Advancements in Molecular Breeding Techniques for Soybeans
Abstract
1. Introduction
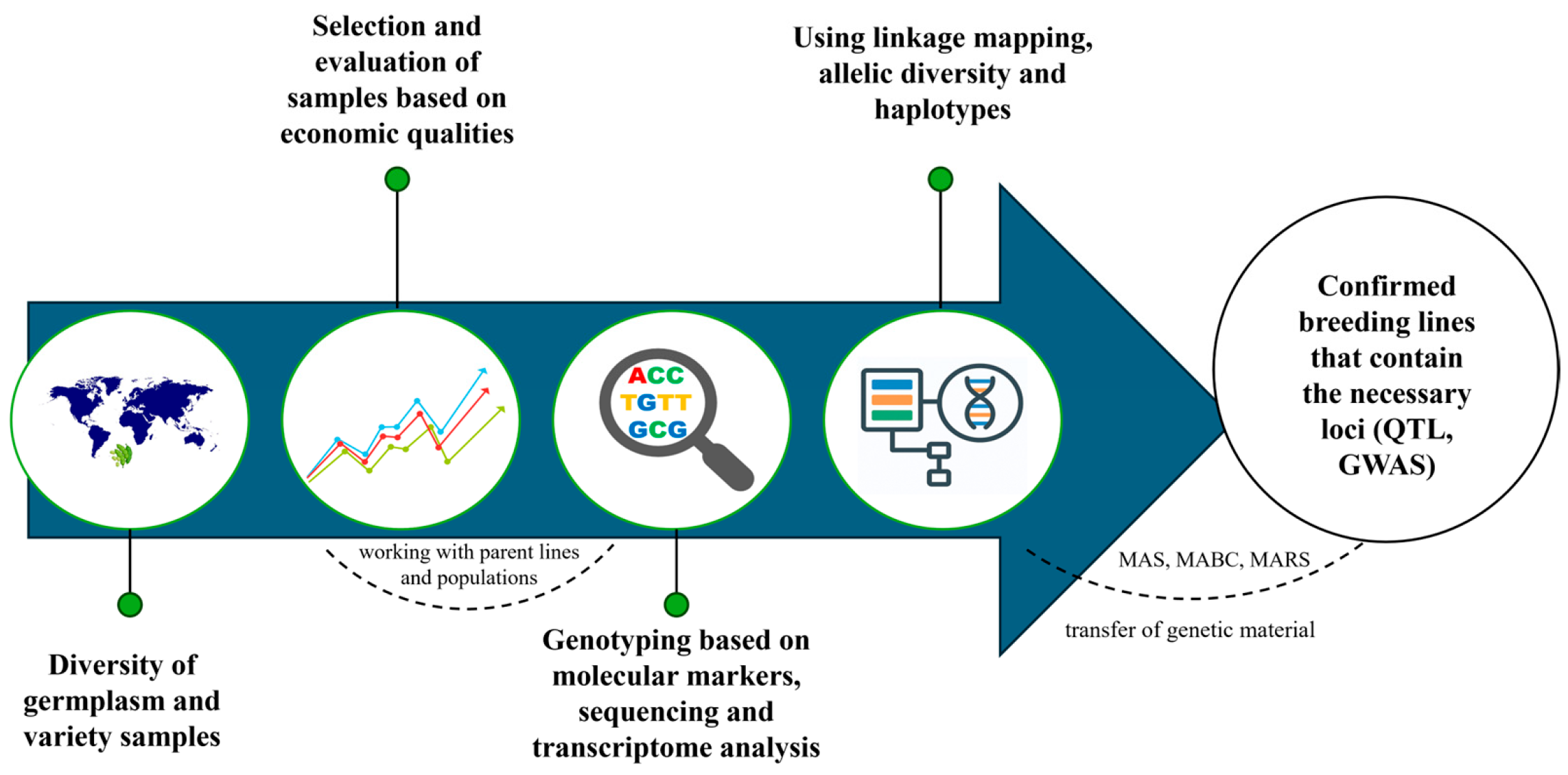
2. Overview of Molecular Breeding Techniques
3. Molecular Markers in Soybean Breeding
4. Marker-Assisted Breeding
5. Genomic Selection
6. CRISPR/Cas9 Technology
7. RNA Interference
- dsRNA processing: The Dicer enzyme cleaves dsRNA into short interfering RNAs (siRNAs), about 21–24 nucleotides long. These small RNAs play a central role in post-transcriptional silencing of genes [56].
- Formation of the RISC complex: siRNAs integrate into the RISC complex (RNA-induced silencing complex), which uses them as a template for searching for complementary mRNAs [57].
- Degradation or blockade of mRNA: After binding to the target mRNA, RISC initiates its cleavage or prevents translation, which leads to a decrease in the expression of the corresponding gene [52].
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jiang, G.-L.; Rajcan, I.; Zhang, Y.-M.; Han, T.; Mian, R. Editorial: Soybean Molecular Breeding and Genetics. Front. Plant Sci. 2023, 14, 1157632. [Google Scholar] [CrossRef]
- Singh, S.; Nanda, S.R. Accelerating Soybean Breeding: A Comprehensive Review of Speed Breeding. Int. J. Adv. Biochem. Res. 2024, 8, 551–556. [Google Scholar] [CrossRef]
- Abdul Aziz, M.; Masmoudi, K. Molecular Breakthroughs in Modern Plant Breeding Techniques. Hortic. Plant J. 2025, 11, 15–41. [Google Scholar] [CrossRef]
- Yao, D.; Zhou, J.; Zhang, A.; Wang, J.; Liu, Y.; Wang, L.; Pi, W.; Li, Z.; Yue, W.; Cai, J.; et al. Advances in CRISPR/Cas9-Based Research Related to Soybean [Glycine max (Linn.) Merr] Molecular Breeding. Front. Plant Sci. 2023, 14, 1247707. [Google Scholar] [CrossRef]
- Qiu, L.-J.; Chen, P.-Y.; Liu, Z.-X.; Li, Y.-H.; Guan, R.-X.; Wang, L.-H.; Chang, R.-Z. The Worldwide Utilization of the Chinese Soybean Germplasm Collection. Plant Genet. Resour. 2011, 9, 109–122. [Google Scholar] [CrossRef]
- Lee, C.; Choi, M.-S.; Kim, H.-T.; Yun, H.-T.; Lee, B.; Chung, Y.-S.; Kim, R.W.; Choi, H.-K. Soybean [Glycine max (L.) Merrill]: Importance as A Crop and Pedigree Reconstruction of Korean Varieties. Plant Breed. Biotechnol. 2015, 3, 179–196. [Google Scholar] [CrossRef]
- Wang, X.; Qi, Y.; Sun, G.; Zhang, S.; Li, W.; Wang, Y. Improving Soybean Breeding Efficiency Using Marker-Assisted Selection. Mol. Plant Breed. 2024, 15, 259–268. [Google Scholar] [CrossRef]
- Patil, G.; Mian, R.; Vuong, T.; Pantalone, V.; Song, Q.; Chen, P.; Shannon, G.J.; Carter, T.C.; Nguyen, H.T. Molecular Mapping and Genomics of Soybean Seed Protein: A Review and Perspective for the Future. Theor. Appl. Genet. 2017, 130, 1975–1991. [Google Scholar] [CrossRef]
- Van, K.; McHale, L. Meta-Analyses of QTLs Associated with Protein and Oil Contents and Compositions in Soybean [Glycine max (L.) Merr.] Seed. Int. J. Mol. Sci. 2017, 18, 1180. [Google Scholar] [CrossRef]
- Fields, J.; Saxton, A.M.; Beyl, C.A.; Kopsell, D.A.; Cregan, P.B.; Hyten, D.L.; Cuvaca, I.; Pantalone, V.R. Seed Protein and Oil QTL in a Prominent Glycine max Genetic Pedigree: Enhancing Stability for Marker Assisted Selection. Agronomy 2023, 13, 567. [Google Scholar] [CrossRef]
- Bhowmik, P.; Konkin, D.; Polowick, P.; Hodgins, C.L.; Subedi, M.; Xiang, D.; Yu, B.; Patterson, N.; Rajagopalan, N.; Babic, V.; et al. CRISPR/Cas9 Gene Editing in Legume Crops: Opportunities and Challenges. Legume Sci. 2021, 3, e96. [Google Scholar] [CrossRef]
- Vinichenko, N.A.; Salina, E.A.; Kochetov, A.V. The scope of use of molecular markers in soybean breeding [in Russian]. Vavilovskii Zhurnal Genet. I Sel. Lett. Vavilov J. Genet. Breed. 2020, 6, 107–125. [Google Scholar]
- Xie, D.; Han, Y.; Zeng, Y.; Chang, W.; Teng, W.; Li, W. SSR- and SNP-Related QTL Underlying Linolenic Acid and Other Fatty Acid Contents in Soybean Seeds Across Multiple Environments. Mol. Breed. 2012, 30, 169–179. [Google Scholar] [CrossRef]
- Perfil’ev, R.N.; Shcherban, A.B.; Salina, E.A. Development of a Marker Panel for Genotyping of Domestic Soybean Cultivars for Genes Controlling the Duration of Vegetation and Response to Photoperiod. Vestn. VOGiS 2021, 25, 761–769. [Google Scholar] [CrossRef]
- Santana, F.A.; Silva, M.F.D.; Guimarães, J.K.F.; Ferreira, M.F.D.S.; Pereira, W.D.; Piovesan, N.D.; Barros, E.G.D. Marker-Assisted Selection Strategies for Developing Resistant Soybean Plants to Cyst Nematode. Crop Breed. Appl. Biotechnol. 2014, 14, 180–186. [Google Scholar] [CrossRef]
- Kim, J.-M.; Kim, K.-H.; Jung, J.; Kang, B.K.; Lee, J.; Ha, B.-K.; Kang, S. Validation of Marker-Assisted Selection in Soybean Breeding Program for Pod Shattering Resistance. Euphytica 2020, 216, 166. [Google Scholar] [CrossRef]
- Kumar, V.; Rani, A.; Shukla, S.; Jha, P. Development of Kunitz Trypsin Inhibitor Free Vegetable Soybean Genotypes Through Marker-Assisted Selection. Int. J. Veg. Sci. 2021, 27, 364–377. [Google Scholar] [CrossRef]
- Rajapakse, S. GENETICS|Molecular Markers. In Encyclopedia of Rose Science; Elsevier: Amsterdam, The Netherlands, 2003; pp. 334–341. ISBN 978-0-12-227620-0. [Google Scholar]
- Maughan, P.J.; Saghai Maroof, M.A.; Buss, G.R.; Huestis, G.M. Amplified Fragment Length Polymorphism (AFLP) in Soybean: Species Diversity, Inheritance, and Near-Isogenic Line Analysis. Theor. Appl. Genet. 1996, 93, 392–401. [Google Scholar] [CrossRef]
- Thiel, T.; Michalek, W.; Varshney, R.; Graner, A. Exploiting EST Databases for the Development and Characterization of Gene-Derived SSR-Markers in Barley (Hordeum vulgare L.). Theor. Appl. Genet. 2003, 106, 411–422. [Google Scholar] [CrossRef]
- Li, A.Q.; Zhao, C.Z.; Wang, X.J.; Liu, Z.J.; Zhang, L.F.; Song, G.Q.; Yin, J.; Li, C.S.; Xia, H.; Bi, Y.P. Identification of SSR Markers Using Soybean (Glycine max) ESTs from Globular Stage Embryos. Electron. J. Biotechnol. 2010, 13, 6–7. [Google Scholar] [CrossRef]
- Zhao, X.; Chang, W.; Han, Y.; Teng, W.; Li, W. A Modified Method for the Development of SSR Molecular Markers Based on Redundant EST Data and Its Application in Soybean. J. Integr. Agric. 2012, 11, 545–555. [Google Scholar] [CrossRef]
- Singh, R.P.; Singh, P.K.; Gupta, R.; Singh, R.L. Biotechnological Tools to Enhance Sustainable Production. In Biotechnology for Sustainable Agriculture; Elsevier: Amsterdam, The Netherlands, 2018; pp. 19–66. ISBN 978-0-12-812160-3. [Google Scholar]
- Chiemeke, F.K.; Olasanmi, B.; Agre, P.A.; Mushoriwa, H.; Chigeza, G.; Abebe, A.T. Genetic Diversity and Population Structure Analysis of Soybean [Glycine max (L.) Merrill] Genotypes Using Agro-Morphological Traits and SNP Markers. Genes 2024, 15, 1373. [Google Scholar] [CrossRef]
- Muñoz-Amatriaín, M.; Cuesta-Marcos, A.; Endelman, J.B.; Comadran, J.; Bonman, J.M.; Bockelman, H.E.; Chao, S.; Russell, J.; Waugh, R.; Hayes, P.M.; et al. The USDA Barley Core Collection: Genetic Diversity, Population Structure, and Potential for Genome-Wide Association Studies. PLoS ONE 2014, 9, e94688. [Google Scholar] [CrossRef]
- Zatybekov, A.; Yermagambetova, M.; Genievskaya, Y.; Didorenko, S.; Abugalieva, S. Genetic Diversity Analysis of Soybean Collection Using Simple Sequence Repeat Markers. Plants 2023, 12, 3445. [Google Scholar] [CrossRef]
- Sudaric, A.; Vrataric, M.; Rajcan, I.; Duvnjak, T.; Volenik, M. Application of Molecular Markers in Parental Selection in Soybean. Acta Agron. Hung 2008, 56, 393–398. [Google Scholar] [CrossRef]
- Qin, J.; Shi, A.; Song, Q.; Li, S.; Wang, F.; Cao, Y.; Ravelombola, W.; Song, Q.; Yang, C.; Zhang, M. Genome Wide Association Study and Genomic Selection of Amino Acid Concentrations in Soybean Seeds. Front. Plant Sci. 2019, 10, 1445. [Google Scholar] [CrossRef]
- Crossa, J.; Pérez, P.; De Los Campos, G.; Mahuku, G.; Dreisigacker, S.; Magorokosho, C. Genomic Selection and Prediction in Plant Breeding. J. Crop Improv. 2011, 25, 239–261. [Google Scholar] [CrossRef]
- Sun, B.; Guo, R.; Liu, Z.; Shi, X.; Yang, Q.; Shi, J.; Zhang, M.; Yang, C.; Zhao, S.; Zhang, J.; et al. Genetic Variation and Marker−Trait Association Affect the Genomic Selection Prediction Accuracy of Soybean Protein and Oil Content. Front. Plant Sci. 2022, 13, 1064623. [Google Scholar] [CrossRef]
- Ravelombola, W.; Qin, J.; Shi, A.; Song, Q.; Yuan, J.; Wang, F.; Chen, P.; Yan, L.; Feng, Y.; Zhao, T.; et al. Genome-Wide Association Study and Genomic Selection for Yield and Related Traits in Soybean. PLoS ONE 2021, 16, e0255761. [Google Scholar] [CrossRef]
- Haider, A. CRISPR-Cas9 Mediated Genome Editing in Soybean for Improving Quality Traits. J. Innov. Agric. 2023, 10, 1–20. [Google Scholar] [CrossRef]
- Zhong, X.; Hong, W.; Shu, Y.; Li, J.; Liu, L.; Chen, X.; Islam, F.; Zhou, W.; Tang, G. CRISPR/Cas9 Mediated Gene-Editing of GmHdz4 Transcription Factor Enhances Drought Tolerance in Soybean (Glycine max [L.] Merr.). Front. Plant Sci. 2022, 13, 988505. [Google Scholar] [CrossRef]
- Sebastian, S.A.; Fader, G.M.; Ulrich, J.F.; Forney, D.R.; Chaleff, R.S. Semidominant Soybean Mutation for Resistance to Sulfonylurea Herbicides. Crop Sci. 1989, 29, 1403–1408. [Google Scholar] [CrossRef]
- Cai, Y.; Chen, L.; Liu, X.; Guo, C.; Sun, S.; Wu, C.; Jiang, B.; Han, T.; Hou, W. CRISPR/Cas9—Mediated Targeted Mutagenesis of GmFT2a Delays Flowering Time in Soya Bean. Plant Biotechnol. J. 2018, 16, 176–185. [Google Scholar] [CrossRef]
- Jacobs, T.B.; LaFayette, P.R.; Schmitz, R.J.; Parrott, W.A. Targeted Genome Modifications in Soybean with CRISPR/Cas9. BMC Biotechnol. 2015, 15, 16. [Google Scholar] [CrossRef]
- Le, H.; Nguyen, N.H.; Ta, D.T.; Le, T.N.T.; Bui, T.P.; Le, N.T.; Nguyen, C.X.; Rolletschek, H.; Stacey, G.; Stacey, M.G.; et al. CRISPR/Cas9-Mediated Knockout of Galactinol Synthase-Encoding Genes Reduces Raffinose Family Oligosaccharide Levels in Soybean Seeds. Front. Plant Sci. 2020, 11, 612942. [Google Scholar] [CrossRef]
- Zhang, P.; Du, H.; Wang, J.; Pu, Y.; Yang, C.; Yan, R.; Yang, H.; Cheng, H.; Yu, D. Multiplex CRISPR/Cas9-Mediated Metabolic Engineering Increases Soya Bean Isoflavone Content and Resistance to Soya Bean Mosaic Virus. Plant Biotechnol. J. 2020, 18, 1384–1395. [Google Scholar] [CrossRef]
- Bao, A.; Chen, H.; Chen, L.; Chen, S.; Hao, Q.; Guo, W.; Qiu, D.; Shan, Z.; Yang, Z.; Yuan, S.; et al. CRISPR/Cas9-Mediated Targeted Mutagenesis of GmSPL9 Genes Alters Plant Architecture in Soybean. BMC Plant Biol. 2019, 19, 131. [Google Scholar] [CrossRef]
- Cheng, Q.; Dong, L.; Su, T.; Li, T.; Gan, Z.; Nan, H.; Lu, S.; Fang, C.; Kong, L.; Li, H.; et al. CRISPR/Cas9-Mediated Targeted Mutagenesis of GmLHY Genes Alters Plant Height and Internode Length in Soybean. BMC Plant Biol. 2019, 19, 562. [Google Scholar] [CrossRef]
- Cai, Y.; Wang, L.; Chen, L.; Wu, T.; Liu, L.; Sun, S.; Wu, C.; Yao, W.; Jiang, B.; Yuan, S.; et al. Mutagenesis of GmFT2a and GmFT5a Mediated by CRISPR/Cas9 Contributes for Expanding the Regional Adaptability of Soybean. Plant Biotechnol. J. 2020, 18, 298–309. [Google Scholar] [CrossRef]
- Wang, J.; Kuang, H.; Zhang, Z.; Yang, Y.; Yan, L.; Zhang, M.; Song, S.; Guan, Y. Generation of Seed Lipoxygenase-Free Soybean Using CRISPR-Cas9. Crop J. 2020, 8, 432–439. [Google Scholar] [CrossRef]
- Han, J.; Guo, B.; Guo, Y.; Zhang, B.; Wang, X.; Qiu, L.-J. Creation of Early Flowering Germplasm of Soybean by CRISPR/Cas9 Technology. Front. Plant Sci. 2019, 10, 1446. [Google Scholar] [CrossRef]
- Cai, Y.; Chen, L.; Sun, S.; Wu, C.; Yao, W.; Jiang, B.; Han, T.; Hou, W. CRISPR/Cas9-Mediated Deletion of Large Genomic Fragments in Soybean. Int. J. Mol. Sci. 2018, 19, 3835. [Google Scholar] [CrossRef]
- Li, M.; Liu, Y.; Wang, C.; Yang, X.; Li, D.; Zhang, X.; Xu, C.; Zhang, Y.; Li, W.; Zhao, L. Identification of Traits Contributing to High and Stable Yields in Different Soybean Varieties Across Three Chinese Latitudes. Front. Plant Sci. 2020, 10, 1642. [Google Scholar] [CrossRef]
- Al Amin, N.; Ahmad, N.; Wu, N.; Pu, X.; Ma, T.; Du, Y.; Bo, X.; Wang, N.; Sharif, R.; Wang, P. CRISPR-Cas9 Mediated Targeted Disruption of FAD2–2 Microsomal Omega-6 Desaturase in Soybean (Glycine max L.). BMC Biotechnol. 2019, 19, 9. [Google Scholar] [CrossRef]
- Do, P.T.; Nguyen, C.X.; Bui, H.T.; Tran, L.T.N.; Stacey, G.; Gillman, J.D.; Zhang, Z.J.; Stacey, M.G. Demonstration of Highly Efficient Dual gRNA CRISPR/Cas9 Editing of the Homeologous GmFAD2–1A and GmFAD2–1B Genes to Yield a High Oleic, Low Linoleic and α-Linolenic acid Phenotype in Soybean. BMC Plant Biol. 2019, 19, 311. [Google Scholar] [CrossRef]
- Wu, N.; Lu, Q.; Wang, P.; Zhang, Q.; Zhang, J.; Qu, J.; Wang, N. Construction and Analysis of GmFAD2-1A and GmFAD2-2A Soybean Fatty Acid Desaturase Mutants Based on CRISPR/Cas9 Technology. Int. J. Mol. Sci. 2020, 21, 1104. [Google Scholar] [CrossRef]
- Ma, J.; Sun, S.; Whelan, J.; Shou, H. CRISPR/Cas9-Mediated Knockout of GmFATB1 Significantly Reduced the Amount of Saturated Fatty Acids in Soybean Seeds. Int. J. Mol. Sci. 2021, 22, 3877. [Google Scholar] [CrossRef]
- Wang, S.; Yokosho, K.; Guo, R.; Whelan, J.; Ruan, Y.-L.; Ma, J.F.; Shou, H. The Soybean Sugar Transporter GmSWEET15 Mediates Sucrose Export from Endosperm to Early Embryo. Plant Physiol. 2019, 180, 2133–2141. [Google Scholar] [CrossRef]
- Sun, L.; Miao, Z.; Cai, C.; Zhang, D.; Zhao, M.; Wu, Y.; Zhang, X.; Swarm, S.A.; Zhou, L.; Zhang, Z.J.; et al. GmHs1-1, Encoding a Calcineurin-Like Protein, Controls Hard-Seededness in Soybean. Nat. Genet. 2015, 47, 939–943. [Google Scholar] [CrossRef]
- Baulcombe, D. RNA Silencing in Plants. Nature 2004, 431, 356–363. [Google Scholar] [CrossRef]
- Zhang, X.; Sato, S.; Ye, X.; Dorrance, A.E.; Morris, T.J.; Clemente, T.E.; Qu, F. Robust RNAi-Based Resistance to Mixed Infection of Three Viruses in Soybean Plants Expressing Separate Short Hairpins from a Single Transgene. Phytopathology 2011, 101, 1264–1269. [Google Scholar] [CrossRef]
- Morozov, S.Y.; Solovyev, A.G.; Kalinina, N.O.; Taliansky, M.E. Double-Stranded RNAs in Plant Protection Against Pathogenic Organisms and Viruses in Agriculture. Acta Nat. 2019, 11, 13–21. [Google Scholar] [CrossRef]
- Hamilton, A.J.; Baulcombe, D.C. A Species of Small Antisense RNA in Posttranscriptional Gene Silencing in Plants. Science 1999, 286, 950–952. [Google Scholar] [CrossRef]
- Voinnet, O. Origin, Biogenesis, and Activity of Plant MicroRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef]
- Mallory, A.C.; Vaucheret, H. Functions of MicroRNAs and Related Small RNAs in Plants. Nat. Genet. 2006, 38, S31–S36. [Google Scholar] [CrossRef]
- Kasai, M.; Tsuchiya, M.; Kanazaw, A. Gene Duplication and RNA Silencing in Soybean. In A Comprehensive Survey of International Soybean Research—Genetics, Physiology, Agronomy and Nitrogen Relationships; Board, J., Ed.; InTech: West Palm Beach, FL, USA, 2013; ISBN 978-953-51-0876-4. [Google Scholar]
- Jiang, Y.; Hu, Y.; Wang, B.; Wu, T. Bivalent RNA Interference to Increase Isoflavone Biosynthesis in Soybean (Glycine max). Braz. Arch. Biol. Technol. 2013, 57, 163–170. [Google Scholar] [CrossRef]
- Subramanian, S.; Graham, M.Y.; Yu, O.; Graham, T.L. RNA Interference of Soybean Isoflavone Synthase Genes Leads to Silencing in Tissues Distal to the Transformation Site and to Enhanced Susceptibility to Phytophthora sojae. Plant Physiol. 2005, 137, 1345–1353. [Google Scholar] [CrossRef]
- Um, J.-H.; Kim, S.; Kim, Y.-K.; Song, S.-B.; Lee, S.-H.; Verma, D.P.S.; Cheon, C.-I. RNA Interference-Mediated Repression of S6 Kinase 1 Impairs Root Nodule Development in Soybean. Mol. Cells 2013, 35, 243–248. [Google Scholar] [CrossRef]

| Application | Target Gene | Mode of Action/ Method | Improved Trait | Reference |
|---|---|---|---|---|
| Increase crop yield (Regulating architecture and flowering period) | SPL9a, SPL9b | Knockout | Influence features of plant architecture | [39] |
| SPL9c, SPL9d | Knockout | Influence features of plant architecture | [39] | |
| GmLHY1a, b, | Knockout | Influence features of plant architecture | [40] | |
| GmLHY2a, 2b, | Knockout | Influence features of plant architecture | [40] | |
| GmFT2a, GmFT4 | Base editing | Affect time of flower | [41] | |
| GmAP1 | Knockout | Affect time of flower | [41] | |
| Gmprr37 | Knockout | Affect time of flower | [42] | |
| GmE1 | Knockout | Affect time of flower | [43] | |
| GmFT2a | Knockout | Affect time of flower | [44] | |
| Enhance crop quality & nutrition (Storage protein/seed oil/sugar content/allergenic properties/bean flavor free Soybean) | Glyma.03g163500 | Knockout | Regulate storage of protein | [45] |
| Glyma.20g148400 | Knockout | Regulate storage of protein | [45] | |
| Glyma.19g164900 | Knockout | Regulate storage of protein | [45] | |
| FAD2-2 | Knockout | Improve content of fatty acid | [46] | |
| GmFAD2-1A | Knockout | Improve content of fatty acid | [47] | |
| GmFAD2-1B | Knockout | Improve content of fatty acid | [47] | |
| GmFAD2-1A | Knockout | Improve content of fatty acid | [48] | |
| GmFAD2-2A | Knockout | Improve content of fatty acid | [48] | |
| Glyma.19G147300 | Knockout | Improve content of fatty acid | [48] | |
| GmFATB1 (GmFATB1a and GmFATB1b) | Knockout | Develop fatty acid content | [49] | |
| GmGOLS1A | Knockout | Develop fatty acid content | [49] | |
| GmGOLS1B | Knockout | Develop fatty acid content | [37] | |
| GmSWEET15a | Knockout | Develop fatty acid content | [50] | |
| Gly m Bd 30K | Knockout | Regulate allergenic | [51] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Fetisov, I.; Eizikovich, O.; Charles Diouf, D.; Romanova, E.; Kezimana, P. Advancements in Molecular Breeding Techniques for Soybeans. Plants 2026, 15, 5. https://doi.org/10.3390/plants15010005
Fetisov I, Eizikovich O, Charles Diouf D, Romanova E, Kezimana P. Advancements in Molecular Breeding Techniques for Soybeans. Plants. 2026; 15(1):5. https://doi.org/10.3390/plants15010005
Chicago/Turabian StyleFetisov, Ivan, Olga Eizikovich, Dominique Charles Diouf, Elena Romanova, and Parfait Kezimana. 2026. "Advancements in Molecular Breeding Techniques for Soybeans" Plants 15, no. 1: 5. https://doi.org/10.3390/plants15010005
APA StyleFetisov, I., Eizikovich, O., Charles Diouf, D., Romanova, E., & Kezimana, P. (2026). Advancements in Molecular Breeding Techniques for Soybeans. Plants, 15(1), 5. https://doi.org/10.3390/plants15010005





