Identification of Genomic Regions Contributing to Protein Accumulation in Wheat under Well-Watered and Water Deficit Growth Conditions
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Field Growth Conditions
2.2. Estimation of Grain Protein Content (GPC)
2.3. Single Nucleotide Polymorphism (SNP)
2.4. Statistical Analysis
| Source | Type III Expected Mean Square |
| Environment (Env) | Var (Error) + 45.372 Var (IBlock (Env × Rep)) + Q (Env, Env × Genotypes) |
| Incomplete block (Env × block) | Var (Error) + 36.829 Var (IBlock (Env × Rep)) |
| Accessions | Var (Error) + Q (Genotypes, Env × Genotypes) |
| Env × Accessions | Var (Error) + Q (Env × Genotypes) |
2.5. Association Mapping
3. Results
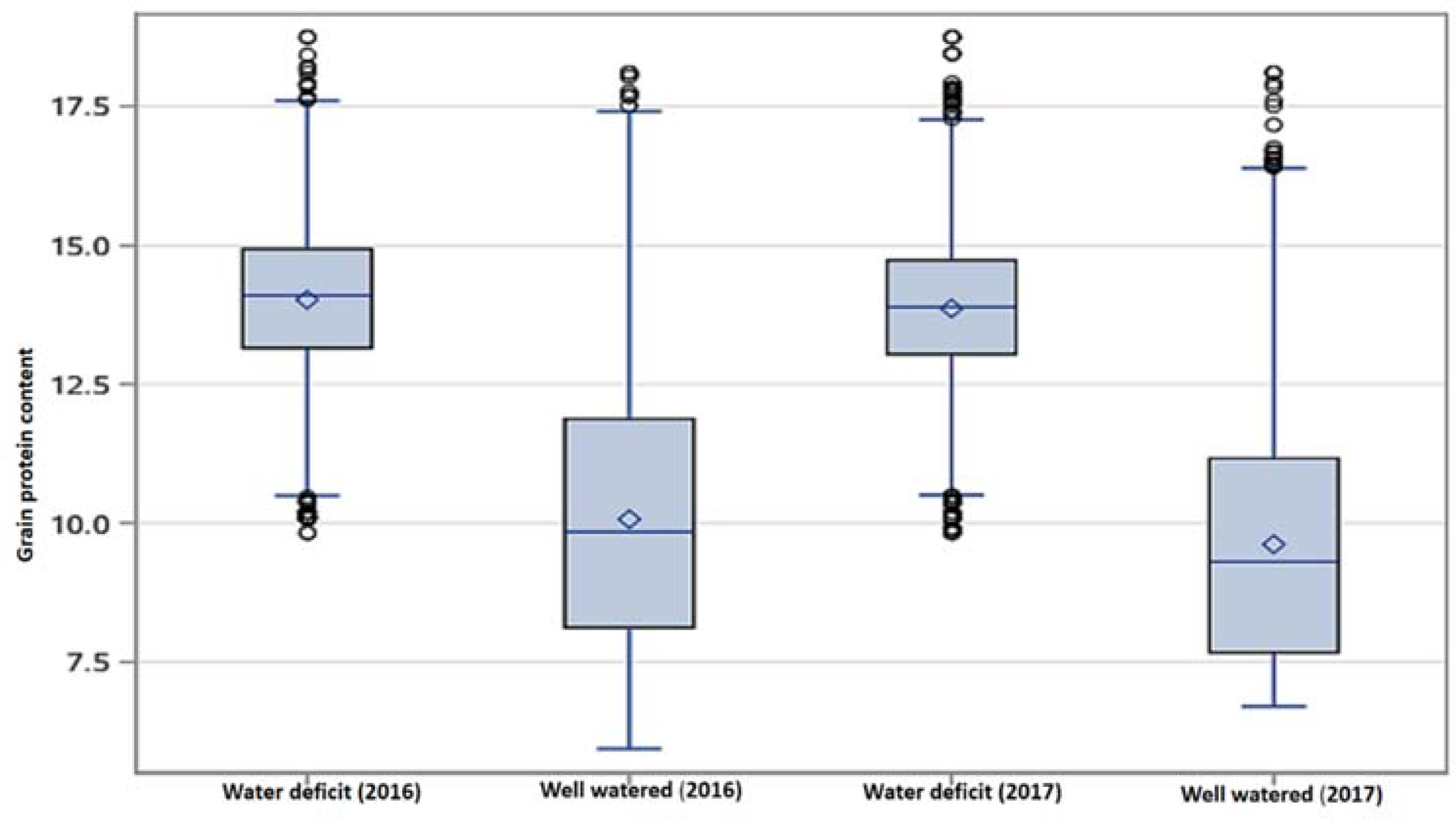
3.1. Grain Protein Content (GPC)
3.2. Association Mapping for Grain Protein Content
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hawkesford, M.J. Reducing the reliance on nitrogen fertilizer for wheat production. J. Cereal Sci. 2014, 59, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Würschum, T.; Leiser, W.L.; Kazman, E.; Longin, C.F.H. Genetic control of protein content and sedimentation volume in European winter wheat cultivars. Theor. Appl. Genet. 2016, 129, 1685–1696. [Google Scholar] [CrossRef] [PubMed]
- Day, L. Proteins from land plants? Potential resources for human nutrition and food security. Trends Food Sci. Technol. 2013, 32, 25–42. [Google Scholar] [CrossRef]
- Cantu, D.; Pearce, S.P.; Distelfeld, A.; Christiansen, M.W.; Uauy, C.; Akhunov, E.; Fahima, T.; Dubcovsky, J. Effect of the down-regulation of the high Grain Protein Content (GPC) genes on the wheat transcriptome during monocarpic senescence. BMC Genome 2011, 12, 492. [Google Scholar] [CrossRef] [PubMed]
- Triboi, E. Environmentally-induced changes in protein composition in developing grains of wheat are related to changes in total protein content. J. Exp. Bot. 2003, 54, 1731–1742. [Google Scholar] [CrossRef] [PubMed]
- Bertheloot, J.; Martre, P.; Andrieu, B. Dynamics of Light and Nitrogen Distribution during Grain Filling within Wheat Canopy. Plant Physiol. 2008, 148, 1707–1720. [Google Scholar] [CrossRef] [PubMed]
- Beta, T.; Nam, S.; Dexter, J.E.; Sapirstein, H.D. Phenolic Content and Antioxidant Activity of Pearled Wheat and Roller-Milled Fractions. Cereal Chem. J. 2005, 82, 390–393. [Google Scholar] [CrossRef]
- Li, X.; Zhou, L.; Liu, F.; Zhou, Q.; Cai, J.; Wang, X.; Dai, T.; Cao, W.; Jiang, D. Variations in Protein Concentration and Nitrogen Sources in Different Positions of Grain in Wheat. Front. Plant Sci. 2016, 7, 942. [Google Scholar] [CrossRef] [PubMed]
- Arduini, I.; Masoni, A.; Ercoli, L.; Mariotti, M. Grain yield, and dry matter and nitrogen accumulation and remobilization in durum wheat as affected by variety and seeding rate. Eur. J. Agron. 2006, 25, 309–318. [Google Scholar] [CrossRef]
- Lantican, M.A.; Braun, H.J.; Payne, T.S.; Singh, R.P.; Sonder, K.; Baum, M.; van Ginkel, M.; Erenstein, O. Impacts of International Wheat Improvement Research, 1994–2014; CIMMYT: Texcoco de Mora, Mexico, 2016; ISBN 9786078263554. [Google Scholar]
- Ashraf, M. Stress-Induced Changes in Wheat Grain Composition and Quality. Crit. Rev. Food Sci. Nutr. 2014, 54, 1576–1583. [Google Scholar] [CrossRef] [PubMed]
- Blanco, A.; Mangini, G.; Giancaspro, A.; Giove, S.; Colasuonno, P.; Simeone, R.; Signorile, A.; De Vita, P.; Mastrangelo, A.M.; Cattivelli, L.; et al. Relationships between grain protein content and grain yield components through quantitative trait locus analyses in a recombinant inbred line population derived from two elite durum wheat cultivars. Mol. Breed 2012, 30, 79–92. [Google Scholar] [CrossRef]
- De Santis, M.A.; Giuliani, M.M.; Giuzio, L.; De Vita, P.; Lovegrove, A.; Shewry, P.R.; Flagella, Z. Differences in gluten protein composition between old and modern durum wheat genotypes in relation to 20th century breeding in Italy. Eur. J. Agron. 2017, 87, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Ravier, C.; Meynard, J.M.; Cohan, J.P.; Gate, P.; Jeuffroy, M.H. Early nitrogen deficiencies favor high yield, grain protein content and N use efficiency in wheat. Eur. J. Agron. 2017, 89, 16–24. [Google Scholar] [CrossRef]
- Gaju, O.; Allard, V.; Martre, P.; Le Gouis, J.; Moreau, D.; Bogard, M.; Hubbart, S.; Foulkes, M.J. Nitrogen partitioning and remobilization in relation to leaf senescence, grain yield and grain nitrogen concentration in wheat cultivars. Field Crops Res. 2014, 155, 213–223. [Google Scholar] [CrossRef]
- Mondal, S.; Rutkoski, J.E.; Velu, G.; Singh, P.K.; Crespo-Herrera, L.A.; Guzmán, C.; Bhavani, S.; Lan, C.; He, X.; Singh, R.P. Harnessing Diversity in Wheat to Enhance Grain Yield, Climate Resilience, Disease and Insect Pest Resistance and Nutrition Through Conventional and Modern Breeding Approaches. Front. Plant Sci. 2016, 7, 991. [Google Scholar] [CrossRef] [PubMed]
- Shewry, P.R.; Hey, S.J. The contribution of wheat to human diet and health. Food Energy Secur. 2015, 4, 178–202. [Google Scholar] [CrossRef] [PubMed]
- Lindeque, R.C. Protein Quality vs. Quantity in South African Commercial Bread Wheat Cultivars. Ph.D. Thesis, University of the Free State, Bloemfontein, South Africa, 2016. [Google Scholar]
- Soriano, J.M.; Villegas, D.; Aranzana, M.J.; García Del Moral, L.F.; Royo, C. Genetic structure of modern durum wheat cultivars and mediterranean landraces matches with their agronomic performance. PLoS ONE 2016, 11, e0160983. [Google Scholar] [CrossRef] [PubMed]
- Shewry, P.R. Improving the protein content and composition of cereal grain. J. Cereal Sci. 2007, 46, 239–250. [Google Scholar] [CrossRef]
- Velu, G.; Singh, R.P.; Cardenas, M.E.; Wu, B.; Guzman, C.; Ortiz-Monasterio, I. Characterization of grain protein content gene (GPC-B1) introgression lines and its potential use in breeding for enhanced grain zinc and iron concentration in spring wheat. Acta Physiol. Plant. 2017, 39, 212. [Google Scholar] [CrossRef]
- Uauy, C.; Brevis, J.C.; Dubcovsky, J. The high grain protein content gene Gpc-B1 accelerates senescence and has pleiotropic effects on protein content in wheat. J. Exp. Bot. 2006, 57, 2785–2794. [Google Scholar] [CrossRef] [PubMed]
- Amiri, R.; Bahraminejad, S.; Sasani, S.; Jalali-Honarmand, S.; Fakhri, R. Bread wheat genetic variation for grain’s protein, iron and zinc concentrations as uptake by their genetic ability. Eur. J. Agron. 2015, 67, 20–26. [Google Scholar] [CrossRef]
- Ravel, C.; Praud, S.; Murigneux, A.; Linossier, L.; Dardevet, M.; Balfourier, F.; Dufour, P.; Brunel, D.; Charmet, G. Identification of Glu-B1-1 as a candidate gene for the quantity of high-molecular-weight glutenin in bread wheat (Triticum aestivum L.) by means of an association study. Theor. Appl. Genet. 2006, 112, 738–743. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Yamamori, M.; Hirano, H.; Hidaka, S. Decrease of Waxy (Wx) Protein in Two Common Wheat Cultivars with Low Amylose Content. Plant Breed 1993, 111, 99–105. [Google Scholar] [CrossRef]
- Giroux, M.J.; Morris, C.F. A glycine to serine change in puroindoline b is associated with wheat grain hardness and low levels of starch-surface friabilin. TAG Theor. Appl. Genet. 1997, 95, 857–864. [Google Scholar] [CrossRef]
- Gupta, R.B.; Singh, N.K.; Shepherd, K.W. The cumulative effect of allelic variation in LMW and HMW glutenin subunits on dough properties in the progeny of two bread wheats. Theor. Appl. Genet. 1989, 77, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Araki, E.; Miura, H.; Sawada, S. Identification of genetic loci affecting amylose content and agronomic traits on chromosome 4A of wheat. TAG Theor. Appl. Genet. 1999, 98, 977–984. [Google Scholar] [CrossRef]
- Payne, P.I.; Nightingale, M.A.; Krattiger, A.F.; Holt, L.M. The relationship between HMW glutenin subunit composition and the bread-making quality of British-grown wheat varieties. J. Sci. Food Agric. 1987, 40, 51–65. [Google Scholar] [CrossRef]
- McCartney, C.A.; Somers, D.J.; Lukow, O.; Ames, N.; Noll, J.; Cloutier, S.; Humphreys, D.G.; McCallum, B.D. QTL analysis of quality traits in the spring wheat cross RL4452 × ‘AC Domain’. Plant Breed 2006, 125, 565–575. [Google Scholar] [CrossRef]
- Sun, H.; Lü, J.; Fan, Y.; Zhao, Y.; Kong, F.; Li, R.; Wang, H.; Li, S. Quantitative trait loci (QTLs) for quality traits related to protein and starch in wheat. Prog. Nat. Sci. 2008, 18, 825–831. [Google Scholar] [CrossRef]
- Fu, Y.-B.; Yang, M.-H.; Zeng, F.; Biligetu, B. Searching for an Accurate Marker-Based Prediction of an Individual Quantitative Trait in Molecular Plant Breeding. Front. Plant Sci. 2017, 8, 1182. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Mette, M.F.; Gowda, M.; Longin, C.F.H.; Reif, J.C. Bridging the gap between marker-assisted and genomic selection of heading time and plant height in hybrid wheat. Heredity 2014, 112, 638–645. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, T.B.; Gurung, S.; Hansen, J.M.; Jackson, E.W.; Bonman, J.M. Association Mapping of Quantitative Trait Loci in Spring Wheat Landraces Conferring Resistance to Bacterial Leaf Streak and Spot Blotch. Plant Genome J. 2012, 5, 1–16. [Google Scholar] [CrossRef]
- Odong, T.L.; van Heerwaarden, J.; Jansen, J.; van Hintum, T.J.L.; van Eeuwijk, F.A. Determination of genetic structure of germplasm collections: Are traditional hierarchical clustering methods appropriate for molecular marker data? Theor. Appl. Genet. 2011, 123, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, D.; Guo, X.; Yang, W.; Sun, J.; Wang, D.; Sourdille, P.; Zhang, A. Investigation of genetic diversity and population structure of common wheat cultivars in northern China using DArT markers. BMC Genet. 2011, 12, 42. [Google Scholar] [CrossRef] [PubMed]
- Hiremath, P.J.; Kumar, A.; Penmetsa, R.V.; Farmer, A.; Schlueter, J.A.; Chamarthi, S.K.; Whaley, A.M.; Carrasquilla-Garcia, N.; Gaur, P.M.; Upadhyaya, H.D.; et al. Large-scale development of cost-effective SNP marker assays for diversity assessment and genetic mapping in chickpea and comparative mapping in legumes. Plant Biotechnol. J. 2012, 10, 716–732. [Google Scholar] [CrossRef] [PubMed]
- Tadesse, W.; Ogbonnaya, F.C.; Jighly, A.; Sanchez-Garcia, M.; Sohail, Q.; Rajaram, S.; Baum, M. Genome-wide association mapping of yield and grain quality traits in winter wheat genotypes. PLoS ONE 2015, 10, e0141339. [Google Scholar] [CrossRef] [PubMed]
- Uauy, C. Wheat genomics comes of age. Curr. Opin. Plant Biol. 2017, 36, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Taulemesse, F.; Gouis, J. Le; Gouache, D.; Gibon, Y.; Allard, V. Bread wheat (Triticum aestivum L.) grain protein concentration is related to early post-flowering nitrate uptake under putative control of plant satiety level. PLoS ONE 2016, 11, e0149668. [Google Scholar] [CrossRef] [PubMed]
- Federer, W.T.; Crossa, J. I.4 Screening Experimental Designs for Quantitative Trait Loci, Association Mapping, Genotype-by Environment Interaction, and Other Investigations. Front. Physiol. 2012, 3, 156. [Google Scholar] [CrossRef] [PubMed]
- Cavanagh, C.R.; Chao, S.; Wang, S.; Huang, B.E.; Stephen, S.; Kiani, S.; Forrest, K.; Saintenac, C.; Brown-Guedira, G.L.; Akhunova, A.; et al. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc. Natl. Acad. Sci. USA 2013, 110, 8057–8062. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Stekhoven, D.J.; Bühlmann, P. MissForest-non-parametric missing value imputation for mixed-type data. Bioinformatics 2012, 28, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L.; et al. Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Federer, W.T.; King, F. Variations on Split Plot and Split Block Experiment Designs; John Wiley & Sons: New York, NY, USA, 2006; ISBN 9780470108581. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017; Available online: https://www.R-project.org/ (accessed on 6 October 2017).
- Lenth, R.V. Least-Squares Means: The R Package lsmeans. J. Stat. Softw. 2016, 69. [Google Scholar] [CrossRef]
- Asadabadi, Y.Z.; Khodarahmi, M.; Nazeri, S.M.; Peyghambari, S.A. Genetic Study of Grain Yield and its Components in Bread Wheat Using Generation Mean Analysis under Water Stress Condition. J. Plant Physiol. Breed 2012, 2, 55–60. [Google Scholar]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef] [PubMed]
- Endelman, J.B.; Jannink, J.-L. Shrinkage Estimation of the Realized Relationship Matrix. Genes Genome Genet. 2012, 2, 1405–1413. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Weir, B.S. Eigenanalysis of SNP data with an identity by descent interpretation. Theor. Popul. Biol. 2016, 107, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [PubMed]
- Wray, N.R.; Yang, J.; Hayes, B.J.; Price, A.L.; Goddard, M.E.; Visscher, P.M. Pitfalls of predicting complex traits from SNPs. Nat. Rev. Genet. 2013, 14, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, G. Estimating the Dimension of a Model. Ann. Stat. 1978, 6, 461–464. [Google Scholar] [CrossRef]
- Singh, S.; Gupta, A.K.; Kaur, N. Influence of drought and sowing time on protein composition, antinutrients, and mineral contents of wheat. Sci. World J. 2012, 2012, 485751. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, A.; Aydin, F. Effect of water stress at various growth stages on some quality characteristics of winter wheat. J. Agron. Crop Sci. 2004, 190, 93–99. [Google Scholar] [CrossRef]
- Grenier, S.; Barre, P.; Litrico, I. Phenotypic Plasticity and Selection: Nonexclusive Mechanisms of Adaptation. Scientifica 2016, 2016, 7021701. [Google Scholar] [CrossRef] [PubMed]
- Dvořáček, V.; Čurn, V. Evaluation of protein fractions as biochemical markers for identification of spelt wheat cultivars (Triticum spelta L.). Plant Soil Environ. 2003, 49, 99–105. [Google Scholar] [CrossRef]
- Jaradat, A.A. Wheat landraces: A mini review. Emir. J. Food Agric. 2013, 25, 20–29. [Google Scholar] [CrossRef]
- Lopes, M.S.; El-Basyoni, I.; Baenziger, P.S.; Singh, S.; Royo, C.; Ozbek, K.; Aktas, H.; Ozer, E.; Ozdemir, F.; Manickavelu, A.; et al. Exploiting genetic diversity from landraces in wheat breeding for adaptation to climate change. J. Exp. Bot. 2015, 66, 3477–3486. [Google Scholar] [CrossRef] [PubMed]
- Baenziger, P.S.; Salah, I.; Little, R.S.; Santra, D.K.; Regassa, T.; Wang, M.Y. Structuring an Efficient Organic Wheat Breeding Program. Sustainability 2011, 3, 1190–1205. [Google Scholar] [CrossRef]
- Chen, X.-Y.; Yang, Y.; Ran, L.-P.; Dong, Z.; Zhang, E.-J.; Yu, X.-R.; Xiong, F. Novel Insights into miRNA Regulation of Storage Protein Biosynthesis during Wheat Caryopsis Development under Drought Stress. Front. Plant Sci. 2017, 8, 1707. [Google Scholar] [CrossRef] [PubMed]
- Endelman, J. Using rrBLUP 4.0. Jeffrey Endelman 17 September 2012. Available online: http://www.afhalifax.ca/bete/revive-eponge/Rblkfct/vignette.pdf (accessed on 8 July 2018).
- Zhao, K.; Aranzana, M.J.; Kim, S.; Lister, C.; Shindo, C.; Tang, C.; Toomajian, C.; Zheng, H.; Dean, C.; Marjoram, P.; et al. An Arabidopsis example of association mapping in structured samples. PLoS Genet. 2007, 3, e4. [Google Scholar] [CrossRef] [PubMed]
- Breseghello, F.; Sorrells, M.M.E. Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 2006, 172, 1165–1177. [Google Scholar] [CrossRef] [PubMed]
- Neumann, K.; Kobiljski, B.; Denčić, S.; Varshney, R.K.; B?rner, A. Genome-wide association mapping: A case study in bread wheat (Triticum aestivum L.). Mol. Breed 2011, 27, 37–58. [Google Scholar] [CrossRef]
- Fiedler, J.D.; Salsman, E.; Liu, Y.; Michalak de Jiménez, M.; Hegstad, J.B.; Chen, B.; Manthey, F.A.; Chao, S.; Xu, S.; Elias, E.M.; et al. Genome-Wide Association and Prediction of Grain and Semolina Quality Traits in Durum Wheat Breeding Populations. Plant Genome 2017, 10. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Feng, B.; Xu, Z.; Fan, X.; Jiang, F.; Jin, X.; Cao, J.; Wang, F.; Liu, Q.; Yang, L.; et al. A genome-wide association study of wheat yield and quality-related traits in southwest China. Mol. Breed 2018, 38, 1. [Google Scholar] [CrossRef]
- Troccoli, A.; Borrelli, G.M.; De Vita, P.; Fares, C.; Di Fonzo, N. Mini Review: Durum Wheat Quality: A Multidisciplinary Concept. J. Cereal Sci. 2000, 32, 99–113. [Google Scholar] [CrossRef]



| Source | DF | Type III SS | Mean Square | F Value |
|---|---|---|---|---|
| Environment | 3 | 70,093.78 | 23,364.59 | 19,188.5 ** |
| IBlock (Replicate Environment) | 256 | 1361.56 | 5.31 | 4.37 |
| Genotypes | 2113 | 26,096.19 | 12.35 | 10.14 ** |
| Environment × Genotypes | 6255 | 26,164.38 | 4.18 | 3.44 ** |
| Error | 9208 | 11,211.99 | 1.21 |
| Well-Watered | Water Deficit | ||||||
|---|---|---|---|---|---|---|---|
| Accession | Origin | Improvement | Mean | Accession | Origin | Improvement | Mean |
| 534,406 | Algeria | landrace | 14.78 | 366,801 | Afghanistan | landrace | 17.965 |
| 534,448 | Algeria | landrace | 15.63 | 350,850 | Austria | landrace | 18.38 |
| 338,364 | Belgium | cultivar | 14.83 | 350,820 | Austria | landrace | 18.1125 |
| 481,731 | Bhutan | landrace | 14.69 | 565,254 | Bolivia | landrace | 17.9275 |
| 14,261 | Canada | breeding | 15.39 | 374,243 | Chad | landrace | 18.135 |
| 313,109 | Colombia | uncertain | 15.13 | 57,825 | India | landrace | 18.0175 |
| 372,434 | Cyprus | landrace | 14.90 | 382,048 | Iran | landrace | 18.535 |
| 428,672 | Czech Republic | cultivar | 15.33 | 625,916 | Iran | landrace | 18.43 |
| 254,023 | Europe | uncertain | 15.27 | 623,758 | Iran | landrace | 18.055 |
| 278,279 | Greece | landrace | 15.09 | 624,992 | Iran | landrace | 18.03 |
| 468,988 | Greece | landrace | 16.11 | 624,124 | Iran | landrace | 17.9125 |
| 15,396 | Lebanon | uncertain | 15.89 | 626,116 | Iran | landrace | 17.9075 |
| 520,369 | Mexico | breeding | 15.80 | 623,968 | Iran | landrace | 17.8525 |
| 525,283 | Morocco | landrace | 15.49 | 70,704 | Iraq | landrace | 18.42 |
| 477,901 | Peru | landrace | 15.05 | 191,987 | Portugal | landrace | 18.3475 |
| 370,724 | Poland | cultivar | 15.03 | 345,474 | Serbia | landrace | 18.3975 |
| 155,119 | Russian Federation | cultivar | 15.68 | 225,424 | Uruguay | breeding | 18.355 |
| 479,700 | South Africa | cultivar | 15.48 | 225,519 | Uruguay | breeding | 17.8375 |
| 241,596 | Taiwan | cultivar | 15.31 | 36,500 | Uzbekistan | landrace | 17.95 |
| 534,366 | Tunisia | landrace | 14.98 | 24,485 | Uzbekistan | landrace | 17.85 |
| Marker | Chrom | Position | Well-Watered | Water Deficit | R2 (%) | Additive Effect | MAF | Marker | Chrom | Position | Well-Watered | Water Deficit | R2 (%) | Additive Effect | MAF | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2016 | 2017 | 2016 | 2017 | 2016 | 2017 | 2016 | 2017 | ||||||||||||
| IWA5150 | 1A | 9.9 | + | + | − | − | 0.893 | −0.007 | 0.19 | IWA8551 | 1D | 32.8 | − | − | + | − | 1.069 | 0.062 | 0.25 |
| IWA6649 | 1A | 11.6 | + | − | + | + | 1.141 | 0.07 | 0.35 | IWA3481 | 1D | 45.1 | − | + | − | + | 1.122 | 0.13 | 0.07 |
| IWA4351 | 1A | 11.6 | + | − | + | + | 1.089 | 0.062 | 0.35 | IWA3446 | 1D | 45.1 | − | − | + | + | 1.001 | 0.086 | 0.07 |
| IWA4643 | 1A | 21 | + | + | − | − | 0.892 | 0.004 | 0.28 | IWA5020 | 1D | 47.7 | − | + | − | − | 0.918 | −0.026 | 0.33 |
| IWA4753 | 1A | 21.7 | − | + | − | − | 0.9 | −0.02 | 0.1 | IWA5019 | 1D | 47.7 | − | + | − | − | 0.918 | −0.026 | 0.33 |
| IWA7191 | 1A | 21.7 | − | − | + | + | 0.965 | 0.053 | 0.13 | IWA5018 | 1D | 47.7 | − | + | − | − | 0.917 | −0.026 | 0.33 |
| IWA4678 | 1A | 22.5 | − | + | − | − | 0.901 | 0.028 | 0.08 | IWA4598 | 1D | 48.6 | − | + | − | − | 0.912 | 0.023 | 0.33 |
| IWA4644 | 1A | 22.9 | − | + | − | − | 0.915 | −0.026 | 0.16 | IWA7007 | 6A | 10 | − | − | + | − | 0.918 | 0.025 | 0.23 |
| IWA4754 | 1A | 23.2 | + | + | − | − | 0.892 | 0.003 | 0.34 | IWA4551 | 6A | 16.2 | + | − | − | − | 1.294 | −0.13 | 0.13 |
| IWA4506 | 1A | 26.9 | + | − | − | − | 0.907 | −0.019 | 0.29 | IWA4552 | 6A | 16.2 | + | − | − | − | 1.313 | −0.131 | 0.13 |
| IWA7050 | 1A | 32.5 | − | − | + | − | 1.305 | 0.092 | 0.38 | IWA7288 | 6A | 17.8 | − | − | + | + | 1.316 | 0.093 | 0.35 |
| IWA4163 | 1A | 32.8 | + | − | − | − | 1.17 | −0.076 | 0.39 | IWA7287 | 6A | 21.9 | − | − | + | + | 1.391 | 0.116 | 0.21 |
| IWA4349 | 1B | 13.2 | + | − | + | − | 1.589 | 0.128 | 0.26 | IWA4962 | 6A | 22.8 | − | + | − | − | 0.923 | −0.027 | 0.24 |
| IWA6787 | 1B | 13.2 | + | − | + | − | 1.433 | 0.105 | 0.28 | IWA4730 | 6B | 48.5 | + | − | − | − | 0.922 | 0.046 | 0.06 |
| IWA7048 | 1B | 22.9 | − | − | + | − | 1.746 | 0.244 | 0.07 | IWA3501 | 6B | 48.8 | + | + | + | + | 2.681 | 0.213 | 0.39 |
| IWA7480 | 1B | 22.9 | − | − | − | + | 1.472 | 0.111 | 0.35 | IWA7937 | 6B | 48.8 | + | + | + | + | 2.654 | 0.187 | 0.37 |
| IWA3169 | 1B | 23.7 | + | + | + | + | 2.027 | 0.161 | 0.31 | IWA3923 | 6B | 48.8 | + | + | − | − | 1.208 | 0.117 | 0.11 |
| IWA8199 | 1B | 27.4 | − | − | + | + | 1.271 | 0.099 | 0.25 | IWA6466 | 6B | 48.8 | + | + | − | − | 1.448 | 0.131 | 0.18 |
| IWA7345 | 1B | 28.1 | − | − | + | + | 1.808 | 0.23 | 0.09 | IWA6467 | 6B | 48.8 | + | + | − | − | 1.455 | 0.132 | 0.18 |
| IWA6611 | 1B | 28.1 | − | − | − | + | 1.636 | 0.131 | 0.27 | IWA5986 | 6B | 50.8 | + | + | − | − | 0.892 | 0.005 | 0.24 |
| IWA6610 | 1B | 28.1 | − | − | − | + | 1.642 | −0.13 | 0.27 | IWA6673 | 6D | 17.2 | − | − | + | − | 1.147 | 0.079 | 0.24 |
| IWA3738 | 1B | 28.2 | − | + | − | − | 1.451 | 0.12 | 0.22 | IWA3624 | 6D | 17.3 | − | − | + | − | 1.073 | 0.068 | 0.41 |
| IWA8275 | 1B | 28.2 | − | − | + | − | 1.618 | 0.128 | 0.27 | IWA7616 | 6D | 29.8 | − | − | − | + | 1.542 | 0.142 | 0.17 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elbasyoni, I.S.; Morsy, S.M.; Ramamurthy, R.K.; Nassar, A.M. Identification of Genomic Regions Contributing to Protein Accumulation in Wheat under Well-Watered and Water Deficit Growth Conditions. Plants 2018, 7, 56. https://doi.org/10.3390/plants7030056
Elbasyoni IS, Morsy SM, Ramamurthy RK, Nassar AM. Identification of Genomic Regions Contributing to Protein Accumulation in Wheat under Well-Watered and Water Deficit Growth Conditions. Plants. 2018; 7(3):56. https://doi.org/10.3390/plants7030056
Chicago/Turabian StyleElbasyoni, Ibrahim S., Sabah M. Morsy, Raghuprakash K. Ramamurthy, and Atef M. Nassar. 2018. "Identification of Genomic Regions Contributing to Protein Accumulation in Wheat under Well-Watered and Water Deficit Growth Conditions" Plants 7, no. 3: 56. https://doi.org/10.3390/plants7030056
APA StyleElbasyoni, I. S., Morsy, S. M., Ramamurthy, R. K., & Nassar, A. M. (2018). Identification of Genomic Regions Contributing to Protein Accumulation in Wheat under Well-Watered and Water Deficit Growth Conditions. Plants, 7(3), 56. https://doi.org/10.3390/plants7030056






