Genome-Wide Analysis of the RbcS Gene Family and Expression Analysis Under Light Response in Brassica napus L.
Abstract
1. Introduction
2. Results
2.1. Genome-Wide Identification of the RbcS Family Gene Members of Brassica Species
2.2. Phylogenetic Analysis and Classification of RbcS Genes
2.3. Gene Structure and Motif Compositions
2.4. Chromosomal Distribution of BnRbcS Gene
2.5. Collinearity Analysis of the RbcS Gene Family
2.6. Physicochemical Properties and Subcellular Localization of BnRbcS Gene
2.7. Analysis of the Secondary Structure of RCBS Gene Family Member Proteins in B. napus
2.8. Potential Interaction Prediction Analysis of BnRbcS Gene
2.9. Analysis of the Cis-Acting Elements in the Promoter Regions of the BnRbcS Genes
2.10. Analysis of BnRbcS Gene Expression Patterns
2.11. Analysis of BnRbcS Gene Expression Patterns Under Light Treatment at 6 h and 12 h
2.12. Haplotype Analysis of RbcSA4-1
3. Discussion
4. Materials and Methods
4.1. Genome-Wide Identification of the RbcS Gene Family
4.2. Multiple Sequence Alignment and Phylogenetic Analysis of the RbcS Gene Family
4.3. Gene Structure and Conserved Motif Analysis of BnRbcS
4.4. Chromosomal Distribution and Collinearity Analysis of BnRbcS
4.5. Physicochemical Properties and Subcellular Localization of BnRbc Proteins
4.6. Cis-Acting Element Analysis in BnRbcS Gene Promoters
4.7. Protein–Protein Interaction Prediction
4.8. Tissue-Specific Expression Analysis of BnRbcS
4.9. qRT-PCR Analysis Under 6-h and 12-h Light Treatments
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| RT-qPCR | Reverse transcription quantitative polymerase chain reaction |
| A. thaliana | Arabidopsis thaliana |
| B. rapa | Brassica rapa L. (AA, 2n = 20) |
| B. nigra | Brassica nigra L. (BB, 2n = 16) |
| B. oleracea | Brassica oleracea L. (CC, 2n = 18) |
| B. juncea | Brassica juncea L. (AABB, 2n = 36) |
| B. napus | Brassica napus L. (AACC, 2n = 38) |
| B. carinata | Brassica carinata L (BBCC, 2n = 34). |
References
- Lohani, N.; Jain, D.; Singh, M.B.; Bhalla, P.L. Engineering Multiple Abiotic Stress Tolerance in Canola, Brassica napus. Front. Plant Sci. 2020, 11, 3. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Anwar, S.; Kuai, J.; Noman, A.; Shahid, M.; Din, M.; Ali, A.; Zhou, G. Alteration in yield and oil quality traits of winter rapeseed by lodging at different planting density and nitrogen rates. Sci. Rep. 2018, 8, 634. [Google Scholar] [CrossRef] [PubMed]
- Pingali, P.L. Green Revolution: Impacts, limits, and the path ahead. Proc. Natl. Acad. Sci. USA 2012, 109, 12302–12308. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Zhang, S.; Shao, S.; Zheng, R.; Yu, Z.; Ye, Q. Translocation and metabolism of the chiral neonicotinoid cycloxaprid in oilseed rape (Brassica napus L.). J. Hazard. Mater. 2022, 426, 128125. [Google Scholar] [CrossRef]
- DeSouza, A.P.; Burgess, S.J.; Doran, L.; Hansen, J.; Manukyan, L.; Maryn, N.; Gotarkar, D.; Leonelli, L.; Niyogi, K.K.; Long, S.P. Soybean photosynthesis and crop yield are improved by accelerating recovery from photoprotection. Science 2022, 377, 851–854. [Google Scholar] [CrossRef]
- Wu, A.; Hammer, G.; Doherty, A.; von Caemmerer, S.; Farquhar, G.D. Quantifying impacts of enhancing photosynthesis on crop yield. Nat. Plants 2019, 5, 380–388. [Google Scholar] [CrossRef]
- Chen, J.-H.; Chen, S.-T.; He, N.-Y.; Wang, Q.; Zhao, Y.; Wei, G.; Guo, F.-Q. Nuclear-encoded synthesis of the D1 subunit of photosystem II increases photosynthetic efficiency and crop yield. Nat. Plants 2020, 6, 570–580. [Google Scholar] [CrossRef]
- Li, X.; Wang, P.; Li, J.; Wei, S.B.; Yan, Y.; Yang, J.; Zhao, M.; Langdale, J.A.; Zhou, W. Maize GOLDEN2-LIKE genes enhance biomass and grain yields in rice by improving photosynthesis and reducing photoinhibition. Commun. Biol. 2020, 3, 151. [Google Scholar] [CrossRef]
- Wang, L.; Zheng, J.D.; Zhou, G.S.; Li, J.; Chen, Q.; Lin, G.B.; Li, Y.; Zuo, Q.S. Moderate nitrogen application improved salt tolerance by enhancing photosynthesis, antioxidants, and osmotic adjustment in rapeseed (Brassica napus L.). Front. Plant Sci. 2023, 14, 1196319. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, M.; Yao, M.; Zhang, X.; Qu, C.; Du, H.; Lu, K.; Li, J.; Wei, L.; Liang, Y. Rapeseed (Brassica napus) Mitogen-Activated Protein Kinase 1 Enhances Shading Tolerance by Regulating the Photosynthesis Capability of Photosystem II. Front. Plant Sci. 2022, 13, 902989. [Google Scholar] [CrossRef]
- Tian, T.; Wang, J.; Wang, H.; Cui, J.; Shi, X.; Song, J.; Li, W.; Zhong, M.; Qiu, Y.; Xu, T. Nitrogen application alleviates salt stress by enhancing osmotic balance, ROS scavenging, and photosynthesis of rapeseed seedlings (Brassica napus). Plant Signal. Behav. 2022, 17, 2081419. [Google Scholar] [CrossRef]
- Gul, H.S.; Ulfat, M.; Zafar, Z.; Haider, W.; Ali, Z.; Manzoor, H.; Afzal, S.; Ashraf, M.; Athar, H.-u.-R. Photosynthesis and Salt Exclusion Are Key Physiological Processes Contributing to Salt Tolerance of Canola (Brassica napus L.): Evidence from Physiology and Transcriptome Analysis. Genes 2023, 14, 3. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Guo, N.; Zhang, M.; Du, Y.; Xu, J.; Li, S.; Wang, J.; Wang, R.; Liu, X.; Qin, M.; et al. Identification of genetic loci and candidate genes regulating photosynthesis and leaf morphology through genome-wide association study in Brassica napus L. Front. Plant Sci. 2024, 15, 1467927. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Li, G.Y.; Islam, A.; Fu, W.M.; Zhou, Y.Q.; Chen, T.T.; Tao, L.; Fu, G. Strengthened antioxidant capacity improves photosynthesis by regulating stomatal aperture and ribulose-1,5-bisphosphate carboxylase/oxygenase activity Plant science: An international journal of experimental. Plant Biol. 2019, 290, 110245. [Google Scholar] [CrossRef]
- Chai, H.; Gao, L.; Zhao, C.; Liu, X.; Jiang, D.; Dai, T.; Tian, Z. Low nitrogen priming enhances Rubisco activation and allocation of nitrogen to the photosynthetic apparatus as an adaptation to nitrogen-deficit stress in wheat seedling. J. Plant Physiol. 2024, 303, 154337. [Google Scholar] [CrossRef]
- Prywes, N.; Phillips, N.R.; Tuck, O.T.; Valentin-Alvarado, L.E.; Savage, D.F. Rubisco Function, Evolution, and Engineering. Annu. Rev. Biochem. 2023, 92, 385–410. [Google Scholar] [CrossRef]
- Taylor-Kearney, L.J.; Wang, R.Z.; Shih, P.M. Evolution and origins of rubisco. Curr. Biol. 2024, 34, R764–R767. [Google Scholar] [CrossRef] [PubMed]
- Iñíguez, C.; Aguiló-Nicolau, P.; Galmés, J. Improving photosynthesis through the enhancement of Rubisco carboxylation capacity. Biochem. Soc. Trans. 2021, 49, 2007–2019. [Google Scholar] [CrossRef]
- South, P.F.; Cavanagh, A.P.; Lopez-Calcagno, P.E.; Raines, C.A.; Ort, D.R. Optimizing photorespiration for improved crop productivity. J. Integr. Plant Biol. 2018, 60, 1217–1230. [Google Scholar] [CrossRef]
- Salvucci, M.E.; Heber, U.; Portis, A.R.; Ogren, W.L. Stimulation of thylakoid energization and ribulose-bisphosphate carboxylase/oxygenase activation in Arabidopsis leaves by methyl viologen. Environ. Sci. Biol. 1987, 221, 215–220. [Google Scholar] [CrossRef]
- Gionfriddo, M.; Zang, K.; Hayer-Hartl, M. The challenge of engineering Rubisco for improving photosynthesis. FEBS Lett. 2023, 597, 1679–1680. [Google Scholar] [CrossRef] [PubMed]
- Amber, K.E.; Hotto, M.; Michel, E.J.S.; Oh, Z.G.; Stern, D.B. Transgenic expression of Rubisco accumulation factor2 and Rubisco subunits increases photosynthesis and growth in maize. J. Exp. Bot. 2024, 75, 4024–4037. [Google Scholar] [CrossRef] [PubMed]
- Qin, K.; Ye, X.; Luo, S.; Alisdair, R.; Fernie, A.R.; Zhang, Y. Engineering carbon assimilation in plants. J. Integr. Plant Biol. 2025, 67, 926–948. [Google Scholar] [CrossRef] [PubMed]
- Spreitzer, R.J. Role of the small subunit in ribulose-1,5-bisphosphate carboxylase/oxygenas. Arch. Biochem. Biophys. 2003, 414, 141–149. [Google Scholar] [CrossRef]
- Shu, H.; Zhao, Q.; Huang, Y.; Shi, Q.; Yang, J. Antihypertensive peptide resources map of ribulose-1,5-bisphosphate carboxylase/oxygenases (RuBisCO) in angiosperms: Revealed by an integrated in silico and in vitro approach. Food Chem. 2024, 433, 137332. [Google Scholar] [CrossRef]
- Shivhare, D.; Ng, J.; Tsai, Y.C.; Mueller-Cajar, O. Probing the rice Rubisco-Rubisco activase interaction via subunit heterooligomerization. Proc. Natl. Acad. Sci. USA 2019, 116, 24041–24048. [Google Scholar] [CrossRef]
- Knight, S.; Andersson, I.; Brändén, C.I. Crystallographic analysis of ribulose 1,5-bisphosphate carboxylase from spinach at 2.4 A resolution. J. Mol. Biol. 1990, 215, 113–160. [Google Scholar] [CrossRef]
- Krebbers, E.; Seurinck, J.; Herdies, L.; Cashmore, A.R.; Timko, M.P. Four genes in two diverged subfamilies encode the ribulose-1,5-bisphosphate carboxylase small subunit polypeptides of Arabidopsis thaliana. Plant Mol. Biol. 1988, 11, 745–759. [Google Scholar] [CrossRef]
- Qin, L.Y.; Xue, Y.X.; Fei, Y.; Zeng, L.F.; Yang, S.S.; Deng, X.P. Identification, evolution and expression analyses of Ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit gene family in wheat (Triticum aestivum L.). Acta Physiol. Plant. 2018, 40, 85. [Google Scholar] [CrossRef]
- Zhou, Y.; Shi, L.; Li, X.; Wei, S.; Ye, X.; Gao, Y.; Zhou, Y.; Cheng, L.; Cheng, L.; Duan, F.; et al. Genetic engineering of RuBisCO by multiplex CRISPR editing small subunits in rice. Plant Biotechnol. J. 2025, 23, 731–749. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, S.; Gong, X.; Chen, J. Toward Accurate Simulation of Coupling between Protein Secondary Structure and Phase Separation. J. Am. Chem. Soc. 2024, 146, 342–357. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Shen, Y.; Cao, H.; He, K.; Chu, Z.; Li, N. OsbHLH057 targets the AATCA cis-element to regulate disease resistance and drought tolerance in rice. Plant Cell Rep. 2022, 41, 1285–1299. [Google Scholar] [CrossRef] [PubMed]
- Berry, J.O.; Yerramsetty, P.; Zielinski, A.M.; Mure, C.M. Photosynthetic gene expression in higher plants. Photosynth. Res. 2013, 117, 91–120. [Google Scholar] [CrossRef] [PubMed]
- Miyazawa, S.I.; Tobita, H.; Ujino-Ihara, T.; Suzuki, Y. Oxygen response of leaf CO2 compensation points used to determine Rubisco specificity factors of gymnosperm species. J. Plant Res. 2020, 133, 205–215. [Google Scholar] [CrossRef]
- Tabita, F.R. The Molecular Biology of the Cyanobacteria. In Plant Physiology; Bryant, D.A., Ed.; Academic Press: San Diego, CA, USA, 1994; Volume 1, pp. 1299–1329. [Google Scholar]
- Suzuki, Y.; Makino, A.; Mae, T. Changes in the turnover of Rubisco and levels of mRNAs of rbcL and RbcS in rice leaves from emergence to senescence. Plant Cell Environ. 2001, 24, 1353–1360. [Google Scholar] [CrossRef]
- Berry-Lowe, S.L.; McKnight, T.D.; Shah, D.M.; Meagher, R.B. The nucleotide sequence, expression and evolution of one member of a multigene family encoding the small subunit of ribulose-l,5-bisphosphate carboxylase in soybean. J. Mol. Appl. Genet. 1982, 1, 483–498. [Google Scholar]
- Suzuki, Y.; Ohkubo, M.; Hatakeyama, H.; Ohashi, K.; Yoshizawa, R.; Kojima, S.; Hayakawa, T.; Yamaya, T.; Mae, T.; Makino, A. Increased Rubisco content in transgenic rice transformed with the ‘sense’ RbcS gene. Plant Cell Physiol. 2007, 48, 626–637. [Google Scholar] [CrossRef]
- Schwender, J.; Goffman, F.; Ohlrogge, J.B.; Shachar-Hill, Y. Rubisco without the Calvin cycle improves the carbon efficiency of developing green seeds. Nature 2004, 432, 779–782. [Google Scholar] [CrossRef]
- Ruuska, S.A.; Schwender, J.; Ohlrogge, J.B. The capacity of green oilseeds to utilize photosynthesis to drive biosynthetic processes. Plant Physiol. 2004, 136, 2700–2709. [Google Scholar] [CrossRef]
- Cheng, F.; Liu, S.; Wu, J.; Fang, L.; Sun, S.; Liu, B.; Li, P.; Hua, W.; Wang, X. BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol. 2011, 11, 136. [Google Scholar] [CrossRef]
- Reiser, L.; Subramaniam, S.; Zhang, P.; Berardini, T. Using the Arabidopsis information resource (TAIR) to find information about Arabidopsis genes. Curr. Protoc. 2022, 2, e574. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11.0: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Shi, T.; Rahmani, R.S.; Gugger, P.F.; Wang, M.; Li, H.; Zhang, Y.; Li, Z.; Wang, Q.; Van de Peer, Y.; Marchal, K.; et al. Distinct expression and methylation patterns for genes with different fates following a single whole-genome duplication in flowering plants. Mol. Biol. Evol. 2020, 37, 2394–2413. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, X.; Tang, H.; Tan, X.; Ficklin, S.P.; Feltus, F.A.; Paterson, A.H. Modes of gene duplication contribute differently to genetic novelty and redundancy, but show parallels across divergent angiosperms. PLoS ONE 2011, 6, e28150. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. Plant-CARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterizationof user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
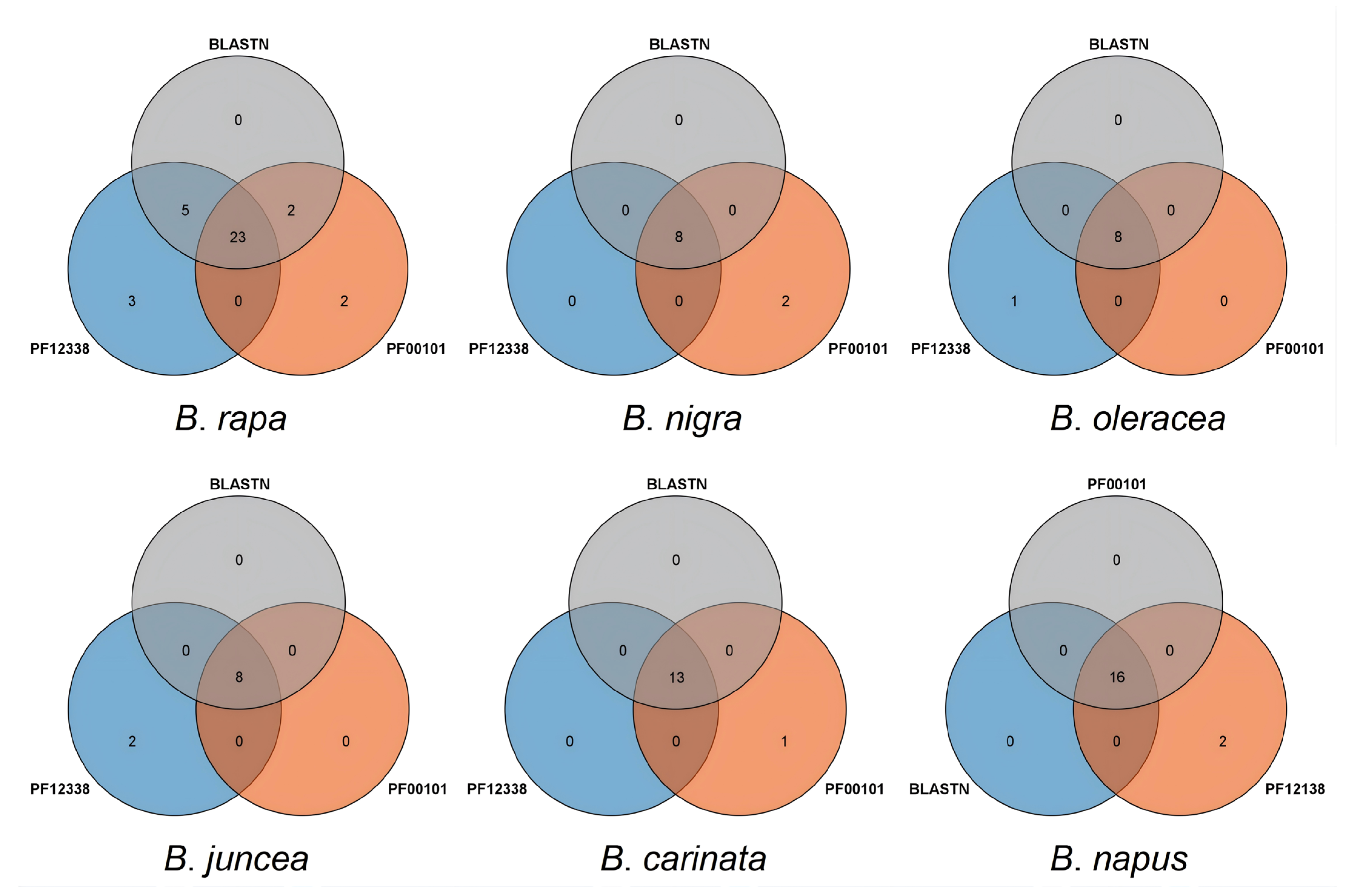

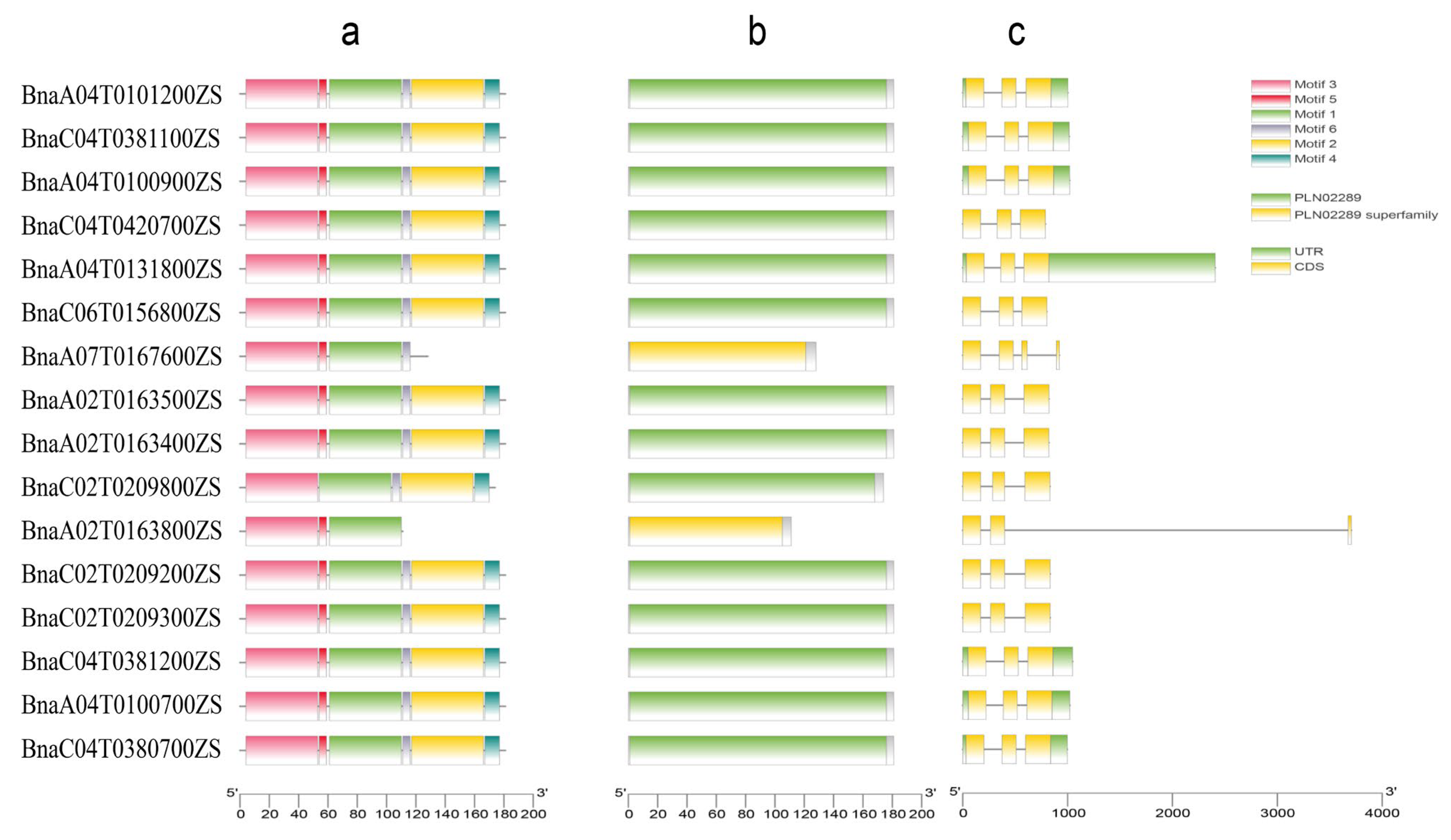
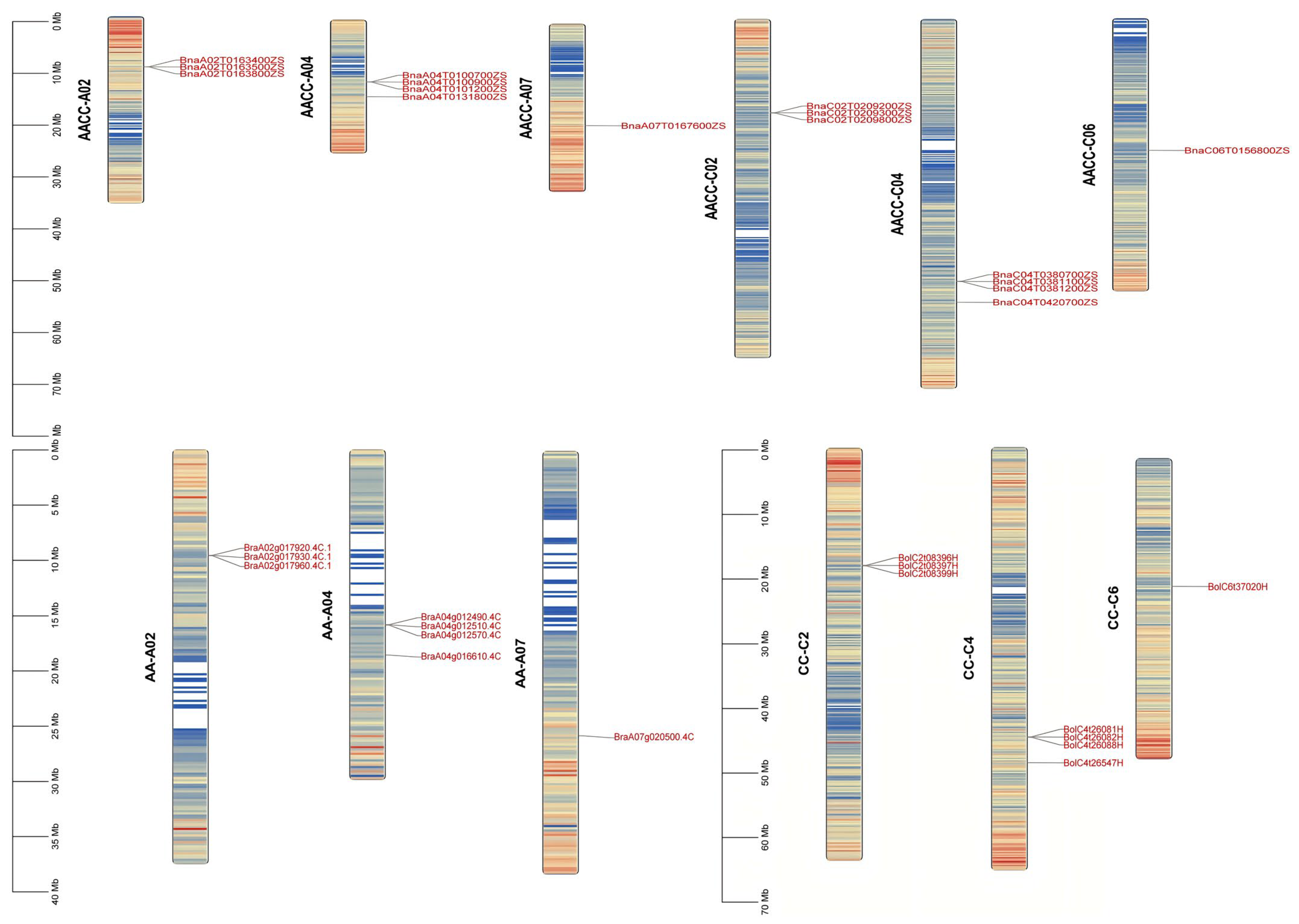
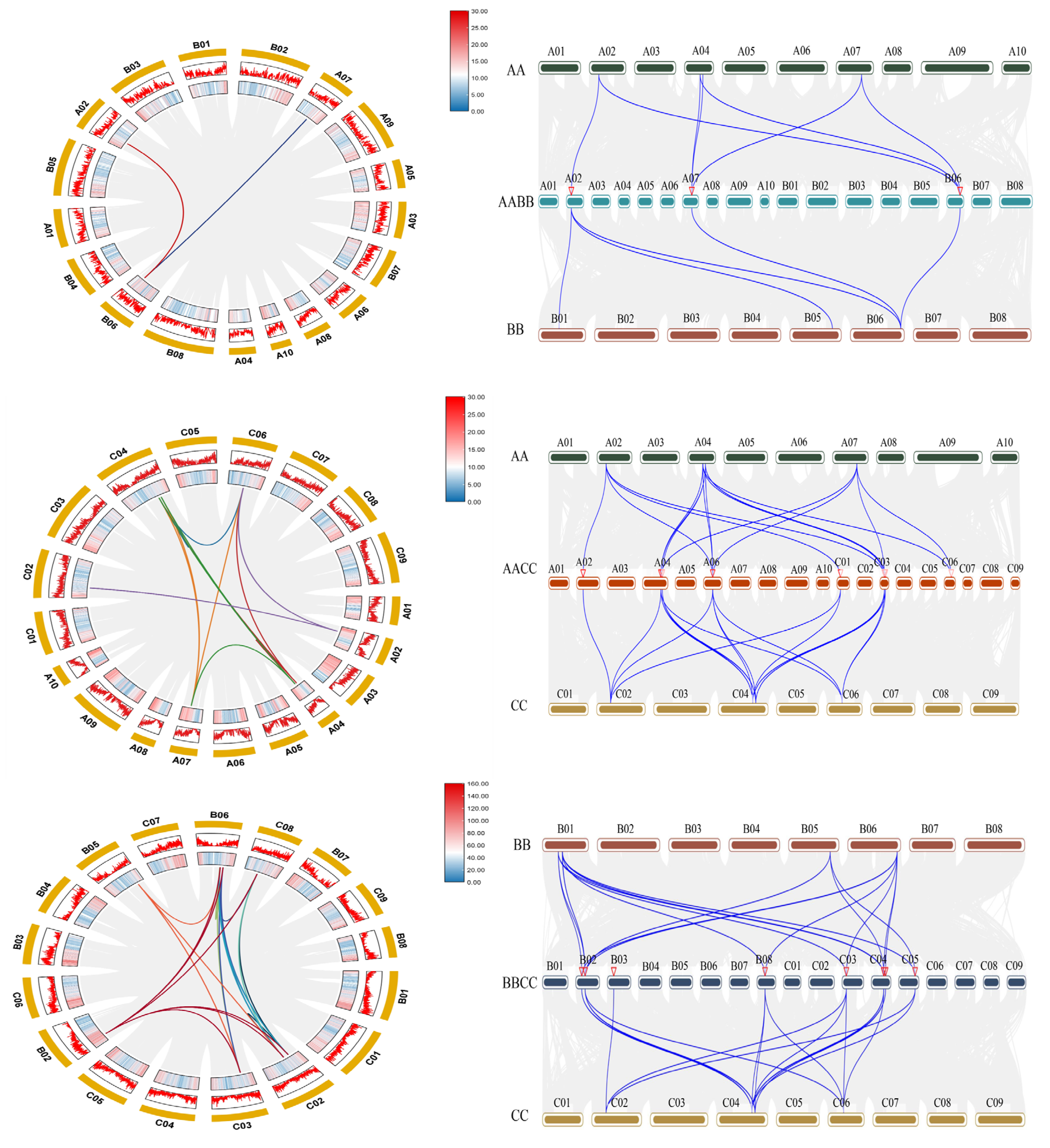





| Genome ID | Plant Species | Genome Type | Genome Size (Kb) | Coding Genes | RcbS Genes |
|---|---|---|---|---|---|
| Chiifu.v4 | Brassica rapa ssp. pekinensis | AA | 113,126 | 83,682 | 8 |
| CN115125v1 | Brassica nigra | BB | 532,791 | 67,021 | 8 |
| HDEM.v0 | Brassica oleracea var. italica | CC | 551,006 | 61,279 | 8 |
| tumida.v1.5 | Brassica juncea var. tumida | AABB | 915,101 | 79,644 | 8 |
| ZS11.v0 | Brassica napus ssp. oleifera | AACC | 987,244 | 100,919 | 16 |
| zd-1.v0 | Brassica carinata | BBCC | 1,082,762 | 97,149 | 13 |
| A. Thaliana Homologues | B. rapa Homologues | B. oleracea Homologues | B. napus Homologues | Chr | Start | End | Length |
|---|---|---|---|---|---|---|---|
| AT1G67090/RbcS-1A | BraA02g017930.4C | BnaA02G0163500ZS | A02 | 9,675,456 | 9,676,279 | 823 | |
| AT1G67090/RbcS-1A | BraA02g017960.4C | BnaA02G0163800ZS | A02 | 9,686,731 | 9,690,436 | 3705 | |
| AT1G67090/RbcS-1A | BolC2t08396H | BnaC02G0209200ZS | C02 | 17,993,607 | 17,994,441 | 834 | |
| AT5G38430/RbcS-1B | BraA02g017920.4C | BnaA02G0163400ZS | A02 | 9,672,337 | 9,673,159 | 822 | |
| AT5G38430/RbcS-1B | BraA04g012510.4C | BnaA04G0100700ZS | A04 | 11,915,596 | 11,916,617 | 1021 | |
| AT5G38430/RbcS-1B | BraA04g012490.4C | BnaA04G0100900ZS | A04 | 11,923,776 | 11,924,795 | 1019 | |
| AT5G38430/RbcS-1B | BraA04g012570.4C | BnaA04G0101200ZS | A04 | 11,938,835 | 11,939,838 | 1003 | |
| AT5G38430/RbcS-1B | BraA04g016610.4C | BnaA04G0131800ZS | A04 | 14,773,357 | 14,775,764 | 2407 | |
| AT5G38430/RbcS-1B | BraA07g020500.4C | BnaA07G0167600ZS | A07 | 19,554,954 | 19,555,875 | 921 | |
| AT5G38430/RbcS-1B | BolC2t08397H | BnaC02G0209300ZS | C02 | 17,997,780 | 17,998,614 | 834 | |
| AT5G38430/RbcS-1B | BolC2t08399H | BnaC02G0209800ZS | C02 | 18,074,145 | 18,074,977 | 832 | |
| AT5G38430/RbcS-1B | BolC4t26088H | BnaC04G0380700ZS | C04 | 50,500,677 | 50,501,674 | 997 | |
| AT5G38430/RbcS-1B | BolC4t26082H | BnaC04G0381200ZS | C04 | 50,539,319 | 50,540,335 | 1016 | |
| AT5G38430/RbcS-1B | BolC4t26081H | BnaC04G0381200ZS | C04 | 50,546,522 | 50,547,569 | 1047 | |
| AT5G38430/RbcS-1B | BolC4t26547H | BnaC04G0420700ZS | C04 | 54,553,194 | 54,553,982 | 788 | |
| AT5G38430/RbcS-1B | BolC6t37020H | BnaC06G0156800ZS | C06 | 25,387,399 | 25,388,201 | 802 | |
| AT5G38420/RbcS-2B | / | / | / | / | / | ||
| AT5G38410/RbcS-3B | / | / | / | / | / |
| Sequence ID | Number of Amino Acid | Molecular Weight | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| BnaC04T0381100ZS | 181 | 20,183.17 | 8.23 | 32.21 | 73.26 | −0.16 | chloroplast |
| BnaA04T0100900ZS | 181 | 20,183.17 | 8.23 | 32.21 | 73.26 | −0.16 | chloroplast |
| BnaC04T0381200ZS | 181 | 20,199.17 | 8.23 | 33.27 | 72.71 | −0.175 | chloroplast |
| BnaA04T0100700ZS | 181 | 20,227.22 | 8.23 | 32.8 | 73.76 | −0.161 | chloroplast |
| BnaA04T0131800ZS | 181 | 20,158.12 | 8.23 | 31.61 | 72.21 | −0.16 | chloroplast |
| BnaC04T0420700ZS | 181 | 20,185.14 | 8.23 | 31.61 | 72.21 | −0.175 | chloroplast |
| BnaA04T0101200ZS | 181 | 20,303.32 | 8.23 | 31.74 | 73.76 | −0.164 | chloroplast |
| BnaC04T0380700ZS | 181 | 20,317.3 | 7.59 | 30.78 | 74.31 | −0.16 | chloroplast |
| BnaC06T0156800ZS | 181 | 20,302.31 | 8.23 | 29.04 | 70.55 | −0.223 | chloroplast |
| BnaA02T0163400ZS | 181 | 20,324.46 | 8.48 | 33.66 | 72.76 | −0.162 | chloroplast |
| BnaA02T0163500ZS | 181 | 20,323.48 | 8.69 | 32.83 | 72.76 | −0.162 | chloroplast |
| BnaC02T0209200ZS | 181 | 20,462.64 | 8.48 | 29.53 | 72.21 | −0.186 | chloroplast |
| BnaC02T0209300ZS | 181 | 20,377.46 | 7.58 | 37.81 | 75.47 | −0.157 | chloroplast |
| BnaC02T0209800ZS | 174 | 19,560.59 | 8.7 | 36.41 | 75.69 | −0.127 | chloroplast |
| BnaA07T0167600ZS | 128 | 13,981.96 | 6.72 | 25.35 | 77.66 | −0.103 | chloroplast |
| BnaA02T0163800ZS | 111 | 12,311.44 | 9.58 | 47.73 | 81.71 | −0.089 | chloroplast |
| Protein ID | Alpha Helix (Hh) | Extended Strand (Ee) | Random Coil (Cc) | |||
|---|---|---|---|---|---|---|
| Quantity | Proportion (%) | Quantity | Proportion (%) | Quantity | Proportion (%) | |
| BnaC04T0381100ZS | 50 | 27.62% | 30 | 16.57% | 101 | 55.80% |
| BnaA04T0100900ZS | 50 | 27.62% | 30 | 16.57% | 101 | 55.80% |
| BnaC04T0381200ZS | 47 | 25.97% | 33 | 18.23% | 101 | 55.80% |
| BnaA04T0100700ZS | 43 | 23.76% | 34 | 18.78% | 104 | 57.46% |
| BnaA04T0131800ZS | 45 | 24.86% | 23 | 12.71% | 113 | 62.43% |
| BnaC04T0420700ZS | 51 | 28.18% | 31 | 17.13% | 99 | 54.70% |
| BnaA04T0101200ZS | 48 | 26.52% | 33 | 18.23% | 100 | 55.25% |
| BnaC04T0380700ZS | 47 | 25.97% | 33 | 18.23% | 101 | 55.80% |
| BnaC06T0156800ZS | 50 | 27.62% | 29 | 16.02% | 102 | 56.35% |
| BnaA02T0163400ZS | 47 | 25.97% | 31 | 17.13% | 103 | 56.91% |
| BnaA02T0163500ZS | 50 | 27.62% | 22 | 12.15% | 109 | 60.22% |
| BnaC02T0209200ZS | 47 | 25.97% | 28 | 15.47% | 106 | 58.56% |
| BnaC02T0209300ZS | 50 | 27.62% | 33 | 18.23% | 98 | 54.14% |
| BnaC02T0209800ZS | 48 | 27.59% | 31 | 17.82% | 95 | 54.60% |
| BnaA07T0167600ZS | 38 | 29.69% | 18 | 14.06% | 72 | 56.25% |
| BnaA02T0163800ZS | 30 | 27.03% | 12 | 10.81% | 69 | 62.16% |
| References allele | G | A | TAT | TCTATGT | T | G |
| Alternative allele | A | - | - | - | - | A |
| SNP annotation | UTR3 | UTR3 | UTR3 | intronic | intronic | exonic |
| SNP positions | 11,938,853 | 11,938,916 | 11,938,973 | 11,939,546 | 11,939,632 | 11,939,677 |
| Hap_1 | A/A | -/- | -/- | -/- | -/- | G/G |
| Hap_2 | G/G | A/A | TAT/TAT | TCTATGT/TCTATGT | T/T | G/G |
| Hap_3 | G/G | A/A | TAT/TAT | TCTATGT/TCTATGT | -/- | A/A |
| Hap_4 | G/G | A/A | -/- | TCTATGT/TCTATGT | -/- | A/A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Li, Y.; Cui, C.; Chai, L.; Zheng, B.; Zhang, K.; Jiang, J.; Zhang, J.; Wu, J.; Lang, J.; Zhang, T.; et al. Genome-Wide Analysis of the RbcS Gene Family and Expression Analysis Under Light Response in Brassica napus L. Plants 2026, 15, 58. https://doi.org/10.3390/plants15010058
Li Y, Cui C, Chai L, Zheng B, Zhang K, Jiang J, Zhang J, Wu J, Lang J, Zhang T, et al. Genome-Wide Analysis of the RbcS Gene Family and Expression Analysis Under Light Response in Brassica napus L. Plants. 2026; 15(1):58. https://doi.org/10.3390/plants15010058
Chicago/Turabian StyleLi, Yanling, Cheng Cui, Liang Chai, Benchuan Zheng, Ka Zhang, Jun Jiang, Jinfang Zhang, Jing Wu, Jing Lang, Tongyun Zhang, and et al. 2026. "Genome-Wide Analysis of the RbcS Gene Family and Expression Analysis Under Light Response in Brassica napus L." Plants 15, no. 1: 58. https://doi.org/10.3390/plants15010058
APA StyleLi, Y., Cui, C., Chai, L., Zheng, B., Zhang, K., Jiang, J., Zhang, J., Wu, J., Lang, J., Zhang, T., Zhou, Y., He, P., Jiang, L., Wang, H., & Li, H. (2026). Genome-Wide Analysis of the RbcS Gene Family and Expression Analysis Under Light Response in Brassica napus L. Plants, 15(1), 58. https://doi.org/10.3390/plants15010058







