Accumulation of Phosphorus-Containing Compounds in Developing Seeds of Low-Phytate Pea (Pisum sativum L.) Mutants
Abstract
:1. Introduction
2. Results
2.1. Agronomic Characteristics of Low-Phytate Pea Genotypes

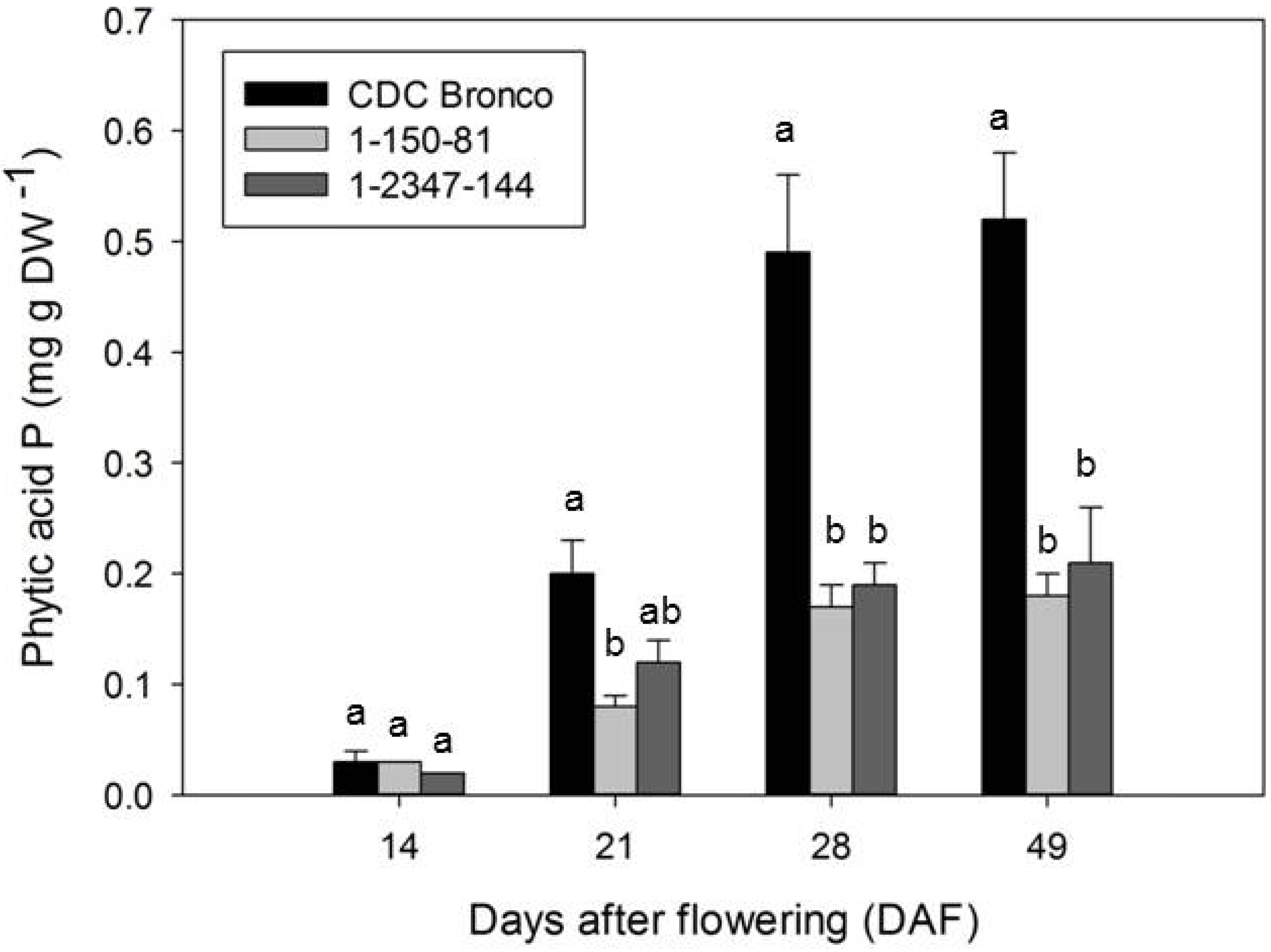
2.2. Accumulation of Phosphorus and Phosphorus-Containing Compounds during Seed Development
| Genotype | Emergence Count (%) d | Days to Flower | Plant Height (cm) e | Mycosphaerella Blight Score (0–9 Scale) f | Lodging Score (1–9 Scale) g | Days to Mature | Grain Yield (t·ha−1) h | 1000 Seed Weight (g) |
|---|---|---|---|---|---|---|---|---|
| CDC Bronco | 56 ± 3.0 a | 58 ± 1.0 a | 79 ± 1.0 a | 5.3 ± 0.1 a | 5.4 ± 0.2 a | 100 ± 1.0 a | 2.83 ± 0.1 a | 219 ± 2.2 a |
| 1-150-81 | 55 ± 3.0 a | 59 ± 1.0 a | 78 ± 2.0 a | 5.2 ± 0.1 a | 5.3 ± 0.1 a | 102 ± 1.0 a | 2.36 ± 0.2 b | 207 ± 1.6 b |
| 1-2347-144 | 53 ± 3.0 a | 59 ± 1.0 a | 76 ± 2.0 a | 5.3 ± 0.1 a | 5.3 ± 0.1 a | 101 ± 1.0 a | 2.33 ± 0.2 b | 205 ± 2.3 b |
2.3. Characterization and Bioinformatic Analyses of PsMIPS

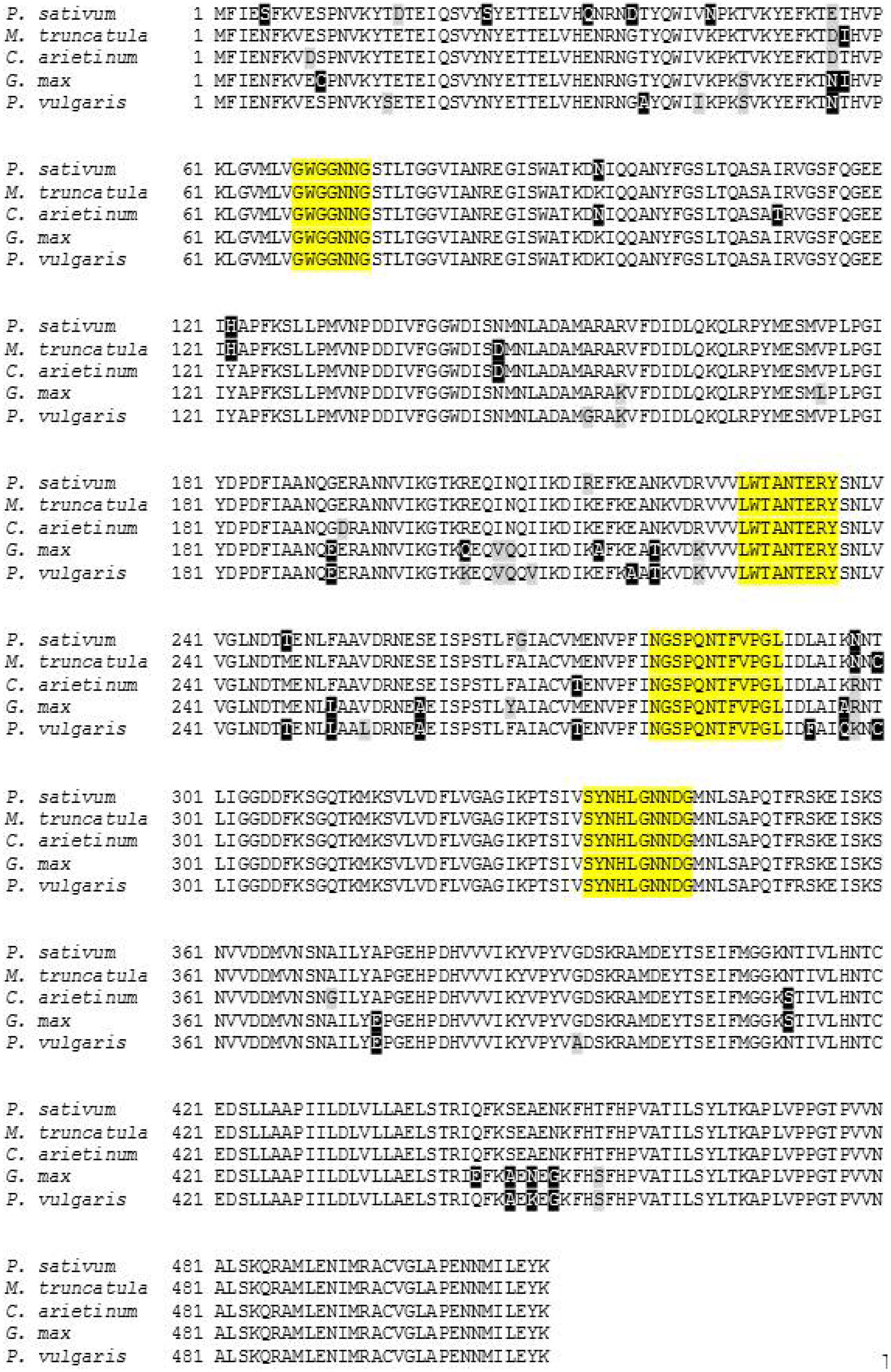
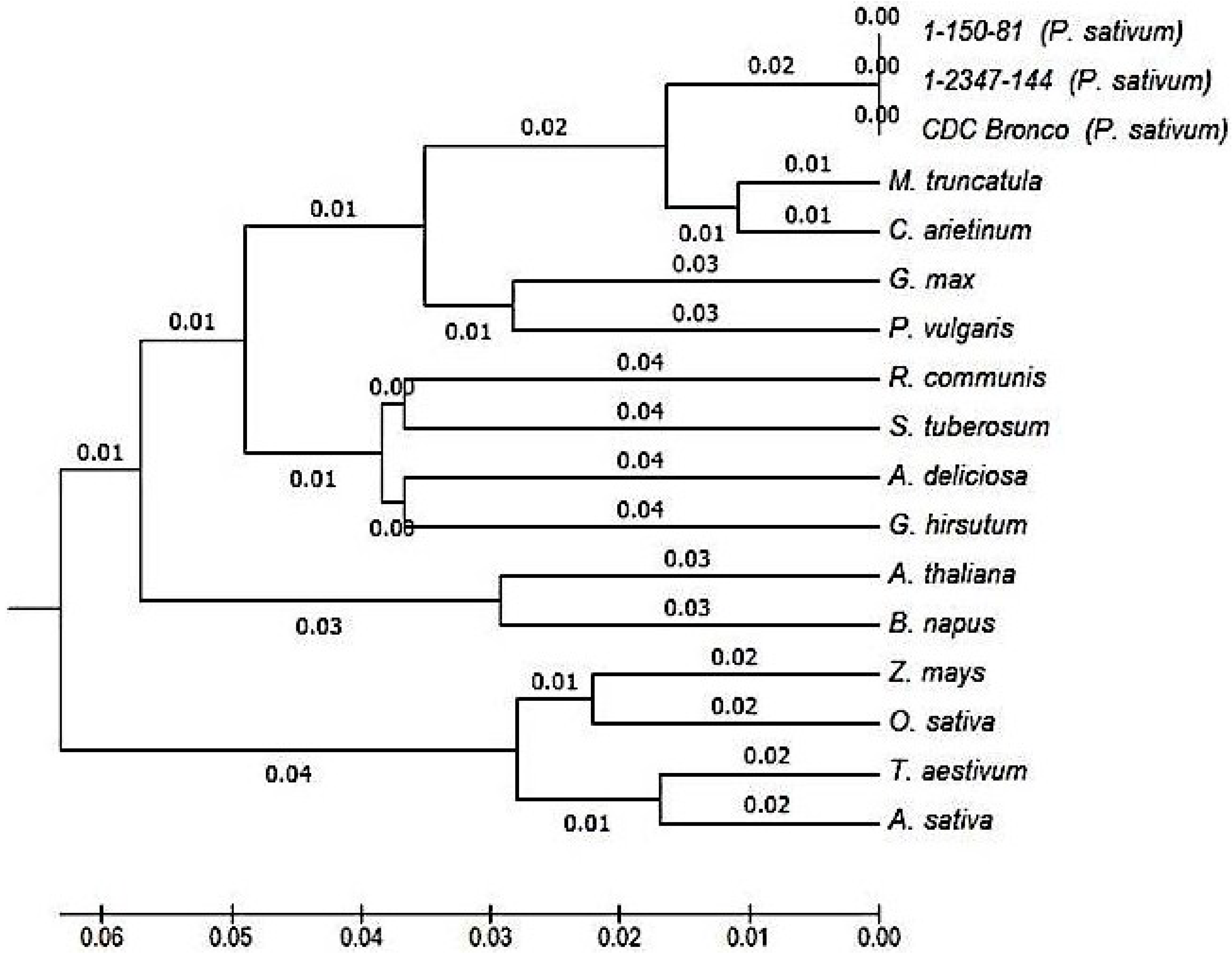
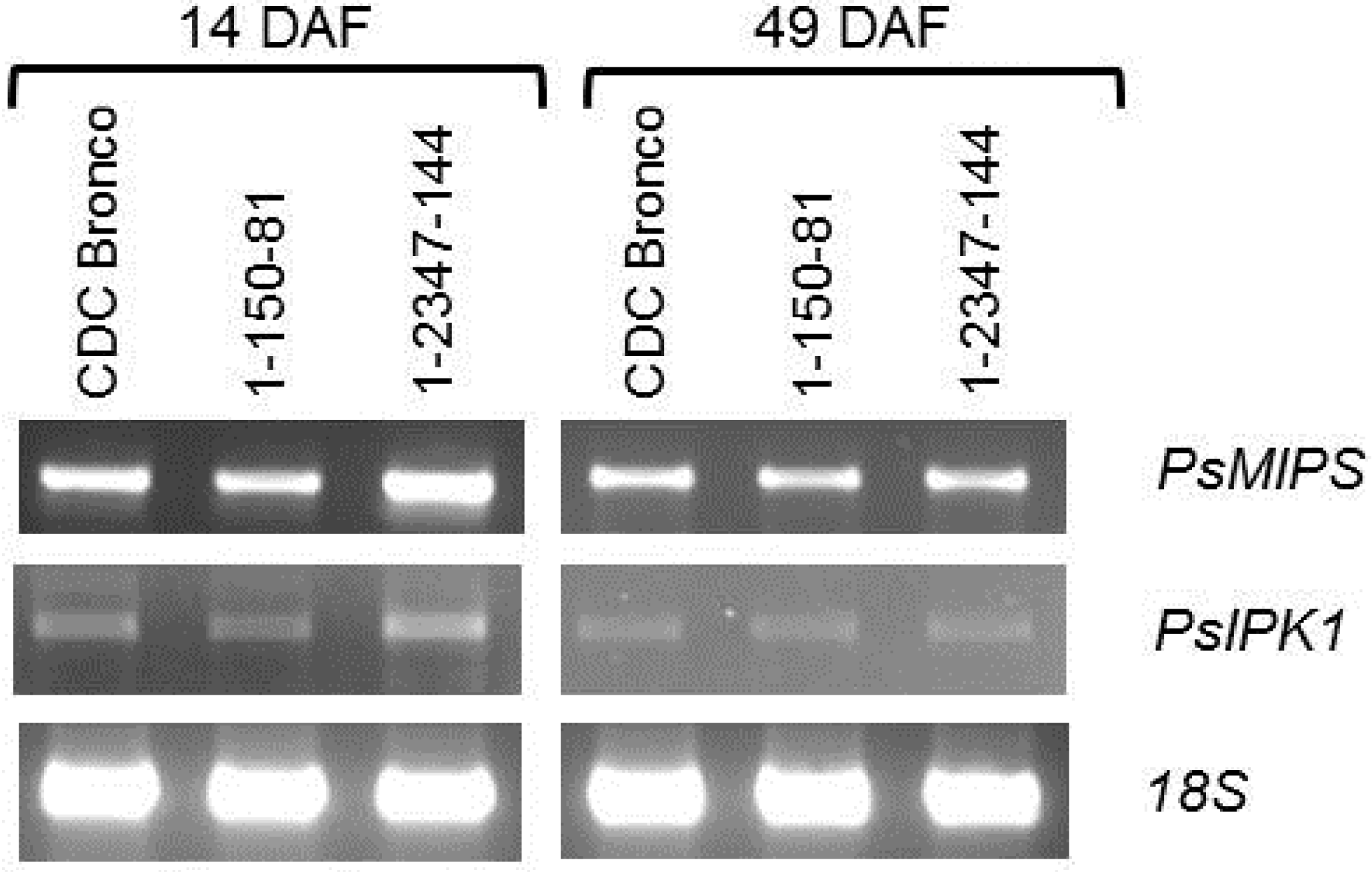
2.4. PsMIPS Gene Expression
3. Discussion
3.1. Agronomic Traits in Low-Phytate Pea
3.2. Redistribution of Phosphorus in Low-Phytate Pea during Seed Development
3.3. Myo-Inositol-3-Phosphate Synthase (MIPS) Is Identical in Normal and Low-Phytate Pea Genotypes
4. Experimental Section
4.1. Plant Material and Growth Conditions
4.2. Extraction and Detection of Inositol Phosphates
4.3. Analysis of Phosphorus Levels
4.4. Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR) and Sequencing
4.5. Semi-Quantitative Reverse Transcriptase-Polymerase Chain Reaction (sqRT-PCR)
4.6. Sequence Analysis
4.7. Statistics
5. Conclusions
Acknowledgments
Author Contributions
Appendix




Conflicts of Interest
References
- Cosgrove, D.J. Inositol Phosphates: Their Chemistry, Biochemistry, and Physiology; Elsevier: New York, NY, USA, 1980. [Google Scholar]
- O’Dell, B.L.; de Boland, A.R.; Koirtyohann, S.R. Distribution of phytate and nutritionally important elements among the morphological components of cereal grains. J. Agric. Food. Chem. 1972, 20, 718–721. [Google Scholar] [CrossRef]
- Lott, J.N.A. Accumulation of seed reserves of phosphorus and other minerals. In Seed Physiology; Murray, D.R., Ed.; Academic Press: Sydney, Australia, 1984; Volume 1, pp. 154–196. [Google Scholar]
- Raboy, V. Seeds for a better future: “Low phytate” grains help to overcome malnutrition and reduce pollution. Trends Plant Sci. 2001, 6, 458–462. [Google Scholar] [CrossRef] [PubMed]
- Reddy, N.R. Occurrence, distribution, content and dietary intake of phytate. In Food Phytates; Reddy, N.R., Sathe, S.K., Eds.; CRC Press: New York, NY, USA, 2002; pp. 25–51. [Google Scholar]
- Erdman, J.W., Jr. Bioavailability of trace minerals from cereals and legumes. Cereal Chem. 1980, 58, 21–26. [Google Scholar]
- Sharpley, A.N.; Charpa, S.C.; Wedepohl, R.; Sims, J.Y.; Daniel, T.C.; Reddy, K.R. Managing agricultural phosphorus for protection of surface waters: Issues and Options. J. Environ. Qual. 1994, 23, 437–451. [Google Scholar] [CrossRef]
- Raboy, V.; Young, K.A.; Dorsch, J.A.; Cook, A. Genetics and breeding of seed phosphorus and phytic acid. J. Plant Physiol. 2001, 158, 489–497. [Google Scholar] [CrossRef]
- Raboy, V.; Gerbasi, P. Genetics of myo-inositol phosphate synthesis and accumulation. In Myo Inositol Phosphates, Phosphoinositides and Signal Transduction; Biswas, B.B., Ed.; Plenum Publ. Co.: New York, NY, USA, 1996; pp. 257–285. [Google Scholar]
- Raboy, V.; Gerbasi, P.F.; Young, K.A.; Stoneberg, S.D.; Pickett, S.G.; Bauman, A.T.; Murthy, P.N.; Sheridan, W.F.; Ertl, D.S. Origin and seed phenotype of maize low phytic acid 1–1 and low phytic acid 2–1. Plant Physiol. 2000, 124, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Larson, S.R.; Young, K.A.; Cook, A.; Blake, T.K.; Raboy, V. Linkage mapping of two mutations that reduce the phytic acid content of barley grain. Theor. Appl. Genet. 1998, 97, 141–146. [Google Scholar] [CrossRef]
- Rasmussen, S.K.; Hatzack, F. Identification of two low-phytate barley (Hordeum vulgare L.) grain mutants by TLC and genetic analysis. Heriditas 1998, 129, 107–112. [Google Scholar]
- Wilcox, J.; Premachandra, G.; Young, K.; Raboy, V. Isolation of high seed inorganic P, low-phytate soybean mutants. Crop Sci. 2000, 40, 1601–1605. [Google Scholar] [CrossRef]
- Warkentin, T.D.; Delgerjav, T.; Arganosa, G.; Rehman, A.U.; Bett, K.E.; Anbessa, Y.K.; Rossnagel, B.; Raboy, V. Development and characterization of low-phytate pea. Crop Sci. 2012, 52, 74–78. [Google Scholar] [CrossRef]
- Ertl, D.S.; Young, K.A.; Raboy, V. Plant genetic approaches to phosphorus management in agricultural production. J. Environ. Qual. 1998, 27, 299–304. [Google Scholar] [CrossRef]
- Loewus, F.A.; Murthy, P.N. Myo-inositol metabolism in plants. Plant Sci. 2000, 150, 1–19. [Google Scholar] [CrossRef]
- Stevenson, J.M.; Perera, I.Y.; Heilmann, I.; Persson, S.; Boss, W.F. Inositol signaling and plant growth. Trends Plant Sci. 2000, 5, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Loewus, F.A.; Loewus, M.W. Myo-inositol: Its biosynthesis and metabolism. Annu. Rev. Plant Physiol. 1983, 34, 137–161. [Google Scholar] [CrossRef]
- Loewus, F.A. Inositol biosynthesis. In Inositol Metabolism in Plants; Morre, D.J., Boss, W.F., Loewus, F.A., Eds.; Wiley-Liss: New York, NY, USA, 1990; pp. 13–19. [Google Scholar]
- Pilu, R.; Panzeri, D.; Gavazzi, G.; Rasmussen, S.K.; Consonni, G.; Nielsen, E. Phenotypic, genetic and molecular characterization of a maize low phytic acid mutant (lpa241). Theor. Appl. Genet. 2003, 107, 980–987. [Google Scholar] [CrossRef] [PubMed]
- Kuwano, M.; Ohyama, A.; Tanaka, Y.; Mimura, T.; Takaiwa, F.; Yoshida, K.T. Molecular breeding for transgenic rice with low-phytic-acid phenotype through manipulating myo-inositol 3-phosphate synthase gene. Mol. Breed. 2006, 18, 263–272. [Google Scholar]
- Kuwano, M.; Mimura, T.; Takaiwa, F. Generation of stable “low phytic acid” transgenic rice through antisense repression of the 1d-myo-inositol 3-phosphate synthase gene (RINO1) using the 18-kDa oleosin promoter. Plant Biotech. J. 2009, 7, 96–105. [Google Scholar] [CrossRef]
- Georges, F.; Hussain, A.A.K.; Keller, W.A. Method for Reducing Phytate in Canola Meal Using Genetic Manipulation Involving Myo-Inositol 1-Phosphate Synthase Gene. Patent WO 00/73473 A1, 2006. [Google Scholar]
- Nunes, A.C.S.; Vianna, G.R.; Cuneo, F.; Amaya-Farfán, J.; de Capdeville, G.; Rech, E.L.; Aragão, F.J.L. RNAi-mediated silencing of the myo-inositol-1-phosphate synthase gene (GmMIPS1) in transgenic soybean inhibited seed development and reduced phytate content. Planta 2006, 224, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Panzeri, D.; Cassani, E.; Doria, E.; Tagliabue, G.; Forti, L.; Campion, B.; Bollini, R.; Brearley, C.A.; Pilu, R.; Nielsen, E.; et al. A defective ABC transporter of the MRP family, responsible for the bean lpa1 mutation, affects the regulation of the phytic acid pathway, reduces seed myo-inositol and alters ABA sensitivity. New Phytol. 2011, 191, 70–83. [Google Scholar]
- Hitz, D.W.; Thomas, J.C.; Phil, S.K.; Scott, A.S. Biochemical and molecular characterization of a mutation that confers a decreased raffinosaccharide and phytic acid phenotype on soybean seeds. Plant Physiol. 2002, 128, 650–660. [Google Scholar] [CrossRef] [PubMed]
- Shunmugam, A.S.K.; Bock, C.; Arganosa, G.C.; Gray, R.G.; Georges, F.; Warkentin, T.D.; Department of Plant Sciences, University of Saskatchewan: Saskatoon, Saskatchewan, Canada. Unpublished raw data. 2014.
- Majumder, A.L.; Johnson, M.D.; Henry, S.A. 1L-myo-inositol-1-phosphate synthase. Biochim. Biophys. Acta 1997, 1348, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Majumder, A.L.; Chatterjee, A.; Dastida, K.G.; Majee, M. Diversification and evolution of L-myo-inositol 1-phosphate synthase. FEBS Lett. 2003, 553, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Gillman, J.D.; Pantalone, V.R.; Bilyeu, K. The low phytic acid phenotype in soybean line CX1834 is due to mutations in two homologs of the maize low phytic acid gene. Plant Genome 2009, 2, 179–190. [Google Scholar] [CrossRef]
- Anderson, B.P.; Fehr, W.R. Seed source affects field emergence of low-phytate soybean lines. Crop Sci. 2008, 48, 929–932. [Google Scholar] [CrossRef]
- Bentsink, K.; Yuan, Y.; Koorneef, M.; Vreugdenhil, D. The genetics of phytate and phosphorus accumulation in seeds and leaves of Arabidopsis thaliana, using natural variation. Theor. Appl. Genet. 2003, 106, 1234–1243. [Google Scholar] [PubMed]
- Shi, J.R.; Wang, H.Y.; Schellin, K.; Li, B.L.; Faller, M.; Stoop, J.M.; Meeley, R.B.; Ertl, D.S.; Ranch, J.P.; Glassman, K. Embryo-specific silencing of a transporter reduces phytic acid content of maize and soybean seeds. Nat. Biotechnol. 2007, 25, 930–937. [Google Scholar] [CrossRef]
- Dong, J.; Yan, W.; Bock, C.; Nokhrina, K.; Keller, W.; Georges, F. Perturbing the metabolic dynamics of myo-inositol in developing Brassica napus seeds through in vivo methylation impacts its utilization as phytate precursor and affects downstream metabolic pathways. BMC Plant Biol. 2013, 13, e84. [Google Scholar] [CrossRef]
- Ali, N.; Paul, S.; Gayen, D.; Sarkar, S.N.; Datta, K.; Datta, S.K. Development of low phytate rice by RNAi mediated seed-specific silencing of inositol 1,3,4,5,6-pentakisphosphate 2-kinase gene (IPK1). PLOS ONE 2013. [Google Scholar] [CrossRef]
- Bregitzer, P.; Raboy, V. Effects of four independent low-phytate mutations on barley agronomic performance. Crop Sci. 2006, 46, 1318–1322. [Google Scholar] [CrossRef]
- Bregitzer, P.; Raboy, V.; Obert, D.E.; Windes, J.; Whitmore, J.C. Registration of “Clearwater” low-phytate hulless spring barley. J. Plant Registr. 2008, 2, 1–4. [Google Scholar] [CrossRef]
- Raboy, V.; Dikinson, D.B.; Neuffer, M.G. A survey of maize kernel mutants for variation in phytic acid. Maydica 1990, 35, 383–390. [Google Scholar]
- Raboy, V. Low Phytic acid Mutants and Selection Thereof. Patent 5,689,054, 1997. [Google Scholar]
- Raboy, V.; Dickinson, D.B. The timing and rate of phytic acid accumulation in developing soybean seeds. Plant Physiol. 1987, 85, 0841–0844. [Google Scholar] [CrossRef]
- Bowen, D.E.; Guttieri, M.J.; Peterson, K.; Peterson, K.; Raboy, V.; Souza, E.J. Phosphorus fractions in developing seeds of four low phytate barley (Hordeum vulgare L.) genotypes. Crop Sci. 2006, 46, 2468–2473. [Google Scholar] [CrossRef]
- Israel, D.W.; Talierico, E.; Kwanyuen, P.; Burton, J.W.; Dean, L. Inositol metabolism in developing seeds of low and normal phytic acid soybean lines. Crop Sci. 2011, 51, 282–289. [Google Scholar] [CrossRef]
- Larson, S.R.; Rutger, J.N.; Young, K.A.; Raboy, V. Isolation and genetic mapping of a non-lethal rice (Oryza sativa L.) low phytic acid 1 mutation. Crop Sci. 2000, 40, 1397–1405. [Google Scholar]
- Yoshida, K.T.; Wada, T.; Koyama, H.; Mizobuchi-Fukuoka, R.; Naito, S. Temporal and spatial patterns of accumulation of the transcript of myo-inositol-1-phosphate synthase and phytin-containing particles during seed development in rice. Plant Physiol. 1999, 119, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Zuo, Y.; Sun, D.; Wang, C.; Guo, T. Characterization of the TaMIPS gene from winter wheat (Triticum aestivum L.) and changes in its expression pattern with phytic acid accumulation in seeds during grain filling. J. Cereal Sci. 2013, 57, 437–443. [Google Scholar]
- Keller, R.; Brearley, C.A.; Trethewey, R.N.; Muller-Rober, B. Reduced inositol content and altered morphology in transgenic potato plants inhibited for 1D-myo-inositol 3-phosphate synthase. Plant J. 1998, 16, 403–410. [Google Scholar] [CrossRef]
- Murphy, A.M.; Otto, B.; Brearley, C.A.; Carr, J.P.; Hanke, D.E. A role for inositol hexakisphosphate in the maintenance of basal resistance to plant pathogens. Plant J. 2008, 56, 638–652. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.H.; Zhao, H.J.; Liu, Q.L.; Frank, T.; Engel, K.H.; An, G.H.; Shu, Q.Y. Mutations of the multi-drug resistance-associated protein ABC transporter gene 5 result in reduction of phytic acid in rice seeds. Theor. Appl. Genet. 2009, 119, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.J.; Zhao, H.J.; Ren, X.L.; Zhu, S.L.; Fu, X.J.; Shu, Q.Y. Generation and characterization of two novel low phytate mutations in soybean (Glycine max L. Merr.). Theor. Appl. Genet. 2007, 115, 945–957. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Wang, H.; Wu, Y.; Hazebroek, J.; Meeley, R.B.; Ertl, D.S. The maize low-phytic acid mutant lpa2 is caused by mutation in an inositol phosphate kinase gene. Plant Physiol. 2003, 131, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Stevenson-Paulik, J.; Bastidas, R.J.; Chiou, S.T.; Frye, R.A.; York, J.D. Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proc. Natl. Acad. Sci. USA 2005, 102, 12612–12617. [Google Scholar]
- Warkentin, T.D.; Vandenberg, A.; Banniza, S.; Slinkard, A. CDC Bronco field pea. Can. J. Plant Sci. 2005, 85, 649–650. [Google Scholar] [CrossRef]
- Talamond, P.; Doulbeau, S.; Rochette, I.; Guyot, J.P. Anion exchange high-performance liquid chromatography with conductivity detection for the analysis of phytic acid in food. J. Chromatogr. A 2000, 871, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Thavarajah, P.; Thavarajah, D.; Vandenberg, A. Low phytic acid lentils (Lens culinaris L.): A potential solution for increased micronutrient bioavailability. J. Agric. Food Chem. 2009, 57, 9044–9049. [Google Scholar]
- Chen, P.S.; Toribara, T.Y.; Warner, H. Microdetermination of phosphorus. Anal. Chem. 1956, 28, 1756–1758. [Google Scholar] [CrossRef]
- Main, D.; Cheng, C-H.; Ficklin, S.P.; Jung, S.; Zheng, P.; Coyne, C.J.; McGee, R.; Mockaitis, K. The Cool Season Food Legume Database: An Integrated Resource for Basic, Translational and Applied Research, Proceedings of the International Plant and Animal Genome Conference, San Diego, CA, USA, 12–16 January 2013.
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, e134. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning, A Laboratory Manual, 3rd ed.; Cold Spring Harbour Laboratory Press: Cold Spring Harbour, New York, NY, USA, 2001; Volume 1–3. [Google Scholar]
- Ozga, J.A.; Yu, J.; Reinecke, D.M. Pollination-, development-, and auxin-specific regulation of gibberellin 3β-hydroxylase gene expression in pea fruit and seeds. Plant Physiol. 2003, 131, 1137–1146. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped Blast and Psi-Blast: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005, 33, 116–120. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shunmugam, A.S.K.; Bock, C.; Arganosa, G.C.; Georges, F.; Gray, G.R.; Warkentin, T.D. Accumulation of Phosphorus-Containing Compounds in Developing Seeds of Low-Phytate Pea (Pisum sativum L.) Mutants. Plants 2015, 4, 1-26. https://doi.org/10.3390/plants4010001
Shunmugam ASK, Bock C, Arganosa GC, Georges F, Gray GR, Warkentin TD. Accumulation of Phosphorus-Containing Compounds in Developing Seeds of Low-Phytate Pea (Pisum sativum L.) Mutants. Plants. 2015; 4(1):1-26. https://doi.org/10.3390/plants4010001
Chicago/Turabian StyleShunmugam, Arun S.K., Cheryl Bock, Gene C. Arganosa, Fawzy Georges, Gordon R. Gray, and Thomas D. Warkentin. 2015. "Accumulation of Phosphorus-Containing Compounds in Developing Seeds of Low-Phytate Pea (Pisum sativum L.) Mutants" Plants 4, no. 1: 1-26. https://doi.org/10.3390/plants4010001
APA StyleShunmugam, A. S. K., Bock, C., Arganosa, G. C., Georges, F., Gray, G. R., & Warkentin, T. D. (2015). Accumulation of Phosphorus-Containing Compounds in Developing Seeds of Low-Phytate Pea (Pisum sativum L.) Mutants. Plants, 4(1), 1-26. https://doi.org/10.3390/plants4010001





