. [Google Scholar] [CrossRef]
- Kosmala, A.; Bocian, A.; Rapacz, M.; Jurczyk, B.; Zwierzykowski, Z. Identification of leaf proteins differentially accumulated during cold acclimation between Festuca pratensis plants with distinct levels of frost tolerance. J. Exp. Bot. 2009, 60, 3595–3609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, L.; Lipavska, H.; Hausman, J.F.; Opatrny, Z. Morphological and molecular characterization of a spontaneously tuberizing potato mutant: An insight into the regulatory mechanisms of tuber induction. BMC Plant Biol. 2008, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Anjum, N.A.; Gill, S.S.; Gill, R.; Hasanuzzaman, M.; Duarte, A.C.; Pereira, E.; Ahmad, I.; Tuteja, R.; Tuteja, N. Metal/metalloid stress tolerance in plants: Role of ascorbate, its redox couple, and associated enzymes. Protoplasma 2014. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Hargett, S.R.; Frankel, L.K.; Bricker, T.M. The effects of simultaneous RNAi suppression of PsbO and PsbP protein expression in photosystem II of Arabidopsis. Photosynth. Res. 2008, 98, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Li, Q.T.; Chu, Y.N.; Reiter, R.J.; Yu, X.M.; Zhu, D.H.; Zhang, W.K.; Ma, B.; Lin, Q.; Zhang, J.S.; et al. Melatonin enhances plant growth and abiotic stress tolerance in soybean plants. J. Exp. Bot. 2015, 66, 695–707. [Google Scholar] [CrossRef] [PubMed]
- Głowacka, K.; Kromdijk, J.; Kucera, K.; Xie, J.; Cavanagh, A.P.; Leonelli, L.; Leakey, A.D.B.; Ort, D.R.; Niyogi, K.K.; Long, S.P. Photosystem II Subunit S overexpression increases the efficiency of water use in a field-grown crop. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Ifuku, K.; Yamamoto, Y.; Ono, T.-A.; Ishihara, S.; Sato, F. PsbP protein, but not PsbQ protein, is essential for the regulation and stabilization of photosystem II in higher plants. Plant Physiol. 2005, 139, 1175–1184. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, S.; Takabayashi, A.; Ido, K.; Endo, T.; Ifuku, K.; Sato, F. Distinct Functions for the Two PsbP-Like Proteins PPL1 and PPL2 in the Chloroplast Thylakoid Lumen. Plant Physiol. 2007, 145, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Allahverdiyeva, Y.; Suorsa, M.; Rossi, F.; Pavesi, A.; Kater, M.M.; Antonacci, A.; Tadini, L.; Pribil, M.; Schneider, A.; Wanner, G.; et al. Arabidopsis plants lacking PsbQ and PsbR subunits of the oxygen-evolving complex show altered PSII super-complex organization and short-term adaptive mechanisms. Plant J. 2013, 671–684. [Google Scholar] [CrossRef] [PubMed]
- Matsui, S.; Ishihara, S.; Ido, K.; Ifuku, K.; Sato, F. Functional analysis of PsbP-like protein 1 (PPL1) in Arabidopsis. In Photosynthesis Research for Food, Fuel and the Future: 15th International Congress on Photosynthesis; Tingyun, K., Congming, L., Lixin, Z., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 415–417. [Google Scholar]
- Yi, X.; Hargett, S.R.; Frankel, L.K.; Bricker, T.M. The PsbP protein, but not the PsbQ protein, is required for normal thylakoid architecture in Arabidopsis thaliana. FEBS Lett. 2009, 583, 2142–2147. [Google Scholar] [CrossRef] [PubMed]
- Roose, J.L.; Frankel, L.K.; Bricker, T.M. Developmental Defects in Mutants of the PsbP Domain Protein 5 in Arabidopsis thaliana. PLoS ONE 2011, 6, e28624. [Google Scholar] [CrossRef] [PubMed]
- Kang, L.; Kim, H.S.; Kwon, Y.S.; Ke, Q.; Ji, C.Y.; Park, S.; Lee, H.; Deng, X.; Kwak, S. IbOr Regulates Photosynthesis under Heat Stress by Stabilizing IbPsbP in Sweetpotato. Front. Plant Sci. 2017, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yabuta, S.; Ifuku, K.; Takabayashi, A.; Ishihara, S.; Ido, K.; Ishikawa, N.; Endo, T.; Sato, F. Three PsbQ-Like Proteins are Required for the Function of the Chloroplast NAD(P)H Dehydrogenase Complex in Arabidopsis Editor-in-Chief’s choice. Plant Cell Physiol. 2018, 1, 866–876. [Google Scholar] [CrossRef]
- Yi, X.; Hargett, S.; Frankel, L.; Bricker, T. The PsbQ protein is required in Arabidopsis for photosystem II assembly/stability and photoautotrophy under low light conditions. J. Biol. Chem. 2006, 281, 26260–26267. [Google Scholar] [CrossRef] [PubMed]
- Stockhaus, J.; Hofer, M.; Renger, G.; Westhoff, P.; Wydrzynski, T.; Willmitzer, L. Anti-sense RNA efficiently inhibits formation of the 10 kd polypeptide of photosystem 11 in transgenic potato plants: Analysis of the role of the 10 kd protein. EMBO J. 1990, 9, 3013–3021. [Google Scholar] [CrossRef] [PubMed]
- Suorsa, M.; Sirpio, S.; Allahverdiyeva, Y.; Paakkarinen, V.; Mamedov, F. PsbR, a Missing Link in the Assembly of the Oxygen-evolving Complex of Plant Photosystem II. J. Biol. Chem. 2006, 281, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Allahverdiyeva, Y.; Mamedov, F.; Suorsa, M.; Styring, S.; Vass, I.; Aro, E.M. Insights into the function of PsbR protein in Arabidopsis thaliana. Biochim. Biophys. ActaBioenerg. 2007, 1767, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Cohen, J.D.; Gardner, G. Low-fluence red light increases the transport and biosynthesis of auxin. Plant Physiol. 2011, 157, 891–904. [Google Scholar] [CrossRef] [PubMed]
- Roose, J.L.; Frankel, L.K.; Bricker, T.M. The PsbP-Domain Protein 1 Functions in the Assembly of Lumenal Domains in Photosystem I. J. Biol. Chem. 2014, 277, 8354–8365. [Google Scholar] [CrossRef] [PubMed]
- Seidler, A. The extrinsic polypeptides of photosystem II. Biochim. Biophys. ActaBioenerg. 1996, 1277, 35–60. [Google Scholar] [CrossRef]
- Tomita, M.; Ifuku, K.; Sato, F.; Noguchi, T. FTIR evidence that the PsbP extrinsic protein induces protein conformational changes around the oxygen-evolving Mn cluster in photosystem II. Biochemistry 2009, 48, 6318–6325. [Google Scholar] [CrossRef] [PubMed]
- Enami, I.; Okumura, A.; Nagao, R.; Suzuki, T.; Iwai, M.; Shen, J.R. Structures and functions of the extrinsic proteins of photosystem II from different species. Photosynth. Res. 2008, 98, 349–363. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, Y.; Schröder, W.P.; Funk, C. Functional analysis of the PsbP-like protein (sll1418) in Synechocystis sp. PCC 6803. Photosynth. Res. 2005, 84, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Sato, N. Molecular Phylogenetics and Evolution Phylogenomic and structural modeling analyses of the PsbP superfamily reveal multiple small segment additions in the evolution of photosystem II-associated PsbP protein in green plants. Mol. Phylogenet. Evol. 2010, 56, 176–186. [Google Scholar] [CrossRef] [PubMed]
- Järvi, S.; Gollan, P.J.; Aro, E. Understanding the roles of the thylakoid lumen in photosynthesis regulation. Front. Plant Sci. 2013, 4, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ifuku, K.; Endo, T.; Shikanai, T.; Aro, E. Structure of the Chloroplast NADH Dehydrogenase-Like Complex: Nomenclature for Nuclear-Encoded Subunits. Plant Cell Physiol. 2011, 52, 1560–1568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopecky, V.K., Jr.; Kohoutova, J.; Lapkouski, M.; Hofbauerova, K.; Dulebo, A.; Kaftan, D.; Kuta, I.; Sovova, Z.; Ettrichova, O.; Gonza, S.; et al. Raman Spectroscopy Adds Complementary Detail to the High-Resolution X-ray Crystal Structure of Photosynthetic PsbP from Spinacia oleracea. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ido, K.; Nield, J.; Fukao, Y.; Nishimura, T.; Sato, F.; Ifuku, K. Cross-linking Evidence for Multiple Interactions of the PsbP and PsbQ Proteins in a Higher Plant Photosystem II. J. Biol. Chem. 2014, 289, 20150–20157. [Google Scholar] [CrossRef] [PubMed]
- Thornton, L.E.; Ohkawa, H.; Roose, J.L.; Kashino, Y.; Keren, N.; Pakrasi, H.B. Homologs of Plant PsbP and PsbQ Proteins Are Necessary for Regulation of Photosystem II Activity in the Cyanobacterium Synechocystis 6803. Plant Cell 2004, 16, 2164–2175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bricker, T.M.; Roose, J.L.; Fagerlund, R.D.; Frankel, L.K.; Eaton-Rye, J.J. The extrinsic proteins of Photosystem II. Biochim. Biophys. Acta 2012, 1817, 121–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, M.; Sauer, U.H.; Schro, W.P. Purification, crystallization communications Purification, crystallization and preliminary X-ray analysis of PPD6, a PsbP-domain protein from Arabidopsis thaliana crystallization communications. Acta crystallogr. Sect. F 2012, 68, 278–280. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yang, H.; Lu, Q.; Wen, X.; Chen, F.; Peng, L.; Zhang, L.; Lu, C. PSBP-DOMAIN PROTEIN1, a Nuclear-Encoded Thylakoid Lumenal Protein, Is Essential for Photosystem I Assembly in Arabidopsis. Plant Cell 2012, 24, 4992–5006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akabori, K. The role of lipids and 17-kDa protein in enhancing the recovery of 02 evolution in cholate-treated thylakoid membranes. FEBS Lett. 1984, 173, 36–40. [Google Scholar] [CrossRef]
- Miyao, M.; Murata, N. The Cl− effect on photosynthetic oxygen evolution: Interaction of Cl− with WkDa, 24-kDa and 33-kDa proteins. FEBS Lett. 1985, 180, 303–308. [Google Scholar] [CrossRef]
- Ohta, H.; Suzuki, T.; Ueno, M.; Okumura, A.; Yoshihara, S.; Shen, J. Extrinsic proteins of photosystem II An intermediate member of the PsbQ protein family in red algal PS II. Eur. J. Biochem. 2003, 4163, 4156–4163. [Google Scholar] [CrossRef]
- Zienkiewicz, M.; Krupnik, T.; Drożak, A.; Wasilewska, W.; Golke, A. Deletion of psbQ’ gene in Cyanidioschyzon merolae reveals the function of extrinsic PsbQ’ in PSII. Plant Mol. Biol. 2018, 96, 135–149. [Google Scholar] [CrossRef] [PubMed]
- Kakiuchi, S.; Uno, C.; Ido, K.; Nishimura, T.; Noguchi, T. The PsbQ protein stabilizes the functional binding of the PsbP protein to photosystem II in higher plants. Biochim. Biophys. Acta Bioenerg. 2012, 1817, 1346–1351. [Google Scholar] [CrossRef] [PubMed]
- Glenn, E.P.; Brown, J.J.; Blumwald, E. Salt Tolerance and Crop Potential of Halophytes Salt Tolerance and Crop Potential of Halophytes. Crit. Rev. Plant Sci. 1999, 18, 227–255. [Google Scholar] [CrossRef]
- Pagliano, C.; La Rocca, N.; Andreucci, F.; Deak, Z.; Vass, I.; Rascio, N.; Barbato, R. The extreme halophyte Salicornia veneta is depleted of the extrinsic PsbQ and PsbP proteins of the oxygen-evolving complex without loss of functional activity. Ann. Bot. 2009, 103, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Boekema, E.; van Breeman, J.; van Roon, H.; Dekker, J. Conformational Changes in Photosystem II Supercomplexes upon Removal of extrinsic subunits. Biochemistry 2000, 39, 12907–12915. [Google Scholar] [CrossRef] [PubMed]
- Ljungberg, U.; Akerlund, H.; Andersson, B. Isolation and characterization of the 10-kDa and 22-kDa polypeptides of higher plant photosystem 2. Eur. J. Biochem. 1986, 482, 477–482. [Google Scholar] [CrossRef]
- Liu, H.; Frankel, L.K.; Bricker, T.M. Characterization and complementation of a psbR mutant in Arabidopsis thaliana. Arch. Biochem. Biophys. 2009, 489, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Ifuku, K. The PsbP and PsbQ family proteins in the photosynthetic machinery of chloroplasts. Plant Physiol. Biochem. 2014, 81, 108–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belhaj, K.; Chaparro-garcia, A.; Kamoun, S.; Patron, N.J.; Nekrasov, V. Editing plant genomes with CRISPR/Cas9. Curr. Opin. Biotechnol. 2015, 32, 76–84. [Google Scholar] [CrossRef] [PubMed]

| S. No. | Plant | Stress | PSII Extrinsic Gene/Protein Mutants | Observation(s) | Reference |
|---|---|---|---|---|---|
| 1 | Arabidopsis thaliana | - | psbO1/psbO2 mutant | PsbO1/PsbO2 are active in photosynthesis and PsbO2 substitutes PsbO1 in PSII | [2] |
| 2 | Solanum tuberosum | Abiotic stress (salt, heavy metal, and osmotic stress) | psbO mutant and psbO overexpressed | Mutant plant showed increased tuberization, chlorophyll content, plant height, leaf number, and increase in ROS-scavenging enzymes | [19,22] |
| 3 | Nicotiana tabacum | High-intensity light | PsbO | No phenotypic change | [42] |
| 4 | Arabidopsis thaliana | - | psbO mutant | Reduced quantum yield | [48] |
| 5 | Spinacia oleracea | - | psbO mutant | Photo-inhibition, accumulation of D1 and CP43, detrimental effect on PSII binding | [58] |
| 6 | Festuca arundinacea and Festuca pratensis | Cold and drought | PsbO expression pattern | Higher stability of PSII during droughts. The difference in photochemical efficiency and PsbO accumulation during cold | [59] |
| 7 | Solanum tuberosum | - | psbO mutant | Reduced rooting, delayed senescence, basal branching, and enhanced tuberization. | [61] |
| 8 | Arabidopsis thaliana | - | psbO mutant | Loss of D1, CP43, and fluorescence yield | [63] |
| 9 | Glycine max | Melatonin treatment | Upregulation of extrinsic and intrinsic proteins | [64] | |
| 10 | Nicotiana tabacum | PsbS overexpressed | Reduced PsbO and increased water efficiency | [65] | |
| 11 | Nicotiana tabacum | - | psbP mutant | Loss of quantum yield and PSII core proteins, loss of manganese cluster | [66] |
| 12 | Arabidopsis thaliana | High-intensity light | PPL1 mutant | Photobleaching | [67] |
| 13 | Arabidopsis thaliana | - | psbq1 and psbq2 mutants | Phenotypic changes, rapid transitions, and low LHCII phosphorylation | [68] |
| 14 | Arabidopsis thaliana | - | psbR mutants | Reduced PSII activity, impaired PSII–LHCII accumulation, and effects on state transitions | [68] |
| 15 | Arabidopsis thaliana | - | psbP1 mutant | Unable to grow photo-autotrophically | [68] |
| 16 | Arabidopsis thaliana | PPL2 mutant | Defective NDH | [69] | |
| 17 | Arabidopsis thaliana | - | psbP mutant | Loss of quantum yield and PSII core proteins | [70] |
| 18 | Arabidopsis thaliana | - | PPD-5 mutant | Decreased NDH with developmental and phenotypical defects | [71] |
| 19 | Ipomea batatas | Environmental stress especially heat stress | IbOr | Higher chlorophyll content and PSII efficiency | [72] |
| 20 | Arabidopsis thaliana | Biotic and Abiotic stress | PQL-1 and PQL-2 mutants | Reduction in NDH accumulation | [73] |
| 21 | Arabidopsis thaliana | Low-light conditions | PsbQ | Exhibited phenotypic changes such as yellowing and death | [74] |
| 22 | Solanum tuberosum | psbR mutant | Retardation of QA- reoxidation | [75] | |
| 23 | Arabidopsis thaliana | - | psbR mutant | Detrimental effects in the binding of psbP and PsbQ | [76,77] |
| 24 | Arabidopsis thaliana | - | PPD-1 mutant | Unable to grow photo-autotrophically. Loss of PSI stability, loss of integrity of PsaA and PsaB incorporation into thylakoid membrane | [78,79] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sasi, S.; Venkatesh, J.; Daneshi, R.F.; Gururani, M.A. Photosystem II Extrinsic Proteins and Their Putative Role in Abiotic Stress Tolerance in Higher Plants. Plants 2018, 7, 100. https://doi.org/10.3390/plants7040100
Sasi S, Venkatesh J, Daneshi RF, Gururani MA. Photosystem II Extrinsic Proteins and Their Putative Role in Abiotic Stress Tolerance in Higher Plants. Plants. 2018; 7(4):100. https://doi.org/10.3390/plants7040100
Chicago/Turabian StyleSasi, Shina, Jelli Venkatesh, Rawya Fatohllah Daneshi, and Mayank Anand Gururani. 2018. "Photosystem II Extrinsic Proteins and Their Putative Role in Abiotic Stress Tolerance in Higher Plants" Plants 7, no. 4: 100. https://doi.org/10.3390/plants7040100
APA StyleSasi, S., Venkatesh, J., Daneshi, R. F., & Gururani, M. A. (2018). Photosystem II Extrinsic Proteins and Their Putative Role in Abiotic Stress Tolerance in Higher Plants. Plants, 7(4), 100. https://doi.org/10.3390/plants7040100






