Multi-Season Genome-Wide Association Study Reveals Loci and Candidate Genes for Fruit Quality and Maturity Traits in Peach
Abstract
1. Introduction
2. Results
2.1. Genotyping Results
2.2. Linkage Disequilibrium
2.3. Phenotypic Characterization of the Peach Germplasm
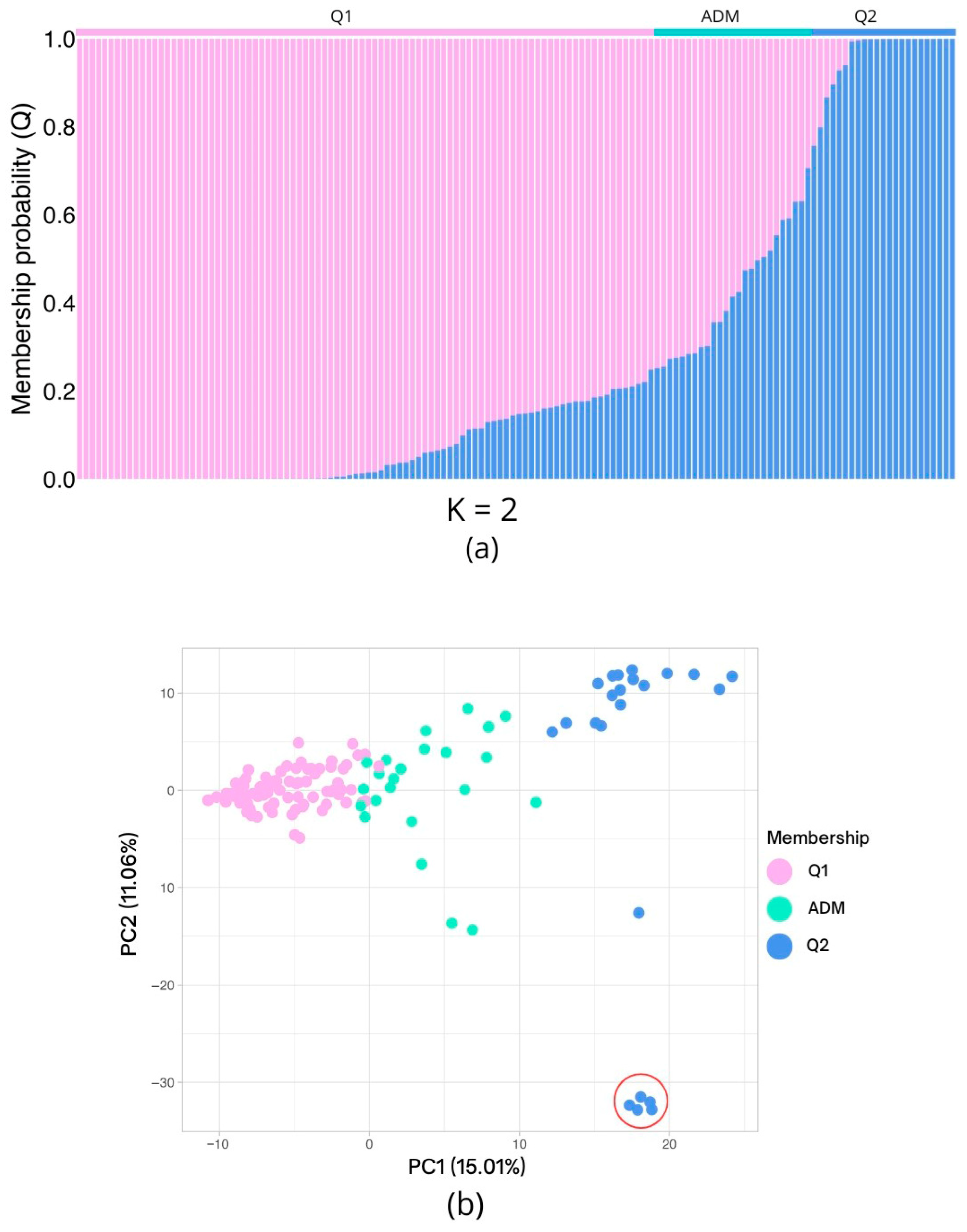
2.4. Population Structure
2.5. Marker–Traits Associations and Discovery of Candidate Genes
3. Discussion
3.1. Genotyping and Linkage Disequilibrium
3.2. Phenotypic Variability
3.3. Population Structure
3.4. Association Analysis
3.4.1. Fruit Weight
3.4.2. Solid Soluble Content
3.4.3. Maturity Date
3.4.4. IAD
4. Materials and Methods
4.1. Plant Material and Genotyping
4.2. Evaluation of Phenology and Fruit Quality Traits
4.3. Linkage Disequilibrium Pattern and Population Structure
4.4. Genome-Wide Association Studies (GWAS) and Annotations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GWAS | Genome-Wide Association Study |
| MTA | Marker–Trait Association |
| SNP | Single-Nucleotide Polymorphism |
| LD | Linkage Disequilibrium |
| FW | Fruit Weight |
| SSC | Soluble Solids Content |
| MD | Maturity Date |
| IAD | Index of Chlorophyll Absorbance Difference |
| BLUEs | Best Linear Unbiased Estimates |
| MLM | Mixed Linear Model |
| QTL | Quantitative Trait Locus/Loci |
| ddRADseq | Double-Digest Restriction-Site Associated DNA Sequencing |
| Mb | Mega-base |
| Kb | Kilo-base |
| GxE | Genotype by Environment |
| PVE | Phenotypic variation explained |
| Q | Membership probability |
| JD | Julian days |
References
- Abbott, A.G.; Lecouls, A.C.; Wang, Y.; Georgi, L.; Scorza, R.; Reighard, G. Peach: The Model Genome for Rosaceae Genomics. Acta Hortic. 2002, 199–209. [Google Scholar] [CrossRef]
- Zheng, Y.; Crawford, G.W.; Chen, X. Archaeological Evidence for Peach (Prunus persica) Cultivation and Domestication in China. PLoS ONE 2014, 9, e106595. [Google Scholar] [CrossRef]
- Li, Y.; Cao, K.; Li, N.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; Guo, J.; Wang, Q.; Ding, T.; et al. Genomic Analyses Provide Insights into Peach Local Adaptation and Responses to Climate Change. Genome Res. 2021, 31, 592–606. [Google Scholar] [CrossRef]
- USDA Foreign Agricultural Service. Fresh Peaches and Cherries: World Markets and Trade; USDA: Washington, DC, USA, 2024. [Google Scholar]
- Frankham, R.; Ballou, J.D.; Ralls, K.; Eldridge, M.D.B.; Dudash, M.R.; Fenster, C.B.; Lacy, R.C.; Sunnucks, P. Loss of Genetic Diversity Reduces Ability to Adapt. In Genetic Management of Fragmented Animal and Plant Populations; Oxford University Press Oxford: Oxford, UK, 2017; pp. 65–86. [Google Scholar]
- Laurens, F.; Aranzana, M.J.; Arus, P.; Bassi, D.; Bink, M.; Bonany, J.; Caprera, A.; Corelli-Grappadelli, L.; Costes, E.; Durel, C.-E.; et al. An Integrated Approach for Increasing Breeding Efficiency in Apple and Peach in Europe. Hortic. Res. 2018, 5, 11. [Google Scholar] [CrossRef]
- Elsadr, H.; Sherif, S.; Banks, T.; Somers, D.; Jayasankar, S. Refining the Genomic Region Containing a Major Locus Controlling Fruit Maturity in Peach. Sci. Rep. 2019, 9, 7522. [Google Scholar] [CrossRef]
- Szabó, Z.; Nyéki, J. Self Pollination in Peach. Int. J. Hortic. Sci. 1999, 5, 76–78. [Google Scholar] [CrossRef]
- Micheletti, D.; Dettori, M.T.; Micali, S.; Aramini, V.; Pacheco, I.; Da Silva Linge, C.; Foschi, S.; Banchi, E.; Barreneche, T.; Quilot-Turion, B.; et al. Whole-Genome Analysis of Diversity and SNP-Major Gene Association in Peach Germplasm. PLoS ONE 2015, 10, e0136803. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Fu, J.; Xu, Y.; Zhang, J.; Ren, F.; Zhao, H.; Tian, S.; Guo, W.; Tu, X.; Zhao, J.; et al. Genome Re-Sequencing Reveals the Evolutionary History of Peach Fruit Edibility. Nat. Commun. 2018, 9, 5404. [Google Scholar] [CrossRef]
- Li, Y.; Cao, K.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; Zhao, P.; Guo, J.; Ding, T.; Guan, L.; et al. Genomic Analyses of an Extensive Collection of Wild and Cultivated Accessions Provide New Insights into Peach Breeding History. Genome Biol. 2019, 20, 36. [Google Scholar] [CrossRef] [PubMed]
- Miller, P.J.; Parfitt, D.E.; Weinbaum, S.A. Outcrossing in Peach. HortScience 1989, 24, 359–360. [Google Scholar] [CrossRef]
- Bouhadida, M.; Moreno, M.Á.; Gonzalo, M.J.; Alonso, J.M.; Gogorcena, Y. Genetic Variability of Introduced and Local Spanish Peach Cultivars Determined by SSR Markers. Tree Genet. Genomes 2011, 7, 257–270. [Google Scholar] [CrossRef]
- Font i Forcada, C.; Oraguzie, N.; Igartua, E.; Moreno, M.Á.; Gogorcena, Y. Population Structure and Marker–Trait Associations for Pomological Traits in Peach and Nectarine Cultivars. Tree Genet. Genomes 2013, 9, 331–349. [Google Scholar] [CrossRef]
- Carrasco, B.; Meisel, L.; Gebauer, M.; Garcia-Gonzales, R.; Silva, H. Breeding in Peach, Cherry and Plum: From a Tissue Culture, Genetic, Transcriptomic and Genomic Perspective. Biol. Res. 2013, 46, 219–230. [Google Scholar] [CrossRef]
- Iglesias, I.; Echeverria, G. Current Situation, Trends and Challenges for Efficient and Sustainable Peach Production. Sci. Hortic. 2022, 296, 110899. [Google Scholar] [CrossRef]
- Luo, C.-X.; Schnabel, G.; Hu, M.; De Cal, A. Global Distribution and Management of Peach Diseases. Phytopathol. Res. 2022, 4, 30. [Google Scholar] [CrossRef]
- Brummer, E.C.; Barber, W.T.; Collier, S.M.; Cox, T.S.; Johnson, R.; Murray, S.C.; Olsen, R.T.; Pratt, R.C.; Thro, A.M. Plant Breeding for Harmony Between Agriculture and the Environment. Front. Ecol. Environ. 2011, 9, 561–568. [Google Scholar] [CrossRef]
- Razzaq, A.; Kaur, P.; Akhter, N.; Wani, S.H.; Saleem, F. Next-Generation Breeding Strategies for Climate-Ready Crops. Front. Plant Sci. 2021, 12, 620420. [Google Scholar] [CrossRef] [PubMed]
- Migicovsky, Z.; Yeats, T.H.; Watts, S.; Song, J.; Forney, C.F.; Burgher-MacLellan, K.; Somers, D.J.; Gong, Y.; Zhang, Z.; Vrebalov, J.; et al. Apple Ripening Is Controlled by a NAC Transcription Factor. Front Genet. 2021, 12, 671300. [Google Scholar] [CrossRef]
- Crump, W.W.; Peace, C.; Zhang, Z.; McCord, P. Detection of Breeding-Relevant Fruit Cracking and Fruit Firmness Quantitative Trait Loci in Sweet Cherry via Pedigree-Based and Genome-Wide Association Approaches. Front Plant Sci. 2022, 13, 823250. [Google Scholar] [CrossRef]
- Matteoli, S.; Diani, M.; Massai, R.; Corsini, G.; Remorini, D. A Spectroscopy-Based Approach for Automated Nondestructive Maturity Grading of Peach Fruits. IEEE Sens. J. 2015, 15, 5455–5464. [Google Scholar] [CrossRef]
- Nascimento, P.A.M.; de Carvalho, L.C.; Júnior, L.C.C.; Pereira, F.M.V.; de Almeida Teixeira, G.H. Robust PLS Models for Soluble Solids Content and Firmness Determination in Low Chilling Peach Using Near-Infrared Spectroscopy (NIR). Postharvest Biol. Technol. 2016, 111, 345–351. [Google Scholar] [CrossRef]
- Onelli, E.; Ghiani, A.; Gentili, R.; Serra, S.; Musacchi, S.; Citterio, S. Specific Changes of Exocarp and Mesocarp Occurring during Softening Differently Affect Firmness in Melting (MF) and Non Melting Flesh (NMF) Fruits. PLoS ONE 2015, 10, e0145341. [Google Scholar] [CrossRef]
- Zhang, B.; Peng, B.; Zhang, C.; Song, Z.; Ma, R. Determination of Fruit Maturity and Its Prediction Model Based on the Pericarp Index of Absorbance Difference (IAD) for Peaches. PLoS ONE 2017, 12, e0177511. [Google Scholar] [CrossRef]
- Ziosi, V.; Noferini, M.; Fiori, G.; Tadiello, A.; Trainotti, L.; Casadoro, G.; Costa, G. A New Index Based on Vis Spectroscopy to Characterize the Progression of Ripening in Peach Fruit. Postharvest Biol. Technol. 2008, 49, 319–329. [Google Scholar] [CrossRef]
- Tijskens, L.M.M.; Zerbini, P.E.; Schouten, R.E.; Vanoli, M.; Jacob, S.; Grassi, M.; Cubeddu, R.; Spinelli, L.; Torricelli, A. Assessing Harvest Maturity in Nectarines. Postharvest Biol. Technol. 2007, 45, 204–213. [Google Scholar] [CrossRef]
- Pinto, C.; Reginato, G.; Shinya, P.; Mesa, K.; Díaz, M.; Atenas, C.; Infante, R. Skin Color and Chlorophyll Absorbance: Indices for Establishing a Harvest Date on Non-Melting Peach. Sci. Hortic. 2015, 192, 231–236. [Google Scholar] [CrossRef]
- Zhang, P.; Wei, Y.; Xu, F.; Wang, H.; Chen, M.; Shao, X. Changes in the Chlorophyll Absorbance Index (IAD) Are Related to Peach Fruit Maturity. N. Z. J. Crop Hortic. Sci. 2020, 48, 34–46. [Google Scholar] [CrossRef]
- Sjöstrand, J.; Tahir, I.; Persson Hovmalm, H.; Garkava-Gustavsson, L.; Stridh, H.; Olsson, M.E. Comparison Between IAD and Other Maturity Indices in Nine Commercially Grown Apple Cultivars. Sci. Hortic. 2024, 324, 112559. [Google Scholar] [CrossRef]
- Breseghello, F.; Sorrells, M.E. Association Mapping of Kernel Size and Milling Quality in Wheat (Triticum Aestivum L.) Cultivars. Genetics 2006, 172, 1165–1177. [Google Scholar] [CrossRef]
- Li, A.; Sun, S.; Wang, H.; Badrunnesa, A.; Meng, J.; Li, X.; Gao, Y.; Niu, L.; Pan, L.; Duan, W.; et al. PeachMD: A Multi-Omics Database for Peach. Mol. Hortic. 2025, 5, 37. [Google Scholar] [CrossRef]
- De Mori, G.; Cipriani, G. Marker-Assisted Selection in Breeding for Fruit Trait Improvement: A Review. Int. J. Mol. Sci. 2023, 24, 8984. [Google Scholar] [CrossRef]
- Aballay, M.M.; Aguirre, N.C.; Filippi, C.V.; Valentini, G.H.; Sánchez, G. Fine-Tuning the Performance of DdRAD-Seq in the Peach Genome. Sci. Rep. 2021, 11, 6298. [Google Scholar] [CrossRef]
- Osorio, M.; Ahumada, S.; Infante, R.; Pacheco, I.; Fiol, A.; Ballesta, P. A Japanese Plum Breeding Core Collection Capturing and Exploiting Genetic Variation. Agriculture 2025, 15, 1369. [Google Scholar] [CrossRef]
- Ballesta, P.; Fiol, A.; Ahumada, S.; Osorio, M.; Ibañez, J.; Fresnedo-Ramírez, J.; Mora-Poblete, F.; Infante, R.; Battistoni, B.; Pacheco, I. Genomic Prediction of Phenological and Fruit-Quality Traits in a Multi-Family Japanese Plum Breeding Population. Hortic. Plant J. 2025; in press. [Google Scholar] [CrossRef]
- Elbasyoni, I.S.; Lorenz, A.J.; Guttieri, M.; Frels, K.; Baenziger, P.S.; Poland, J.; Akhunov, E. A Comparison Between Genotyping-by-Sequencing and Array-Based Scoring of SNPs for Genomic Prediction Accuracy in Winter Wheat. Plant Sci. 2018, 270, 123–130. [Google Scholar] [CrossRef]
- Cao, K.; Zheng, Z.; Wang, L.; Liu, X.; Zhu, G.; Fang, W.; Cheng, S.; Zeng, P.; Chen, C.; Wang, X.; et al. Comparative Population Genomics Reveals the Domestication History of the Peach, Prunus persica, and Human Influences on Perennial Fruit Crops. Genome Biol. 2014, 15, 415. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, Y.; Wang, X.; Zhao, G.; Zhu, G.; Cao, K.; Fang, W.; Wu, J.; Ma, K.; Chen, C.; et al. Genomic Analysis Provides Insights into the Westward Expansion of Domesticated Peaches in China. Hortic. Plant J. 2024, 10, 367–375. [Google Scholar] [CrossRef]
- Yu, Y.; Guan, J.; Xu, Y.; Ren, F.; Zhang, Z.; Yan, J.; Fu, J.; Guo, J.; Shen, Z.; Zhao, J.; et al. Population-Scale Peach Genome Analyses Unravel Selection Patterns and Biochemical Basis Underlying Fruit Flavor. Nat. Commun. 2021, 12, 3604. [Google Scholar] [CrossRef] [PubMed]
- Mas-Gómez, J.; Cantín, C.M.; Moreno, M.Á.; Martínez-García, P.J. Genetic Diversity and Genome-Wide Association Study of Morphological and Quality Traits in Peach Using Two Spanish Peach Germplasm Collections. Front. Plant Sci. 2022, 13, 854770. [Google Scholar] [CrossRef]
- Vodiasova, E.; Pronozin, A.; Rozanova, I.; Tsiupka, V.; Vasiliev, G.; Plugatar, Y.; Dolgov, S.; Smykov, A. Genetic Diversity and Population Structure of Prunus persica Cultivars Revealed by Genotyping-by-Sequencing (GBS). Horticulturae 2025, 11, 189. [Google Scholar] [CrossRef]
- Rawandoozi, Z.; Hartmann, T.; Byrne, D.; Carpenedo, S. Heritability, Correlation, and Genotype by Environment Interaction of Phenological and Fruit Quality Traits in Peach. J. Am. Soc. Hortic. Sci. 2021, 146, 56–67. [Google Scholar] [CrossRef]
- Biscarini, F.; Nazzicari, N.; Bink, M.; Arús, P.; Aranzana, M.J.; Verde, I.; Micali, S.; Pascal, T.; Quilot-Turion, B.; Lambert, P.; et al. Genome-Enabled Predictions for Fruit Weight and Quality from Repeated Records in European Peach Progenies. BMC Genom. 2017, 18, 432. [Google Scholar] [CrossRef]
- da Silva Linge, C.; Bassi, D.; Bianco, L.; Pacheco, I.; Pirona, R.; Rossini, L. Genetic Dissection of Fruit Weight and Size in an F2 Peach (Prunus persica (L.) Batsch) Progeny. Mol. Breed. 2015, 35, 71. [Google Scholar] [CrossRef]
- Dirlewanger, E.; Quero-García, J.; Le Dantec, L.; Lambert, P.; Ruiz, D.; Dondini, L.; Illa, E.; Quilot-Turion, B.; Audergon, J.-M.; Tartarini, S.; et al. Comparison of the Genetic Determinism of Two Key Phenological Traits, Flowering and Maturity Dates, in Three Prunus Species: Peach, Apricot and Sweet Cherry. Heredity 2012, 109, 280–292. [Google Scholar] [CrossRef]
- Hernández Mora, J.R.; Micheletti, D.; Bink, M.; Van de Weg, E.; Cantín, C.; Nazzicari, N.; Caprera, A.; Dettori, M.T.; Micali, S.; Banchi, E.; et al. Integrated QTL Detection for Key Breeding Traits in Multiple Peach Progenies. BMC Genom. 2017, 18, 404. [Google Scholar] [CrossRef] [PubMed]
- Salazar, J.A.; Pacheco, I.; Zapata, P.; Shinya, P.; Ruiz, D.; Martínez-Gómez, P.; Infante, R. Identification of Loci Controlling Phenology, Fruit Quality and Post-Harvest Quantitative Parameters in Japanese Plum (Prunus salicina Lindl.). Postharvest Biol. Technol. 2020, 169, 111292. [Google Scholar] [CrossRef]
- Khadivi-Khub, A.; Khalili, Z. A Breeding Project: The Selection of Promising Apricot (Prunus Armeniaca L.) Genotypes with Late Blooming Time and High Fruit Quality. Sci. Hortic. 2017, 216, 93–102. [Google Scholar] [CrossRef]
- Aranzana, M.J.; Decroocq, V.; Dirlewanger, E.; Eduardo, I.; Gao, Z.S.; Gasic, K.; Iezzoni, A.; Jung, S.; Peace, C.; Prieto, H.; et al. Prunus Genetics and Applications after de Novo Genome Sequencing: Achievements and Prospects. Hortic. Res. 2019, 6, 58. [Google Scholar] [CrossRef]
- Shin, J.S.; Park, H.S.; Lee, K.W.; Song, J.S.; Han, H.Y.; Kim, H.W.; Cho, T.J. Advances in the Strategic Approaches of Pre- and Post-Harvest Treatment Technologies for Peach Fruits (Prunus persica). Horticulturae 2023, 9, 315. [Google Scholar] [CrossRef]
- Ksouri, N.; Sánchez, G.; Font i Forcada, C.; Contreras-Moreira, B.; Gogorcena, Y. A Reproducible DdRAD-Seq Protocol Reveals Novel Genomic Association Signatures for Fruit-Related Traits in Peach. Plant Methods 2025, 21, 101. [Google Scholar] [CrossRef]
- Dirlewanger, E.; Moing, A.; Rothan, C.; Svanella, L.; Pronier, V.; Guye, A.; Plomion, C.; Monet, R. Mapping QTLs Controlling Fruit Quality in Peach (Prunus persica (L.) Batsch). Theor. Appl. Genet. 1999, 98, 18–31. [Google Scholar] [CrossRef]
- Eduardo, I.; Pacheco, I.; Chietera, G.; Bassi, D.; Pozzi, C.; Vecchietti, A.; Rossini, L. QTL Analysis of Fruit Quality Traits in Two Peach Intraspecific Populations and Importance of Maturity Date Pleiotropic Effect. Tree Genet. Genomes 2011, 7, 323–335. [Google Scholar] [CrossRef]
- Jiang, Q.; Xu, Q.; Pan, J.; Yao, X.; Cheng, Z. Impacts of Chronic Habitat Fragmentation on Genetic Diversity of Natural Populations of Prunus persica in China. Plants 2022, 11, 1458. [Google Scholar] [CrossRef]
- Font i Forcada, C.; Guajardo, V.; Chin-Wo, S.R.; Moreno, M.Á. Association Mapping Analysis for Fruit Quality Traits in Prunus persica Using SNP Markers. Front. Plant Sci. 2019, 9, 2005. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software STRUCTURE: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Cirilli, M.; Rossini, L.; Geuna, F.; Palmisano, F.; Minafra, A.; Castrignanò, T.; Gattolin, S.; Ciacciulli, A.; Babini, A.R.; Liverani, A.; et al. Genetic Dissection of Sharka Disease Tolerance in Peach (P. persica L. Batsch). BMC Plant Biol. 2017, 17, 192. [Google Scholar] [CrossRef]
- McCarthy, M.I.; Abecasis, G.R.; Cardon, L.R.; Goldstein, D.B.; Little, J.; Ioannidis, J.P.A.; Hirschhorn, J.N. Genome-Wide Association Studies for Complex Traits: Consensus, Uncertainty and Challenges. Nat. Rev. Genet. 2008, 9, 356–369. [Google Scholar] [CrossRef] [PubMed]
- Hamblin, M.T.; Buckler, E.S.; Jannink, J.-L. Population Genetics of Genomics-Based Crop Improvement Methods. Trends Genet. 2011, 27, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Ziyatdinov, A.; Kim, J.; Prokopenko, D.; Privé, F.; Laporte, F.; Loh, P.-R.; Kraft, P.; Aschard, H. Estimating the Effective Sample Size in Association Studies of Quantitative Traits. G3 Genes Genomes Genet. 2021, 11, jkab057. [Google Scholar] [CrossRef]
- Cao, K.; Chen, C.; Yang, X.; Bie, H.; Wang, L. Genomic Selection for Fruit Weight and Soluble Solid Contents in Peach. Sci. Agric. Sin. 2023, 56, 951–963. [Google Scholar]
- Beavis, W.; Mahama, A.A.; Suza, W. Multi Environment Trials: Linear Mixed Models. In Quantitative Genetics for Plant Breeding; Iowa State University Digital Press: Ames, IA, USA, 2023. [Google Scholar]
- Kaler, A.S.; Purcell, L.C. Estimation of a Significance Threshold for Genome-Wide Association Studies. BMC Genom. 2019, 20, 618. [Google Scholar] [CrossRef]
- Johnson, R.C.; Nelson, G.W.; Troyer, J.L.; Lautenberger, J.A.; Kessing, B.D.; Winkler, C.A.; O’Brien, S.J. Accounting for Multiple Comparisons in a Genome-Wide Association Study (GWAS). BMC Genom. 2010, 11, 724. [Google Scholar] [CrossRef]
- Shi, P.; Xu, Z.; Zhang, S.; Wang, X.; Ma, X.; Zheng, J.; Xing, L.; Zhang, D.; Ma, J.; Han, M.; et al. Construction of a High-Density SNP-Based Genetic Map and Identification of Fruit-Related QTLs and Candidate Genes in Peach [Prunus persica (L.) Batsch]. BMC Plant Biol. 2020, 20, 438. [Google Scholar] [CrossRef]
- Cirilli, M.; Baccichet, I.; Chiozzotto, R.; Silvestri, C.; Rossini, L.; Bassi, D. Genetic and Phenotypic Analyses Reveal Major Quantitative Loci Associated to Fruit Size and Shape Traits in a Non-Flat Peach Collection (P. persica L. Batsch). Hortic. Res. 2021, 8, 232. [Google Scholar] [CrossRef]
- Cao, K.; Li, Y.; Deng, C.H.; Gardiner, S.E.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; Wang, L. Comparative Population Genomics Identified Genomic Regions and Candidate Genes Associated with Fruit Domestication Traits in Peach. Plant Biotechnol. J. 2019, 17, 1954–1970. [Google Scholar] [CrossRef]
- Li, H.; Dong, Q.; Zhu, X.; Zhao, Q.; Ran, K. Genome-Wide Identification, Expression, and Interaction Analysis for Ovate Family Proteins in Peach. Mol. Biol. Rep. 2019, 46, 3755–3764. [Google Scholar] [CrossRef] [PubMed]
- Veerappan, K.; Natarajan, S.; Chung, H.; Park, J. Molecular Insights of Fruit Quality Traits in Peaches, Prunus persica. Plants 2021, 10, 2191. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Ma, R.; Gao, L.; Zhang, J.; Zhang, A.; Zhang, X.; Ren, F.; Zhang, W.; Liao, L.; Yang, Q.; et al. A 1.7-Mb Chromosomal Inversion Downstream of a PpOFP1 Gene Is Responsible for Flat Fruit Shape in Peach. Plant Biotechnol. J. 2021, 19, 192–205. [Google Scholar] [CrossRef] [PubMed]
- Jurado, S.; Abraham, Z.; Manzano, C.; López-Torrejón, G.; Pacios, L.F.; Del Pozo, J.C. The Arabidopsis Cell Cycle F-Box Protein SKP2A Binds to Auxin. Plant Cell 2011, 22, 3891–3904. [Google Scholar] [CrossRef]
- Zhang, J.; Wei, B.; Yuan, R.; Wang, J.; Ding, M.; Chen, Z.; Yu, H.; Qin, G. The Arabidopsis RING-Type E3 Ligase TEAR1 Controls Leaf Development by Targeting the TIE1 Transcriptional Repressor for Degradation. Plant Cell 2017, 29, 243–259. [Google Scholar] [CrossRef]
- Wang, X.; Kota, U.; He, K.; Blackburn, K.; Li, J.; Goshe, M.B.; Huber, S.C.; Clouse, S.D. Sequential Transphosphorylation of the BRI1/BAK1 Receptor Kinase Complex Impacts Early Events in Brassinosteroid Signaling. Dev. Cell 2008, 15, 220–235. [Google Scholar] [CrossRef]
- Rose, J.K.C.; Bennett, A.B. Cooperative Disassembly of the Cellulose–Xyloglucan Network of Plant Cell Walls: Parallels Between Cell Expansion and Fruit Ripening. Trends Plant Sci. 1999, 4, 176–183. [Google Scholar] [CrossRef]
- Micheli, F. Pectin Methylesterases: Cell Wall Enzymes with Important Roles in Plant Physiology. Trends Plant Sci. 2001, 6, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Nuñez-Lillo, G.; Balladares, C.; Pavez, C.; Urra, C.; Sanhueza, D.; Vendramin, E.; Dettori, M.T.; Arús, P.; Verde, I.; Blanco-Herrera, F.; et al. High-Density Genetic Map and QTL Analysis of Soluble Solid Content, Maturity Date, and Mealiness in Peach Using Genotyping by Sequencing. Sci. Hortic. 2019, 257, 108734. [Google Scholar] [CrossRef]
- Bemer, M.; Karlova, R.; Ballester, A.R.; Tikunov, Y.M.; Bovy, A.G.; Wolters-Arts, M.; Rossetto, P.d.B.; Angenent, G.C.; de Maagd, R.A. The Tomato FRUITFULL Homologs TDR4/FUL1 and MBP7/FUL2 Regulate Ethylene-Independent Aspects of Fruit Ripening. Plant Cell 2012, 24, 4437–4451. [Google Scholar] [CrossRef] [PubMed]
- Cirilli, M.; Bassi, D.; Ciacciulli, A. Sugars in Peach Fruit: A Breeding Perspective. Hortic. Res. 2016, 3, 15067. [Google Scholar] [CrossRef]
- Cao, K.; Zhou, Z.; Wang, Q.; Guo, J.; Zhao, P.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; Wang, X.; et al. Genome-Wide Association Study of 12 Agronomic Traits in Peach. Nat. Commun. 2016, 7, 13246. [Google Scholar] [CrossRef]
- da Silva Linge, C.; Cai, L.; Fu, W.; Clark, J.; Worthington, M.; Rawandoozi, Z.; Byrne, D.H.; Gasic, K. Corrigendum: Multi-Locus Genome-Wide Association Studies Reveal Fruit Quality Hotspots in Peach Genome. Front. Plant Sci. 2022, 13, 879112. [Google Scholar] [CrossRef]
- Chen, Z.; Guo, X.; Du, J.; Yu, M. ALA Promotes Sucrose Accumulation in Early Peach Fruit by Regulating SPS Activity. Curr. Issues Mol. Biol. 2024, 46, 7944–7954. [Google Scholar] [CrossRef]
- Pirona, R.; Eduardo, I.; Pacheco, I.; Da Silva Linge, C.; Miculan, M.; Verde, I.; Tartarini, S.; Dondini, L.; Pea, G.; Bassi, D.; et al. Fine Mapping and Identification of a Candidate Gene for a Major Locus Controlling Maturity Date in Peach. BMC Plant Biol. 2013, 13, 166. [Google Scholar] [CrossRef]
- Salazar, J.A.; Ruiz, D.; Campoy, J.A.; Tartarini, S.; Dondini, L.; Martínez-Gómez, P. Inheritance of Reproductive Phenology Traits and Related QTL Identification in Apricot. Tree Genet. Genomes 2016, 12, 71. [Google Scholar] [CrossRef]
- Salazar, J.A.; Pacheco, I.; Shinya, P.; Zapata, P.; Silva, C.; Aradhya, M.; Velasco, D.; Ruiz, D.; Martínez-Gómez, P.; Infante, R. Genotyping by Sequencing for SNP-Based Linkage Analysis and Identification of QTLs Linked to Fruit Quality Traits in Japanese Plum (Prunus Salicina Lindl.). Front. Plant Sci. 2017, 8, 476. [Google Scholar] [CrossRef]
- Calle, A.; Wünsch, A. Multiple-Population QTL Mapping of Maturity and Fruit-Quality Traits Reveals LG4 Region as a Breeding Target in Sweet Cherry (Prunus Avium L.). Hortic. Res. 2020, 7, 127. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Liu, Y.; Zhang, X.; Chen, X.; Sun, J.; Zhao, Y.; Zhang, J.; Yao, J.; Liao, L.; Zhou, H.; et al. Two Adjacent NAC Transcription Factors Regulate Fruit Maturity Date and Flavor in Peach. New Phytol. 2024, 241, 632–649. [Google Scholar] [CrossRef] [PubMed]
- Romeu, J.F.; Monforte, A.J.; Sánchez, G.; Granell, A.; García-Brunton, J.; Badenes, M.L.; Ríos, G. Quantitative Trait Loci Affecting Reproductive Phenology in Peach. BMC Plant Biol. 2014, 14, 52. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zhao, L.; Yang, Q.; Amar, M.H.; Ogutu, C.; Peng, Q.; Liao, L.; Zhang, J.; Han, Y. Identification of EIL and ERF Genes Related to Fruit Ripening in Peach. Int. J. Mol. Sci. 2020, 21, 2846. [Google Scholar] [CrossRef]
- Baranov, D.; Timerbaev, V. Recent Advances in Studying the Regulation of Fruit Ripening in Tomato Using Genetic Engineering Approaches. Int. J. Mol. Sci. 2024, 25, 760. [Google Scholar] [CrossRef]
- Rose, J.K.C.; Cosgrove, D.J.; Albersheim, P.; Darvill, A.G.; Bennett, A.B. Detection of Expansin Proteins and Activity during Tomato Fruit Ontogeny. Plant Physiol. 2000, 123, 1583–1592. [Google Scholar] [CrossRef]
- Tijskens, L.M.M.; Rodis, P.S.; Hertog, M.L.A.T.M.; Kalantzi, U.; van Dijk, C. Kinetics of Polygalacturonase Activity and Firmness of Peaches during Storage. J. Food Eng. 1998, 35, 111–126. [Google Scholar] [CrossRef]
- Uluisik, S.; Seymour, G.B. Pectate Lyases: Their Role in Plants and Importance in Fruit Ripening. Food Chem. 2020, 309, 125559. [Google Scholar] [CrossRef]
- Aranzana, M.J.; Illa, E.; Howad, W.; Arús, P. A First Insight into Peach [Prunus persica (L.) Batsch] SNP Variability. Tree Genet. Genomes 2012, 8, 1359–1369. [Google Scholar] [CrossRef]
- Zita, W.; Bressoud, S.; Glauser, G.; Kessler, F.; Shanmugabalaji, V. Chromoplast Plastoglobules Recruit the Carotenoid Biosynthetic Pathway and Contribute to Carotenoid Accumulation during Tomato Fruit Maturation. PLoS ONE 2022, 17, e0277774. [Google Scholar] [CrossRef]
- Kim, I.; Kim, E.-H.; Choi, Y.; Kim, H.U. Fibrillin2 in Chloroplast Plastoglobules Participates in Photoprotection and Jasmonate-Induced Senescence. Plant Physiol. 2022, 189, 1363–1379. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zeng, W.; Ding, Y.; Wang, Y.; Niu, L.; Yao, J.-L.; Pan, L.; Lu, Z.; Cui, G.; Li, G.; et al. PpERF3 Positively Regulates ABA Biosynthesis by Activating PpNCED2/3 Transcription during Fruit Ripening in Peach. Hortic. Res. 2019, 6, 19. [Google Scholar] [CrossRef] [PubMed]
- Mou, W.; Li, D.; Luo, Z.; Mao, L.; Ying, T. Transcriptomic Analysis Reveals Possible Influences of ABA on Secondary Metabolism of Pigments, Flavonoids and Antioxidants in Tomato Fruit during Ripening. PLoS ONE 2015, 10, e0129598. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, X.; Meng, J.; Li, A.; Duan, W.; Sun, S.; Pan, L.; Zeng, W.; Wang, Z.; Niu, L. Expression Analysis of Chlorophyll-Degradation-Related Genes in Prunus persica L. Peel and the Functional Verification of Key Genes. Plants 2025, 14, 312. [Google Scholar] [CrossRef]
- Healey, A.; Furtado, A.; Cooper, T.; Henry, R.J. Protocol: A Simple Method for Extracting next-Generation Sequencing Quality Genomic DNA from Recalcitrant Plant Species. Plant Methods 2014, 10, 21. [Google Scholar] [CrossRef]
- Peterson, B.K.; Weber, J.N.; Kay, E.H.; Fisher, H.S.; Hoekstra, H.E. Double Digest RADseq: An Inexpensive Method for De Novo SNP Discovery and Genotyping in Model and Non-Model Species. PLoS ONE 2012, 7, e37135. [Google Scholar] [CrossRef]
- Catchen, J.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An Analysis Tool Set for Population Genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef]
- Verde, I.; Jenkins, J.; Dondini, L.; Micali, S.; Pagliarani, G.; Vendramin, E.; Paris, R.; Aramini, V.; Gazza, L.; Rossini, L.; et al. The Peach v2.0 Release: High-Resolution Linkage Mapping and Deep Resequencing Improve Chromosome-Scale Assembly and Contiguity. BMC Genom. 2017, 18, 225. [Google Scholar] [CrossRef]
- Md, V.; Misra, S.; Li, H.; Aluru, S. Efficient Architecture-Aware Acceleration of BWA-MEM for Multicore Systems. In Proceedings of the 2019 IEEE International Parallel and Distributed Processing Symposium (IPDPS), Rio de Janeiro, Brazil, 20–24 May 2019; IEEE: New York, NY, USA, 2019; pp. 314–324. [Google Scholar]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The Variant Call Format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Koné, S. R Core Team R: A Language and Environment for Statistical Computing. Int. J. Mod. Nonlinear Theory Appl. 2024, 13, 53–69. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, H.; Tang, Z.; Xu, J.; Yin, D.; Zhang, Z.; Yuan, X.; Zhu, M.; Zhao, S.; Li, X.; et al. RMVP: A Memory-Efficient, Visualization-Enhanced, and Parallel-Accelerated Tool for Genome-Wide Association Study. Genom. Proteom. Bioinform. 2021, 19, 619–628. [Google Scholar] [CrossRef] [PubMed]
- García-Abadillo, J.; Barba, P.; Carvalho, T.; Sosa-Zuñiga, V.; Lozano, R.; Carvalho, H.F.; Garcia-Rojas, M.; Salazar, E.; y Sánchez, J.I. Dissecting the Complex Genetic Basis of Pre- and Post-Harvest Traits in Vitis Vinifera L. Using Genome-Wide Association Studies. Hortic. Res. 2024, 11, uhad283. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for Association Mapping of Complex Traits in Diverse Samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Hill, W.G.; Weir, B.S. Maximum-Likelihood Estimation of Gene Location by Linkage Disequilibrium. Am. J. Hum. Genet. 1994, 54, 705–714. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A Fast, Scalable and User-Friendly Tool for Maximum Likelihood Phylogenetic Inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Vroh Bi, I.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A Unified Mixed-Model Method for Association Mapping That Accounts for Multiple Levels of Relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Törönen, P.; Medlar, A.; Holm, L. PANNZER2: A Rapid Functional Annotation Web Server. Nucleic Acids Res. 2018, 46, W84–W88. [Google Scholar] [CrossRef] [PubMed]
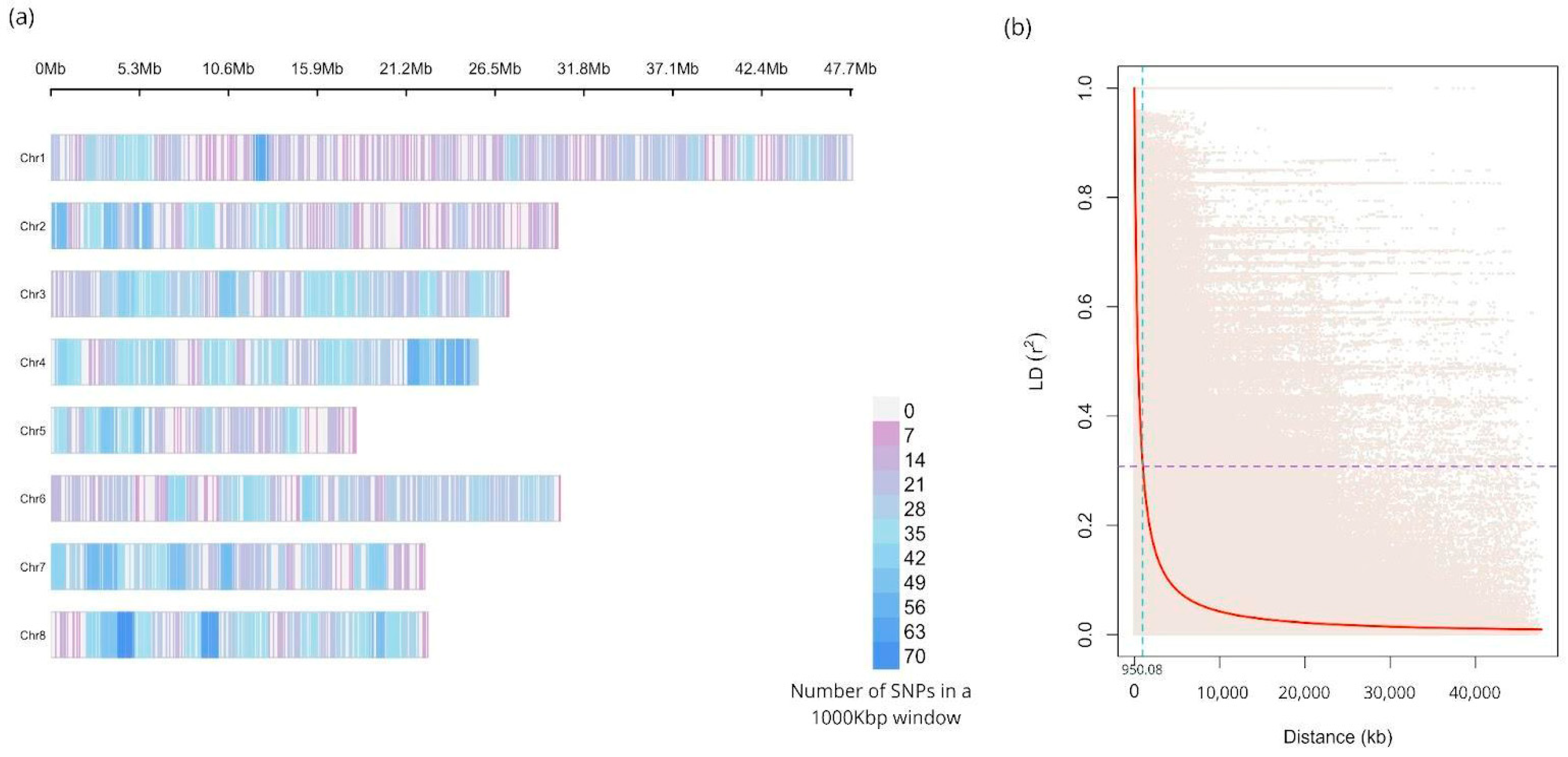



| Chromosome | Chr. Size (Mbp) | N° Markers | Marker Density | N° Cross-Paired Markers | r2 |
|---|---|---|---|---|---|
| Chr1 | 47.86 | 956 | 2.00 | 456,490 | 0.1245 |
| Chr2 | 30.43 | 695 | 2.28 | 241,165 | 0.1512 |
| Chr3 | 27.39 | 769 | 2.81 | 295,296 | 0.1222 |
| Chr4 | 25.86 | 859 | 3.32 | 368,511 | 0.1527 |
| Chr5 | 18.50 | 452 | 2.44 | 101,926 | 0.2573 |
| Chr6 | 30.79 | 715 | 2.32 | 255,255 | 0.1265 |
| Chr7 | 22.39 | 683 | 3.05 | 232,903 | 0.1164 |
| Chr8 | 22.58 | 732 | 3.24 | 267,546 | 0.1293 |
| Trait | Marker | Window | p-Value | Gene | Protein Description | Annotation |
|---|---|---|---|---|---|---|
| FW | Pp01_26499374 | 26024374:26974374 | 0.00339 | Prupe.1G255100 | Xyloglucan endotransglucosylase/ hydrolase | xyloglucan:xyloglucosyl transferase activity; xyloglucan metabolic process, multidimensional cell growth, plant-type cell wall organization or biogenesis |
| Pp01_26611800 | 26136800:27086800 | 0.00339 | ||||
| Pp01_26952521 | 26477521:27427521 | 0.00297 | ||||
| Pp01_31541184 | 31066184:32016184 | 0.00412 | Prupe.1G337000 | Xyloglucan endotransglucosylase/hydrolase | xyloglucan:xyloglucosyl transferase activity, xyloglucan metabolic process | |
| Pp01_31546083 | 31071083:32021083 | 0.00412 | ||||
| Pp01_31568771 | 31093771:32043771 | 0.00412 | ||||
| Pp01_31601793 | 31126793:32076793 | 0.00410 | Prupe.1G417900 | Pectin methylesterase CGR3 | pectin metabolic process; methyltransferase activity | |
| Pp03_5240083 | 4765083:5715083 | 0.00482 | Prupe.3G069200 | OVATE domain-containing protein | Helicase activity | |
| Pp03_5246023 | 4771023:5721023 | 0.00239 | ||||
| Pp03_5361469 | 4886469:5836469 | 0.00160 | ||||
| Pp06_3223902 | 2748902:3698902 | 5.52 × 10−5 | Prupe.6G050800 | endo-1,3-beta-glucosidase 9 | glucan endo-1,3-beta-D-glucosidase activity | |
| Pp08_8024146 | 7549146:8499146 | 0.00205 | Prupe.8G152500 | Xyloglucan endotransglucosylase/hydrolase | xyloglucan:xyloglucosyl transferase activity; xyloglucan metabolic process | |
| Pp08_15930625 | 15455625:16405625 | 4.34 × 10−4 | Prupe.8G122300; Prupe.8G140300; Prupe.8G140900 | F-box (SKIP22/FBXO-like, SCF E3 ligase); RING-type E3 ubiquitin (RHY1A-like); BAK1-like (BRI1 LRR-RLK co-receptor) | Cyclin-dependent protein serine/threonine kinase inhibitor activity; ubiquitin protein ligase activity; protein binding, brassinosteroid signalling | |
| Pp08_21623883 | 21148883:22098883 | 0.00491 | Prupe.8G247700 | Auxin-binding protein T85 | auxin binding; positive regulation of cell size | |
| Pp08_21685700 | 21210700:22160700 | 0.00491 | ||||
| Pp08_21717973 | 21242973:22192973 | 0.00563 | ||||
| Pp08_21725549 | 21250549:22200549 | 0.00513 | ||||
| SSC | Pp01_12736773 | 12261773:13211773 | 0.00625 | Prupe.1G159700 | Sucrose-phosphate synthase | sucrose-phosphate synthase activity; sucrose biosynthetic process |
| Pp04_16076936 | 15601936:16551936 | 0.00336 | Prupe.4G240300 | Sorbitol dehydrogenase | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | |
| Pp05_16778038 | 16328038:17228038 | 3.75 × 10−5 | Prupe.5G208500 | Agamous-like MADS-box protein AGL8 homolog (CMB1-like) | RNA polymerase II transcription regulatory region sequence-specific DNA binding, protein dimerization activity, 2-alkenal reductase [NAD(P)+] activity, positive regulation of transcription by RNA polymerase II | |
| MD | Pp03_26053887 | 25578887:26528887 | 0.00316 | Prupe.3G287200 | Polygalacturonase | polygalacturonase activity |
| Pp03_26061543 | 25586543:26536543 | 0.00270 | ||||
| Pp03_26442013 | 25967013:26917013 | 0.00211 | Prupe.3G296600 | Pectate lyase | pectate lyase activity; pectin catabolic process | |
| Pp03_26631520 | 26156520:27106520 | 0.00463 | ||||
| Pp03_26643124 | 26168124:27118124 | 0.00460 | ||||
| Pp03_26785134 | 26310134:27260134 | 0.00316 | Prupe.3G309400 | Omega-hydroxypalmitate O-feruloyl transferase | fruit ripening, climacteric; response to ethylene | |
| Pp03_2995684 | 2520684:3470684 | 0.00608 | Prupe.3G043600 | Putative polygalacturonase (Fragment) | polygalacturonase activity | |
| Pp03_3093068 | 2618068:3568068 | 0.00488 | ||||
| Pp04_9841821 | 9366821:10316821 | 0.00243 | Prupe.4G165100 | Pectate lyase superfamily protein domain-containing protein | polygalacturonase activity | |
| Pp04_9871961 | 9396961:10346961 | 0.00528 | ||||
| Pp04_11112825 | 10662825:11562825 | 2.53 × 10−6 | Prupe.4G186800, Prupe.4G187100 | NAC domain-containing protein 72, NAC transcription factor 25 | Sequence-specific DNA binding, regulation of DNA-templated transcription | |
| Pp06_2988972 * | 2538972:3438972 | 1.97 × 10−5 | Prupe.6G039700, Prupe.6G046900, Prupe.6G042000 | ERF/PTI6-like; WRKY33-like; Expansin-A8 (ExpA8) | Regulation of DNA-templated transcription, ethylene-activated signaling pathway and DNA-binding transcription factor activity; Regulation of DNA-templated transcription and DNA-binding transcription factor activity; plant-type cell wall organization and anatomical structure morphogenesis | |
| Pp06_24178736 | 23703736:24653736 | 0.00107 | Prupe.6G242400 | Ethylene-responsive transcription factor RAP2-7 | DNA-binding transcription factor activity | |
| Pp06_24240967 | 23765967:24715967 | 0.00104 | ||||
| Pp06_24315671 | 23840671:24790671 | 0.00101 | ||||
| Pp06_24318377 | 23843377:24793377 | 0.00048 | Prupe.6G247100 | Pectate lyase | pectate lyase activity; pectin catabolic process | |
| Pp06_25309512 | 24834512:25784512 | 0.00413 | ||||
| IAD | Pp01_33303400 | 32828400:33778400 | 0.00389 | Prupe.1G353100 | Pheophytinase | chlorophyllase activity |
| Pp02_19688605 | 19688605:20138605 | 6.83 × 10−6 | Prupe.2G135300 | Plastid-lipid associated protein PAP/fibrillin family protein | Chromoplast remodelling, plastid localization | |
| Pp04_4119665 | 3644665:4594665 | 2.44 × 10−4 | Prupe.4G082000 | 9-cis-epoxycarotenoid dioxygenase NCED2, chloroplastic | carotene catabolic process; abscisic acid biosynthetic process; chloroplast stroma; carotenoid dioxygenase activity; metal ion binding | |
| Pp06_8576293 | 8101293:9051293 | 0.00589 | Prupe.6G113600 | Pheophorbide a oxygenase, chloroplastic | pheophorbide a oxygenase activity; chlorophyllide a oxygenase activity | |
| Pp06_8587833 | 8112833:9062833 | 0.00589 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Osorio, M.; Fiol, A.; Ballesta, P.; Ahumada, S.; Marambio, P.; Martínez-Carrasco, P.; Infante, R.; Pacheco, I. Multi-Season Genome-Wide Association Study Reveals Loci and Candidate Genes for Fruit Quality and Maturity Traits in Peach. Plants 2026, 15, 189. https://doi.org/10.3390/plants15020189
Osorio M, Fiol A, Ballesta P, Ahumada S, Marambio P, Martínez-Carrasco P, Infante R, Pacheco I. Multi-Season Genome-Wide Association Study Reveals Loci and Candidate Genes for Fruit Quality and Maturity Traits in Peach. Plants. 2026; 15(2):189. https://doi.org/10.3390/plants15020189
Chicago/Turabian StyleOsorio, María, Arnau Fiol, Paulina Ballesta, Sebastián Ahumada, Pilar Marambio, Pamela Martínez-Carrasco, Rodrigo Infante, and Igor Pacheco. 2026. "Multi-Season Genome-Wide Association Study Reveals Loci and Candidate Genes for Fruit Quality and Maturity Traits in Peach" Plants 15, no. 2: 189. https://doi.org/10.3390/plants15020189
APA StyleOsorio, M., Fiol, A., Ballesta, P., Ahumada, S., Marambio, P., Martínez-Carrasco, P., Infante, R., & Pacheco, I. (2026). Multi-Season Genome-Wide Association Study Reveals Loci and Candidate Genes for Fruit Quality and Maturity Traits in Peach. Plants, 15(2), 189. https://doi.org/10.3390/plants15020189







