Network Pharmacology of the Phytochemical Content of Sunflower Seed (Helianthus annuus L.) Extract from LC-MS on Wound-Healing Activity and the In Vitro Wound Scratch Assay
Abstract
1. Introduction
2. Results
2.1. Antioxidant Activity of the Sunflower Seed Extract
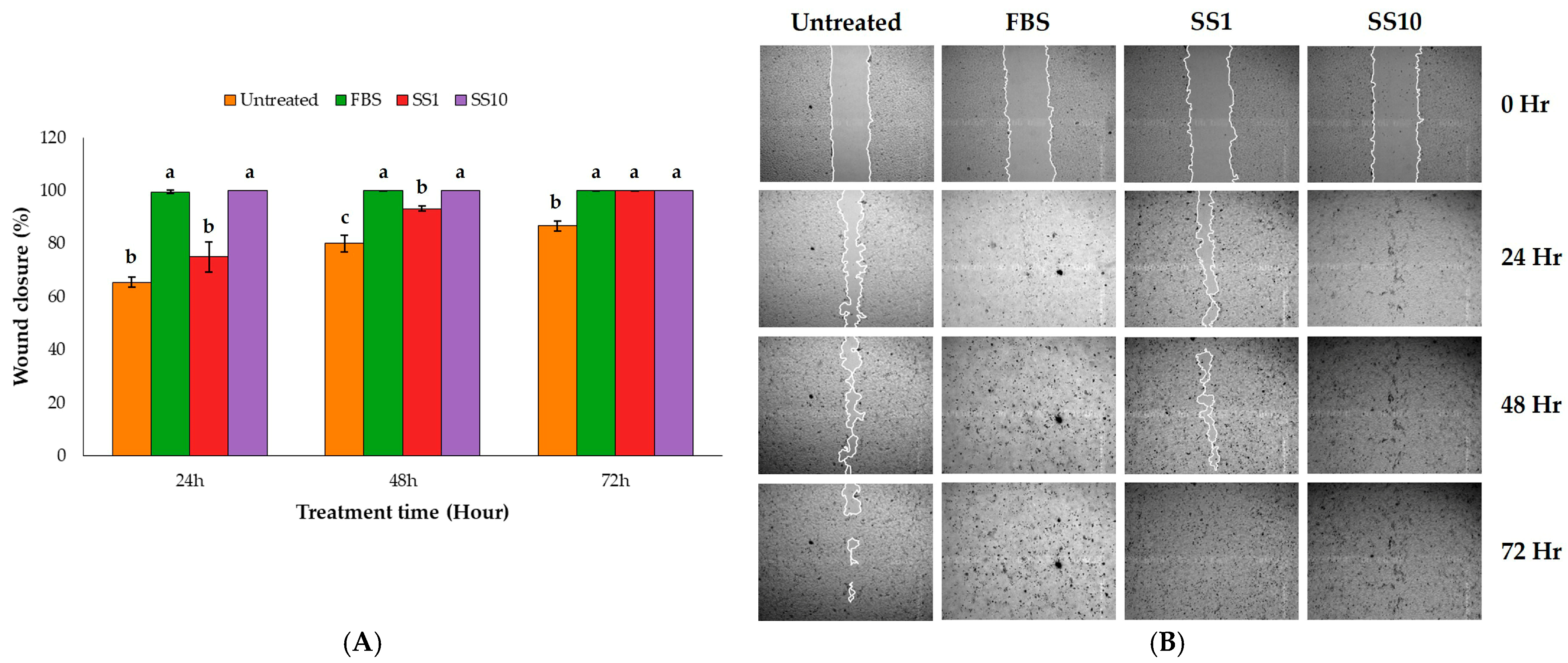
2.2. Wound-Healing Assay
2.3. Phytochemical Profiling
2.3.1. Analysis of the Sunflower Seed Extract via Liquid Chromatography–Mass Spectrometry (LC-MS)
2.3.2. Quantification of Phenolic and Indolamine Compounds
2.4. Network Pharmacology Was Used to Determine the Wound-Healing Effect of the Sunflower Seed Extract
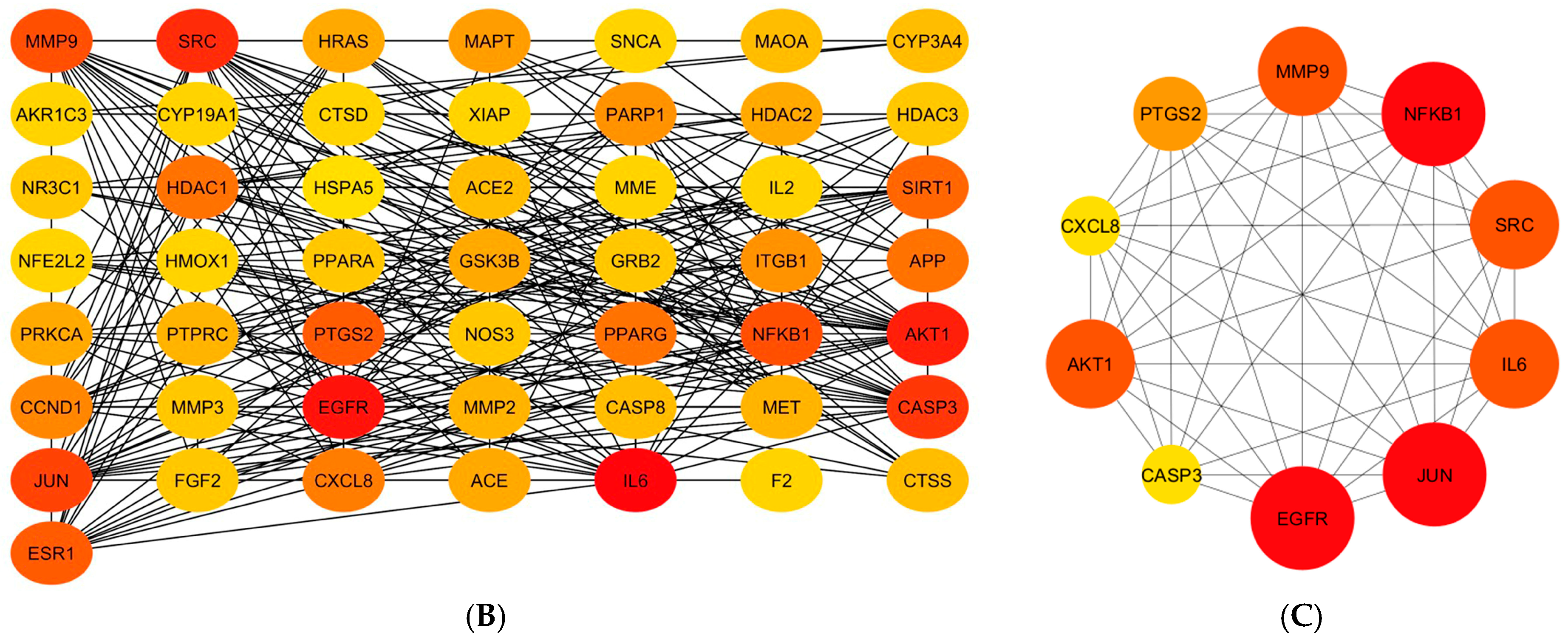
2.4.1. Screening of Wound-Healing-Associated Gene Targets
2.4.2. Protein–Protein Interactions (PPIs)
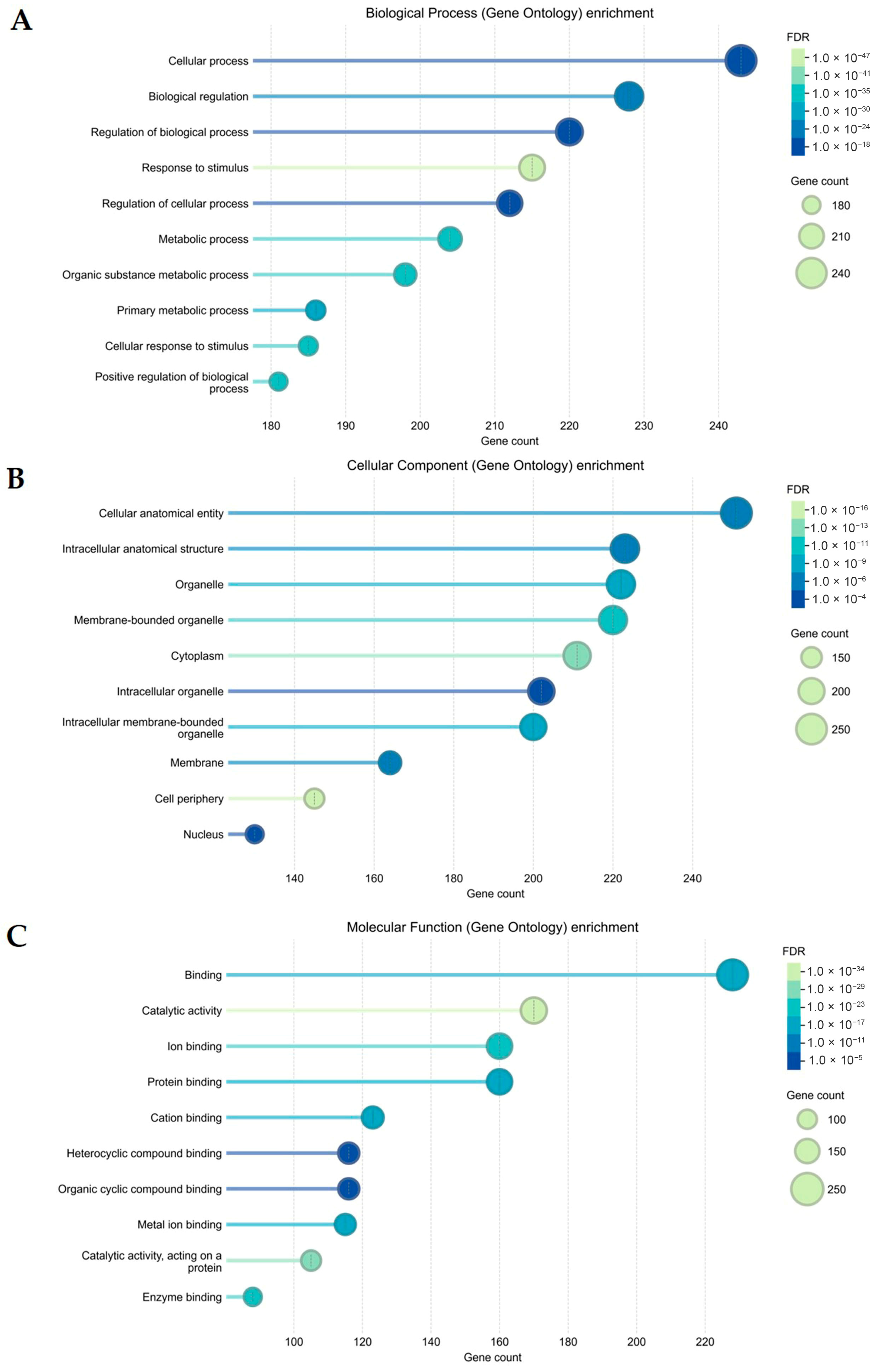
2.4.3. Gene Ontology (GO) and Kyoto Encyclopaedia of Genes and Genomes (KEGG) Pathway Analyses
2.5. Molecular Docking
2.6. Pharmacokinetic Properties
3. Discussion
4. Materials and Methods
4.1. Preparation of the Sunflower Seed Extract
4.2. Antioxidant Activity
4.2.1. DPPH Assay
4.2.2. ABTS Assay
4.2.3. FRAP Assay
4.3. Evaluation of Wound Healing
4.3.1. Cell Culture
4.3.2. Cell Viability Assay
4.3.3. Wound Scratch Assay
4.4. Phytochemical Profiling Analysis
4.4.1. LC-MS
4.4.2. Determination of the TPC and the TFC
4.4.3. HPLC
4.5. Network Pharmacology
4.5.1. Screening of Gene Targets of Compounds in the Sunflower Seed Extract
4.5.2. PPI Network Construction
4.5.3. GO and KEGG Pathway Analyses
4.6. Molecular Docking Study
4.7. Pharmacokinetic Predictions
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pastar, I.; Stojadinovic, O.; Yin, N.C.; Ramirez, H.; Nusbaum, A.G.; Sawaya, A.; Patel, S.B.; Khalid, L.; Isseroff, R.R.; Tomic-Canic, M. Epithelialization in Wound Healing: A Comprehensive Review. Adv. Wound Care 2014, 3, 445–464. [Google Scholar] [CrossRef]
- Cañedo-Dorantes, L.; Cañedo-Ayala, M. Skin Acute Wound Healing: A Comprehensive Review. Int. J. Inflamm. 2019, 2019, 3706315. [Google Scholar] [CrossRef]
- Enoch, S.; Leaper, D.J. Basic Science of Wound Healing. Surgery 2007, 26, 31–37. [Google Scholar]
- Guo, S.; DiPietro, L.A. Factors Affecting Wound Healing. J. Dent. Res. 2010, 89, 219–229. [Google Scholar] [CrossRef]
- González-Pérez, S.; Vereijken, J.M. Sunflower Proteins: Overview of Their Physicochemical, Structural and Functional Properties. J. Sci. Food Agric. 2007, 87, 2173–2191. [Google Scholar] [CrossRef]
- Phillips, K.M.; Ruggio, D.M.; Ashraf-Khorassani, M. Phytosterol Composition of Nuts and Seeds Commonly Consumed in the United States. J. Agric. Food Chem. 2005, 53, 9436–9445. [Google Scholar] [CrossRef] [PubMed]
- Thepthanee, C.; Li, H.; Wei, H.; Prakitchaiwattana, C.; Siriamornpun, S. Effect of Soaking, Germination, and Roasting on Phenolic Composition, Antioxidant Activities, and Fatty Acid Profile of Sunflower (Helianthus annuus L.) Seeds. Horticulturae 2024, 10, 387. [Google Scholar] [CrossRef]
- De Oliveira Filho, J.G.; Egea, M.B. Sunflower Seed Byproduct and Its Fractions for Food Application: An Attempt to Improve the Sustainability of the Oil Process. J. Food Sci. 2021, 86, 1497–1510. [Google Scholar] [CrossRef]
- Gai, F.; Karamać, M.; Janiak, M.A.; Amarowicz, R.; Peiretti, P.G. Sunflower (Helianthus annuus L.) Plants at Various Growth Stages Subjected to Extraction—Comparison of the Antioxidant Activity and Phenolic Profile. Antioxidants 2020, 9, 535. [Google Scholar] [CrossRef]
- Guo, S.; Ge, Y.; Na Jom, K. A Review of Phytochemistry, Metabolite Changes, and Medicinal Uses of the Common Sunflower Seed and Sprouts (Helianthus annuus L.). Chem. Cent. J. 2017, 11, 95. [Google Scholar] [CrossRef]
- Rehman, A.; Saeed, A.; Kanwal, R.; Ahmad, S.; Changazi, S.H. Therapeutic Effect of Sunflower Seeds and Flax Seeds on Diabetes. Cureus 2021, 13, e17256. [Google Scholar] [CrossRef]
- Comino-Sanz, I.M.; López-Franco, M.D.; Castro, B.; Pancorbo-Hidalgo, P.L. The Role of Antioxidants on Wound Healing: A Review of the Current Evidence. J. Clin. Med. 2021, 10, 3558. [Google Scholar] [CrossRef] [PubMed]
- Kotha, R.R.; Tareq, F.S.; Yildiz, E.; Luthria, D.L. Oxidative Stress and Antioxidants—A Critical Review on In Vitro Antioxidant Assays. Antioxidants 2022, 11, 2388. [Google Scholar] [CrossRef] [PubMed]
- Bagdas, D.; Etoz, B.C.; Gul, Z.; Ziyanok, S.; Inan, S.; Turacozen, O.; Gul, N.Y.; Topal, A.; Cinkilic, N.; Tas, S.; et al. In Vivo Systemic Chlorogenic Acid Therapy under Diabetic Conditions: Wound Healing Effects and Cytotoxicity/Genotoxicity Profile. Food Chem. Toxicol. 2015, 81, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Xie, M.; He, L.; Song, X.; Cao, T. Chlorogenic Acid: A Review on Its Mechanisms of Anti-Inflammation, Disease Treatment, and Related Delivery Systems. Front. Pharmacol. 2023, 14, 1218015. [Google Scholar] [CrossRef]
- Wang, L.; Pan, X.; Jiang, L.; Chu, Y.; Gao, S.; Jiang, X.; Zhang, Y.; Chen, Y.; Luo, S.; Peng, C. The Biological Activity Mechanism of Chlorogenic Acid and Its Applications in Food Industry: A Review. Front. Nutr. 2022, 9, 943911. [Google Scholar] [CrossRef]
- Filippo, P.A.D.; Ribeiro, L.M.F.; Gobbi, F.P.; Lemos, G.B.; Rodrigues, R.B.R.; Jerdy, H.; Silva, L.C.D.; Viana, I.S.; Quirino, C.R. Effects of Pure and Ozonated Sunflower Seed Oil (Helianthus annuus) on Hypergranulation Tissue Formation, Infection and Healing of Equine Lower Limb Wounds. Braz. J. Vet. Med. 2020, 42, e113520. [Google Scholar] [CrossRef]
- Maliza, R.; Syaidah, R.; Agusta, I. Incision-Wound Healing Activity of Sunflower Seed Oil (Helianthus annuus L.): In Vivo and in Silico Study. Farmacia 2023, 71, 1001–1012. [Google Scholar] [CrossRef]
- Walton, E.W. Topical Phytochemicals: Applications for Wound Healing. Adv. Skin Wound Care 2014, 27, 328–332. [Google Scholar] [CrossRef]
- Budovsky, A.; Yarmolinsky, L.; Ben-Shabat, S. Effect of Medicinal Plants on Wound Healing. Wound Repair Regen. 2015, 23, 171–183. [Google Scholar] [CrossRef]
- Kusumawati, A.H.; Farhamzah, F.; Alkandahri, M.Y.; Sadino, A.; Agustina, L.S.; Apriana, S.D. Antioxidant Activity and Sun Protection Factor of Black Glutinous Rice (Oryza sativa Var. Glutinosa). Trop. J. Nat. Prod. Res. 2021, 5, 1958–1961. [Google Scholar] [CrossRef]
- ISO 10993-5; Biological Evaluation of Medical Devices—Part 5: Tests for in Vitro Cytotoxicity. International Organization for Standardization: Geneve, Switzerland, 2009.
- Kurilung, A.; Limjiasahapong, S.; Kaewnarin, K.; Wisanpitayakorn, P.; Jariyasopit, N.; Wanichthanarak, K.; Sartyoungkul, S.; Wong, S.C.C.; Sathirapongsasuti, N.; Kitiyakara, C.; et al. Measurement of Very Low-Molecular Weight Metabolites by Traveling Wave Ion Mobility and Its Use in Human Urine Samples. J. Pharm. Anal. 2024, 14, 100921. [Google Scholar] [CrossRef]
- Inwongwan, S.; Sriwari, S.; Pumas, C. Metabolomic Insights into the Adaptations and Biotechnological Potential of Euglena gracilis Under Different Trophic Conditions. Plants 2025, 14, 1580. [Google Scholar] [CrossRef] [PubMed]
- Siriparu, P.; Panyatip, P.; Pota, T.; Ratha, J.; Yongram, C.; Srisongkram, T.; Sungthong, B.; Puthongking, P. Effect of Germination and Illumination on Melatonin and Its Metabolites, Phenolic Content, and Antioxidant Activity in Mung Bean Sprouts. Plants 2022, 11, 2990. [Google Scholar] [CrossRef]
- Pires, D.E.V.; Blundell, T.L.; Ascher, D.B. pkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef]
- Hwang, E.; Gao, W.; Xiao, Y.; Ngo, H.T.; Yi, T. Helianthus annuus L. Flower Prevents UVB-induced Photodamage in Human Dermal Fibroblasts by Regulating the MAPK/AP-1, NFAT, and Nrf2 Signaling Pathways. J. Cell Biochem. 2019, 120, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Žilić, S.; Maksimović Dragišić, J.; Maksimović, V.; Maksimović, M.; Basić, Z.; Crevar, M.; Stanković, G. The Content of Antioxidants in Sunflower Seed and Kernel. Helia 2010, 33, 75–84. [Google Scholar] [CrossRef]
- Alexandrino, T.D.; Da Silva, M.G.; Ferrari, R.A.; Ruiz, A.L.T.G.; Duarte, R.M.T.; Simabuco, F.M.; Bezerra, R.M.N.; Pacheco, M.T.B. Evaluation of Some in Vitro Bioactivities of Sunflower Phenolic Compounds. Curr. Res. Food Sci. 2021, 4, 662–669. [Google Scholar] [CrossRef] [PubMed]
- Yun, J.M.; Jung, J.; Park, S.-H.; Seo, Y.H.; Lee, J.K.; Bae, M.H.; Eun, S.; Kim, O.-K.; Lee, J. Sunflower (Helianthus annuus) Seed Extract Suppresses the Lipogenesis Pathway and Stimulates the Lipolysis Pathway in High-Fat Diet-Induced Obese Mice. Food Nutr. Res. 2022, 66, 8587. [Google Scholar] [CrossRef]
- Zoumpoulakis, P.; Sinanoglou, V.; Siapi, E.; Heropoulos, G.; Proestos, C. Evaluating Modern Techniques for the Extraction and Characterisation of Sunflower (Hellianthus annus L.) Seeds Phenolics. Antioxidants 2017, 6, 46. [Google Scholar] [CrossRef]
- Fisk, I.D.; White, D.A.; Carvalho, A.; Gray, D.A. Tocopherol—An Intrinsic Component of Sunflower Seed Oil Bodies. J. Am. Oil Chem. Soc. 2006, 83, 341–344. [Google Scholar] [CrossRef]
- Amakura, Y.; Yoshimura, M.; Yamakami, S.; Yoshida, T. Isolation of Phenolic Constituents and Characterization of Antioxidant Markers from Sunflower (Helianthus annuus) Seed Extract. Phytochem. Lett. 2013, 6, 302–305. [Google Scholar] [CrossRef]
- Gagour, J.; Ahmed, M.N.; Bouzid, H.A.; Oubannin, S.; Bijla, L.; Ibourki, M.; Hajib, A.; Koubachi, J.; Harhar, H.; Gharby, S. Proximate Composition, Physicochemical, and Lipids Profiling and Elemental Profiling of Rapeseed (Brassica napus L.) and Sunflower (Helianthus annuus L.) Grown in Morocco. Evid.-Based Complement. Altern. Med. 2022, 2022, 3505943. [Google Scholar] [CrossRef] [PubMed]
- Pedrosa, M.M.; Muzquiz, M.; García-Vallejo, C.; Burbano, C.; Cuadrado, C.; Ayet, G.; Robredo, L.M. Determination of Caffeic and Chlorogenic Acids and Their Derivatives in Different Sunflower Seeds. J. Sci. Food Agric. 2000, 80, 459–464. [Google Scholar] [CrossRef]
- Weisz, G.M.; Kammerer, D.R.; Carle, R. Identification and Quantification of Phenolic Compounds from Sunflower (Helianthus annuus L.) Kernels and Shells by HPLC-DAD/ESI-MSn. Food Chem. 2009, 115, 758–765. [Google Scholar] [CrossRef]
- Maeda, H.; Dudareva, N. The Shikimate Pathway and Aromatic Amino Acid Biosynthesis in Plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef] [PubMed]
- Sripad, G.; Prakash, V.; Rao, M.S.N. Extractability of Polyphenols of Sunflower Seed in Various Solvents. J. Biosci. 1982, 4, 145–152. [Google Scholar] [CrossRef]
- Sastry, M.C.S.; Murray, D.R. The Tryptophan Content of Extractable Seed Proteins from Cultivated Legumes, Sunflower and Acacia. J. Sci. Food Agric. 1986, 37, 535–538. [Google Scholar] [CrossRef]
- Volicer, L. Can Dietary Intervention Help in Management of Problem Behaviors in Dementia? J. Nutr. Health Aging 2009, 13, 499–501. [Google Scholar] [CrossRef]
- Verde, A.; Míguez, J.M.; Leao-Martins, J.M.; Gago-Martínez, A.; Gallardo, M. Melatonin Content in Walnuts and Other Commercial Nuts. Influence of Cultivar, Ripening and Processing (Roasting). J. Food Compost. Anal. 2022, 105, 104180. [Google Scholar] [CrossRef]
- Manchester, L.C.; Tan, D.-X.; Reiter, R.J.; Park, W.; Monis, K.; Qi, W. High Levels of Melatonin in the Seeds of Edible Plants Possible Function in Germ Tissue Protection. Life Sci. 2000, 67, 3023–3029. [Google Scholar] [CrossRef] [PubMed]
- Pugazhenthi, K.; Kapoor, M.; Clarkson, A.N.; Hall, I.; Appleton, I. Melatonin Accelerates the Process of Wound Repair in Full-thickness Incisional Wounds. J. Pineal Res. 2008, 44, 387–396. [Google Scholar] [CrossRef]
- Bandeira, L.G.; Bortolot, B.S.; Cecatto, M.J.; Monte-Alto-Costa, A.; Romana-Souza, B. Exogenous Tryptophan Promotes Cutaneous Wound Healing of Chronically Stressed Mice through Inhibition of TNF-α and IDO Activation. PLoS ONE 2015, 10, e0128439. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, L.; Li, X.; Miao, Y.; Li, D. Integrating Network Pharmacology, Molecular Docking and Experimental Validation to Explore the Pharmacological Mechanisms of Quercetin Against Diabetic Wound. Int. J. Med. Sci. 2024, 21, 2837–2850. [Google Scholar] [CrossRef]
- John, S.; Chengamaraju, N.S.; Mahesh, A.R.; Raj, B. Integrated In Silico and In Vivo Evaluation of Cassytha filiformis: Molecular Insights into Caspase-3/8 Binding and Wound Healing Effect. Pharmacol. Res.-Mod. Chin. Med. 2025, 16, 100650. [Google Scholar] [CrossRef]
- Luthfiana, D.; Utomo, D.H. Network Pharmacology Reveals the Potential of Dolastatin 16 as a Diabetic Wound Healing Agent. In Silico Pharmacol. 2023, 11, 23. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Guarino, M.; Hernández-Bule, M.L.; Bacci, S. Cellular and Molecular Processes in Wound Healing. Biomedicines 2023, 11, 2526. [Google Scholar] [CrossRef]
- Landén, N.X.; Li, D.; Ståhle, M. Transition from Inflammation to Proliferation: A Critical Step during Wound Healing. Cell. Mol. Life Sci. 2016, 73, 3861–3885. [Google Scholar] [CrossRef]
- Oeckinghaus, A.; Ghosh, S. The NF- B Family of Transcription Factors and Its Regulation. Cold Spring Harb. Perspect. Biol. 2009, 1, a000034. [Google Scholar] [CrossRef]
- Nakamura, Y.; Sotozono, C.; Kinoshita, S. The Epidermal Growth Factor Receptor (EGFR): Role in Corneal Wound Healing and Homeostasis. Exp. Eye Res. 2001, 72, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Rushendran, R.; Singh, S.A.; Begum, R.F.; Chitra, V.; Ali, N.; Prajapati, B.G. Bioinformatics Exploration of the Therapeutic Potential of Lotus Seed Compounds in Multiple Sclerosis: A Network Analysis of c-Jun Pathway. Drug Dev. Res. 2025, 86, e70038. [Google Scholar] [CrossRef]
- Wu, L.; Chen, Y.; Chen, M.; Yang, Y.; Che, Z.; Li, Q.; You, X.; Fu, W. Application of Network Pharmacology and Molecular Docking to Elucidate the Potential Mechanism of Astragalus–Scorpion against Prostate Cancer. Andrologia 2021, 53, e14165. [Google Scholar] [CrossRef] [PubMed]
- Espada, J.; Martín-Pérez, J. An Update on Src Family of Nonreceptor Tyrosine Kinases Biology. In International Review of Cell and Molecular Biology; Elsevier: Amsterdam, The Netherlands, 2017; Volume 331, pp. 83–122. ISBN 978-0-12-812469-7. [Google Scholar]
- Veerappan, K.; Natarajan, S.; Ethiraj, P.; Vetrivel, U.; Samuel, S. Inhibition of IKKβ by Celastrol and Its Analogues—An in Silico and in Vitro Approach. Pharm. Biol. 2017, 55, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Huo, Y.; Tan, D.-X.; Liang, Z.; Zhang, W.; Zhang, Y. Melatonin in Chinese Medicinal Herbs. Life Sci. 2003, 73, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Arabshahi-Delouee, S.; Urooj, A. Antioxidant Properties of Various Solvent Extracts of Mulberry (Morus indica L.) Leaves. Food Chem. 2007, 102, 1233–1240. [Google Scholar] [CrossRef]
- Kim, D.-O.; Chun, O.K.; Kim, Y.J.; Moon, H.-Y.; Lee, C.Y. Quantification of Polyphenolics and Their Antioxidant Capacity in Fresh Plums. J. Agric. Food Chem. 2003, 51, 6509–6515. [Google Scholar] [CrossRef]
- Iqbal, M.J.; Hanif, S.; Mahmood, Z.; Anwar, F.; Jamil, A. Antioxidant and Antimicrobial Activities of Chowlai (Amaranthus viridis L.) Leaf and Seed Extracts. J. Med. Plants Res. 2012, 6, 4450–4455. [Google Scholar] [CrossRef]
- Chiong, H.S.; Yong, Y.K.; Ahmad, Z.; Sulaiman, M.R.; Zakaria, Z.A.; Yuen, K.H.; Nazrul-Hakim, M.; Tang, S. Cytoprotective and Enhanced Anti-Inflammatory Activities of Liposomal Piroxicam Formulation in Lipopolysaccharide-Stimulated RAW 264.7 Macrophages. Int. J. Nurs. 2013, 8, 1245–1255. [Google Scholar] [CrossRef]
- Loggenberg, S.R.; Twilley, D.; De Canha, M.N.; Meyer, D.; Mabena, E.C.; Lall, N. Evaluation of Wound Healing and Antibacterial Potential of Greyia radlkoferi Szyszyl. Ethanolic Leaf Extract. Front. Pharmacol. 2022, 13, 806285. [Google Scholar] [CrossRef]
- Panyatip, P.; Padumanonda, T.; Yongram, C.; Kasikorn, T.; Sungthong, B.; Puthongking, P. Impact of Tea Processing on Tryptophan, Melatonin, Phenolic and Flavonoid Contents in Mulberry (Morus alba L.) Leaves: Quantitative Analysis by LC-MS/MS. Molecules 2022, 27, 4979. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING Database in 2023: Protein–Protein Association Networks and Functional Enrichment Analyses for Any Sequenced Genome of Interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A Free Online Platform for Data Visualization and Graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Kencana, M.V.S.A.; Widyantara, K.A.; Saputra, B.W.; Setiawati, A. Towards a Predicted Anti-Aging Molecular Targets of Asiaticoside Based on Bioinformatics Analysis. Int. J. Med. Biochem. 2025, 8, 78–88. [Google Scholar] [CrossRef]
- Rasyid, H.; Armunanto, R.; Purwono, B. Molecular Docking Analysis on Epidermal Growth Factor Receptor Wild Type (EGFRwt) with Quinazoline Derivative Compounds as Tyrosine Kinase Inhibitors. Appl. Sci. Eng. Prog. 2017, 10, 293–299. [Google Scholar]
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An Automated Pipeline for the Setup of Poisson-Boltzmann Electrostatics Calculations. Nucleic Acids Res. 2004, 32, W665–W667. [Google Scholar] [CrossRef]
- Hevener, K.E.; Zhao, W.; Ball, D.M.; Babaoglu, K.; Qi, J.; White, S.W.; Lee, R.E. Validation of Molecular Docking Programs for Virtual Screening against Dihydropteroate Synthase. J. Chem. Inf. Model. 2009, 49, 444–460. [Google Scholar] [CrossRef] [PubMed]




| Retention Time (min) | Compound | Adduct | Theoretical (m/z) | Observed (m/z) | Mass Error (ppm) | Intensity (×105) | Relative (%) |
|---|---|---|---|---|---|---|---|
| 0.657 | D-Gluconic acid | [M − H]− | 196.16 | 195.05 | −2.31 | 0.73 | 0.11 |
| 1.238 | Pyroglutamic acid | [M − H]− | 129.11 | 128.03 | 0.08 | 9.32 | 1.37 |
| 2.646 | Xanthine | [M − H]− | 152.11 | 151.03 | −6.89 | 9.09 | 1.34 |
| 2.656 | Xanthosine | [M − H]− | 284.23 | 283.07 | −0.64 | 16.7 | 2.46 |
| 2.764 | L-(-)-Phenylalanine | [M − H]− | 165.19 | 164.07 | −1.1 | 0.90 | 0.13 |
| 3.250 | Chlorogenic acid | [M − H]− | 354.31 | 353.09 | 1.47 | 157 | 23.21 |
| 3.392 | Tryptophan | [M − H]− | 204.22 | 203.08 | 2.27 | 2.30 | 0.34 |
| 3.667 | Bergenin | [M − H]− | 328.27 | 327.07 | −3.00 | 0.11 | 0.02 |
| 3.776 | 2-Isopropylmalic acid | [M − H]− | 176.17 | 175.06 | −1.31 | 3.11 | 0.46 |
| 3.839 | D-(-)-Quinic acid | [M − H]− | 192.17 | 191.06 | −8.43 | 319 | 47.03 |
| 3.941 | Glutamylphenylalanine | [M − H]− | 294.30 | 293.12 | −0.41 | 2.86 | 0.42 |
| 4.024 | Caffeic acid | [M − H]− | 180.16 | 179.03 | −1.06 | 17.7 | 2.60 |
| 4.597 | Agnuside | [M − H]− | 466.40 | 465.14 | −6.11 | 4.01 | 0.59 |
| 4.732 | Tricin 5-glucoside | [M − H]− | 492.40 | 491.12 | 2.69 | 22.5 | 3.32 |
| 5.056 | Umbelliferone | [M − H]− | 162.14 | 161.02 | 0.93 | 2.93 | 0.43 |
| 5.368 | Plantaginin | [M − H]− | 448.40 | 447.09 | 0.34 | 0.34 | 0.05 |
| 5.584 | 4-Hydroxyquinoline | [M − H]− | 145.16 | 144.05 | 0.42 | 1.28 | 0.19 |
| 5.634 | Phenylacetic acid | [M − H]− | 136.15 | 135.05 | −8.74 | 2.80 | 0.38 |
| 5.907 | 3,4-Di-O-caffeoylquinic acid | [M − H]− | 516.40 | 515.12 | −7.11 | 17.3 | 9.89 |
| 5.966 | Eriodictyol | [M − H]− | 288.25 | 287.06 | 0.63 | 1.66 | 0.25 |
| 5.976 | Isookanin-7-O-glucoside | [M − H]− | 450.40 | 449.11 | 1.22 | 17.9 | 2.64 |
| 6.007 | Azelaic acid | [M − H]− | 188.22 | 187.10 | 1.98 | 3.13 | 0.46 |
| 12.632 | α-Linolenic acid | [M − H]− | 278.40 | 277.22 | 4.73 | 4.53 | 0.67 |
| 14.214 | 16-Hydroxyhexadecanoic acid | [M − H]− | 272.42 | 271.23 | −16.41 | 2.27 | 0.33 |
| 14.567 | Linoelaidic acid | [M − H]− | 280.40 | 279.23 | 2.08 | 8.91 | 1.31 |
| Retention Time (min) | Compound | Adduct | Theoretical (m/z) | Observed (m/z) | Mass Error (ppm) | Intensity (×105) | Relative (%) |
|---|---|---|---|---|---|---|---|
| 0.702 | Trigonelline | [M + H]+ | 137.14 | 138.06 | −15.94 | 38.3 | 5.13 |
| 0.737 | Arginine | [M + H]+ | 174.20 | 175.12 | −1.83 | 1.14 | 0.15 |
| 0.780 | Isoleucine | [M + H]+ | 131.17 | 132.10 | −6.59 | 4.56 | 0.61 |
| 0.934 | Valine | [M + H]+ | 117.15 | 118.09 | −3.13 | 11.1 | 1.48 |
| 0.982 | 4-Guanidinobutyric acid | [M + H]+ | 145.10 | 146.09 | 4.04 | 1.17 | 0.16 |
| 1.514 | Tyrosine | [M + H]+ | 181.19 | 182.08 | 2.58 | 3.65 | 0.49 |
| 2.654 | 4-Methyl-5-thiazoleethanol | [M + H]+ | 143.21 | 144.05 | −2.22 | 34.9 | 4.67 |
| 2.778 | 7,8-Dimethoxycoumarin | [M + H]+ | 206.19 | 207.07 | −3.82 | 285 | 38.09 |
| 2.998 | 1,2,3,9-Tetrahydro-4H- carbazol-4-one | [M + Na]+ | 185.22 | 208.07 | 4.95 | 21.0 | 2.81 |
| 3.409 | p-Coumaric acid | [M + H]+ | 164.16 | 165.05 | 3.76 | 3.49 | 0.47 |
| 3.422 | 2-Naphthylamine | [M + H]+ | 143.18 | 144.08 | −10.9 | 19.4 | 2.59 |
| 3.864 | Chlorogenic acid | [M + Na]+ | 354.31 | 377.09 | 0.74 | 145 | 19.37 |
| 3.936 | Caffeoylcholine | [M]+ | 266.31 | 266.14 | −9.17 | 85.4 | 11.43 |
| 3.974 | 4-Coumaroylcholine | [M]+ | 250.31 | 250.14 | −6.96 | 18.9 | 2.53 |
| 5.250 | (+)Catechin | [M + H]+ | 290.27 | 291.09 | 12.06 | 1.02 | 0.14 |
| 5.307 | 1-Ethyl-9H-pyrido [3,4-b] indole | [M + H]+ | 196.25 | 197.11 | −0.71 | 3.52 | 0.47 |
| 5.503 | N-Acetyltryptophan | [M + H]+ | 246.26 | 247.11 | 1.17 | 2.61 | 0.35 |
| 5.925 | 3,4-Di-O-caffeoylquinic acid | [M + Na]+ | 516.40 | 539.12 | 5.79 | 10.5 | 1.40 |
| 5.990 | Eriodictyol | [M + H]+ | 288.25 | 289.07 | −4.22 | 9.58 | 1.28 |
| 6.159 | Cimifugin 4′-O-beta-D- glucopyranoside | [M + Na]+ | 468.40 | 491.15 | −7.78 | 17.0 | 2.28 |
| 6.453 | Skimmin | [M + H]+ | 324.28 | 325.09 | −0.74 | 10.9 | 1.45 |
| 8.426 | Pinoresinol 4-O-glucoside | [M + Na]+ | 520.50 | 543.18 | −6.74 | 2.33 | 0.31 |
| 12.454 | Phosphocholine | [M + H]+ | 183.14 | 184.07 | 2.01 | 17.6 | 2.35 |
| Phytochemical | Retention Time (min) | Standard Equation (R2) | Content |
|---|---|---|---|
| Phenolic acids | |||
| Chlorogenic acid | 19.739 | y = 3428.8x − 591.22 (R2 = 0.9999) | 1.38 ± 0.02 mg/g·DW |
| Caffeic acid | 23.633 | y = 9810.5x − 3571.8 (R2 = 0.9995) | 5.17 ± 0.06 µg/g·DW |
| p–Coumaric acid | 30.890 | y = 7749.8x − 4117.6 (R2 = 0.9994) | NQ |
| Indolamines | |||
| Tryptophan | 8.338 | y = 1.8051x + 0.0064 (R2 = 0.9993) | 15.52 ± 0.04 µg/g·DW |
| Melatonin | 30.793 | y = 6.5972x + 0.1177 (R2 = 0.9973) | 0.02 ± 0.00 µg/g·DW |
| KEGG ID | Signalling Pathway | False Discovery Rate p-Value | Target Genes |
|---|---|---|---|
| hsa04510 | Focal adhesion | 1.15 × 10−9 | CCND1, ITGA2B, PRKCG, BIRC3, EGFR, MET, ROCK2, GSK3B, XIAP, JUN, SRC, GRB2, ITGB1, ITGA4, ROCK1, HRAS, PRKCA, AKT1, ITGB3 |
| hsa04657 | IL-17 signalling pathway | 9.82 × 10−8 | NFκB1, MMP13, MMP3, CXCL8, CASP3, MMP1, GSK3B, CASP8, PTGS2, JUN, MMP9, IL6 |
| hsa01521 | EGFR tyrosine kinase inhibitor resistance | 1.84 × 10−7 | PRKCG, FGF2, EGFR, MET, GSK3B, SRC, GRB2, IL6, HRAS, PRKCA, AKT1 |
| hsa00590 | Arachidonic acid metabolism | 2.11 × 10−7 | LTA4H, ALOX12, CBR1, PTGS1, PLA2G4A, PTGS2, CYP2C19, ALOX5, AKR1C3, PLA2G2A |
| hsa04015 | Rap1 signalling pathway | 2.11 × 10−7 | ITGA2B, PRKCG, FGF2, EGFR, MET, DRD2, CNR1, LPAR3, SRC, ITGB1, HRAS, PRKCA, AKT1, ITGB3, FPR1, FGF1 |
| hsa04668 | TNF signalling pathway | 5.18 × 10−7 | NFκB1, BIRC3, MMP3, CASP3, SELE, CASP8, PTGS2, CASP7, JUN, MMP9, IL6, AKT1 |
| hsa04370 | VEGF signalling pathway | 1.05 × 10−6 | PRKCG, NOS3, MAPKAPK2, PLA2G4A, PTGS2, SRC, HRAS, PRKCA, AKT1 |
| hsa04611 | Platelet activation | 1.08 × 10−6 | ITGA2B, NOS3, F2, ROCK2, PTGS1, PLA2G4A, SRC, ITGB1, ROCK1, TBXA2R, AKT1, ITGB3 |
| hsa04218 | Cellular senescence | 1.20 × 10−6 | SIRT1, NFKB1, CCND1, CDC25A, CXCL8, MAPKAPK2, IGFBP3, HLA-A, IL6, HRAS, CAPN1, AKT1, CCNA2 |
| hsa04151 | PI3K–Akt signalling pathway | 1.77 × 10−6 | NFKB1, IL2, CCND1, ITGA2B, FGF2, EGFR, NOS3, MET, GSK3B, LPAR3, GRB2, ITGB1, ITGA4, IL6, HRAS, PRKCA, AKT1, ITGB3, FGF1 |
| hsa04010 | MAPK signalling pathway | 2.34 × 10−6 | NFκB1, PRKCG, FGF2, EGFR, CASP3, MET, MAPT, MAPKAPK2, PLA2G4A, JUN, HSPA1B, RPS6KA3, GRB2, HRAS, PRKCA, AKT1, FGF1 |
| hsa04066 | HIF-1 signalling pathway | 9.33 × 10−6 | HMOX1, NFκB1, PRKCG, EGFR, NOS3, NOS2, EGLN1, IL6, PRKCA, AKT1 |
| hsa04012 | ErbB signalling pathway | 1.17 × 10−5 | PRKCG, EGFR, GSK3B, JUN, SRC, GRB2, HRAS, PRKCA, AKT1 |
| hsa04540 | Gap junction | 1.92 × 10−5 | HTR2B, PRKCG, EGFR, DRD2, SRC, GRB2, HRAS, PRKCA, HTR2A |
| hsa04014 | Ras signalling pathway | 4.8 × 10−5 | NFκB1, PRKCG, FGF2, EGFR, MET, PLA2G4A, GRB2, PLA2G2A, HRAS, PRKCA, AKT1, FGF1, PTPN11 |
| hsa04062 | Chemokine signalling pathway | 1.60 × 10−4 | GSK3A, NFKB1, CXCL8, ROCK2, GSK3B, PRKCD, SRC, GRB2, ROCK1, HRAS, AKT1 |
| hsa04310 | Wnt signalling pathway | 1.70 × 10−4 | CCND1, MMP7, PRKCG, PPARD, CTBP2, ROCK2, GSK3B, PSEN1, JUN, PRKCA |
| hsa04630 | JAK–STAT signalling pathway | 2.00 × 10−4 | IL2, CCND1, EGFR, PTPN2, GRB2, IL6, PTPN6, HRAS, AKT1, PTPN11 |
| hsa04064 | NF-kappa B signalling pathway | 5.50 × 10−3 | NFκB1, BIRC3, CXCL8, PARP1, PTGS2, XIAP |
| Compound | Compound Name | ΔG (kcal/mol) | ||
|---|---|---|---|---|
| NF-κB | EGFR | MMP9 | ||
| 1 | Pyroglutamic acid | −4.34 | −3.63 | −3.89 |
| 2 | Xanthine | −4.56 | −3.92 | −5.17 |
| 3 | Xanthosine | −5.98 | −5.84 | −6.34 |
| 4 | D-(-)-quinic acid | −3.77 | −3.84 | −3.81 |
| 5 | Caffeic acid | −5.34 | −4.97 | −5.24 |
| 6 | Tricin 5-glucoside | −10.32 | −6.93 | −6.97 |
| 7 | 3,4-Di-O-caffeoylquinic acid | −9.16 | −6.43 | −7.08 |
| 8 | Isookanin-7-O-glucoside | −10.06 | −6.82 | −8.65 |
| 9 | Linoelaidic acid | −6.90 | −5.60 | −5.63 |
| 10 | Trigonelline | −4.21 | −4.03 | −3.89 |
| 11 | Valine | −4.04 | −4.23 | −4.65 |
| 12 | 4-Methyl-5-thiazoleethanol | −4.25 | −4.33 | −5.18 |
| 13 | 7,8-Dimethoxycoumarin | −5.92 | −5.30 | −6.49 |
| 14 | 1,2,3,9-Tetrahydro-4H-carbazol-4-one | −6.84 | −5.75 | −8.14 |
| 15 | 2-Naphthylamine | −5.45 | −5.38 | −6.35 |
| 16 | Chlorogenic acid | −8.12 | −6.73 | −8.90 |
| 17 | Caffeoylcholine | −6.37 | −5.54 | −7.85 |
| 18 | 4-Coumaroylcholine | −6.19 | −5.62 | −8.21 |
| 19 | Eriodictyol | −8.71 | −7.12 | −8.29 |
| 20 | Cimifugin 4′-O-beta-D-glucopyranoside | −7.43 | −7.12 | −9.41 |
| 21 | Skimmin | −3.30 | −2.76 | −4.00 |
| - | Co-ligands * | −11.90 | −7.00 | −7.87 |
| Protein | Co-Ligand | Grid Box Size (Å) | Spacing (Å) | Root-Mean-Square Deviation (Å) |
|---|---|---|---|---|
| NF-κB | KSA or K-252A | X = 48.383 Y = 30.057 Z = −56.809 | 0.375 | 0.951 |
| EGFR | AQ4 or [6,7-Bis(2-methoxy-ethoxy)quinazoline-4-yl]- (3-ethynylphenyl)amine | X = 22.013 Y = 0.252 Z = 52.794 | 0.477 | 1.775 |
| MMP9 | NFH or N~2~-[(2R)-2-{[formyl(hydroxy)amino]methyl}-4-methylpentanoyl]-N,3-dimethyl-L-valinamide | X = 65.607 Y = 31.083 Z = 117.697 | 0.408 | 1.651 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Ratha, J.; Padumanonda, T.; Yongram, C.; Siriparu, P.; Datham, S.; Subhan, M.; Chenboonthai, C.; Puthongking, P. Network Pharmacology of the Phytochemical Content of Sunflower Seed (Helianthus annuus L.) Extract from LC-MS on Wound-Healing Activity and the In Vitro Wound Scratch Assay. Plants 2026, 15, 187. https://doi.org/10.3390/plants15020187
Ratha J, Padumanonda T, Yongram C, Siriparu P, Datham S, Subhan M, Chenboonthai C, Puthongking P. Network Pharmacology of the Phytochemical Content of Sunflower Seed (Helianthus annuus L.) Extract from LC-MS on Wound-Healing Activity and the In Vitro Wound Scratch Assay. Plants. 2026; 15(2):187. https://doi.org/10.3390/plants15020187
Chicago/Turabian StyleRatha, Juthamat, Tanit Padumanonda, Chawalit Yongram, Pimolwan Siriparu, Suthida Datham, Muhammad Subhan, Chatchavarn Chenboonthai, and Ploenthip Puthongking. 2026. "Network Pharmacology of the Phytochemical Content of Sunflower Seed (Helianthus annuus L.) Extract from LC-MS on Wound-Healing Activity and the In Vitro Wound Scratch Assay" Plants 15, no. 2: 187. https://doi.org/10.3390/plants15020187
APA StyleRatha, J., Padumanonda, T., Yongram, C., Siriparu, P., Datham, S., Subhan, M., Chenboonthai, C., & Puthongking, P. (2026). Network Pharmacology of the Phytochemical Content of Sunflower Seed (Helianthus annuus L.) Extract from LC-MS on Wound-Healing Activity and the In Vitro Wound Scratch Assay. Plants, 15(2), 187. https://doi.org/10.3390/plants15020187






