Melatonin Mitigates Cd-Induced Growth Repression and RNA m6A Hypermethylation by Triggering MMR-Mediated DNA Damage Response
Abstract
1. Introduction
2. Methods and Materials
2.1. Plant Materials, Growth, and Treatment Conditions
2.2. Morphological Measurement
2.3. RNA Extraction, First-Strand cDNA Synthesis, and qRT-PCR Analysis
2.4. RNA-m6A-Level Measurement
2.5. Statistical Analysis
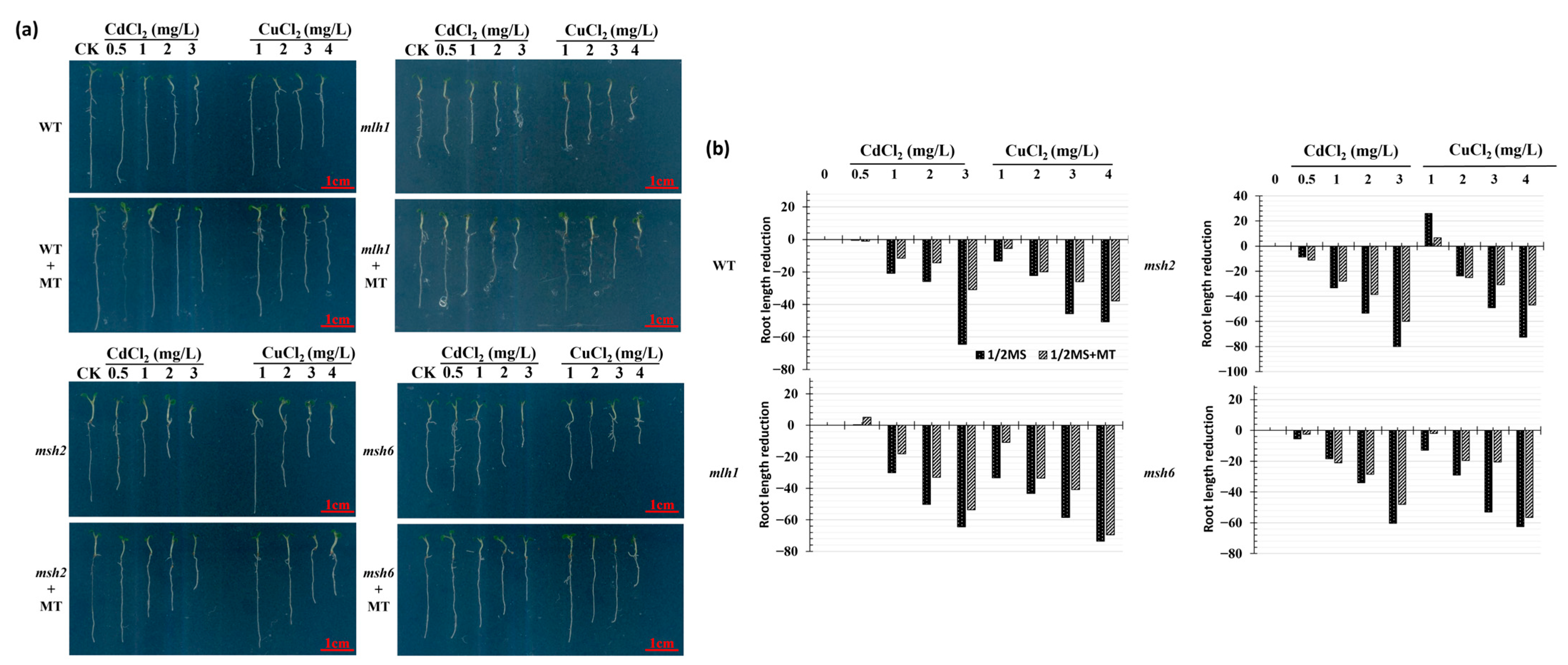
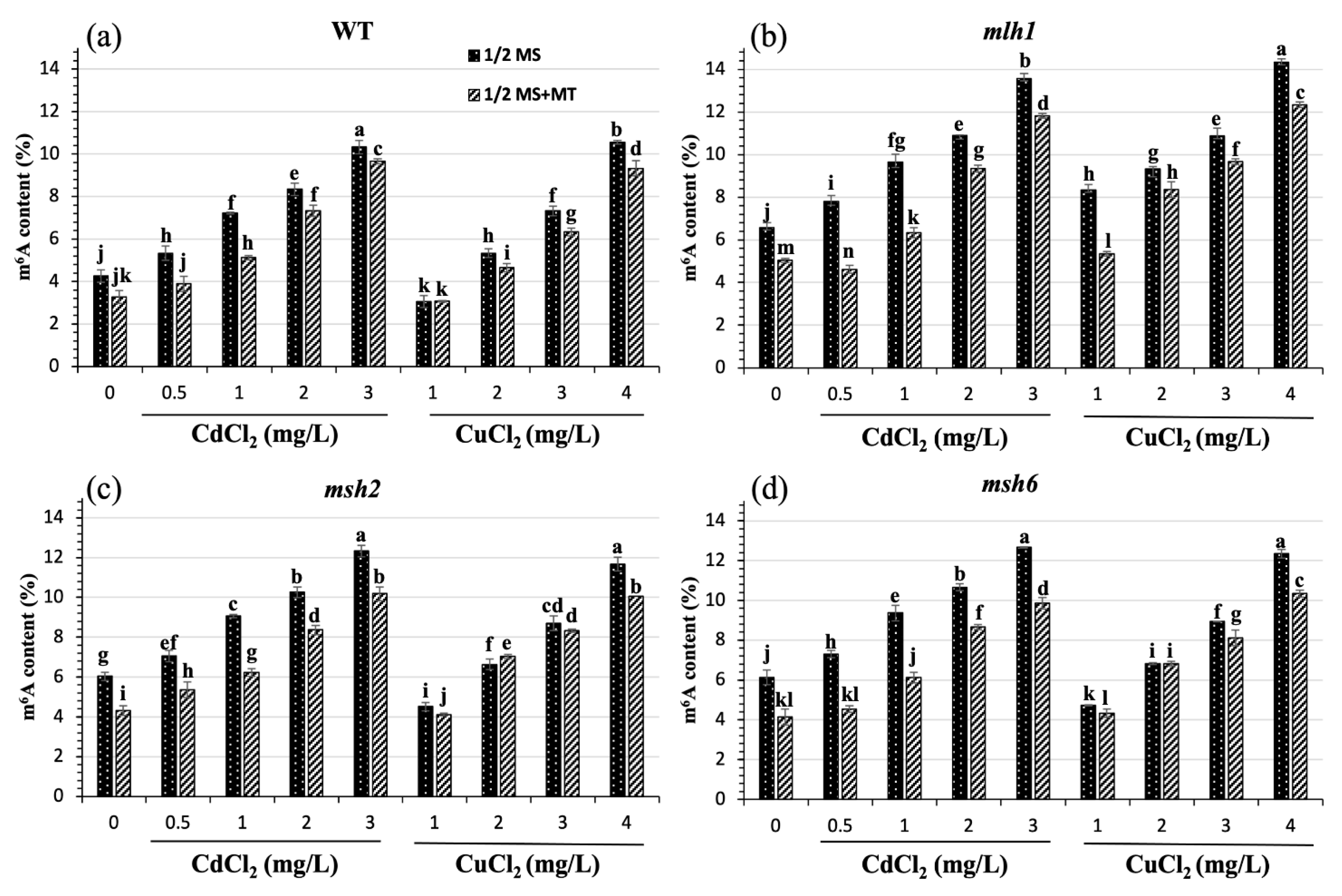
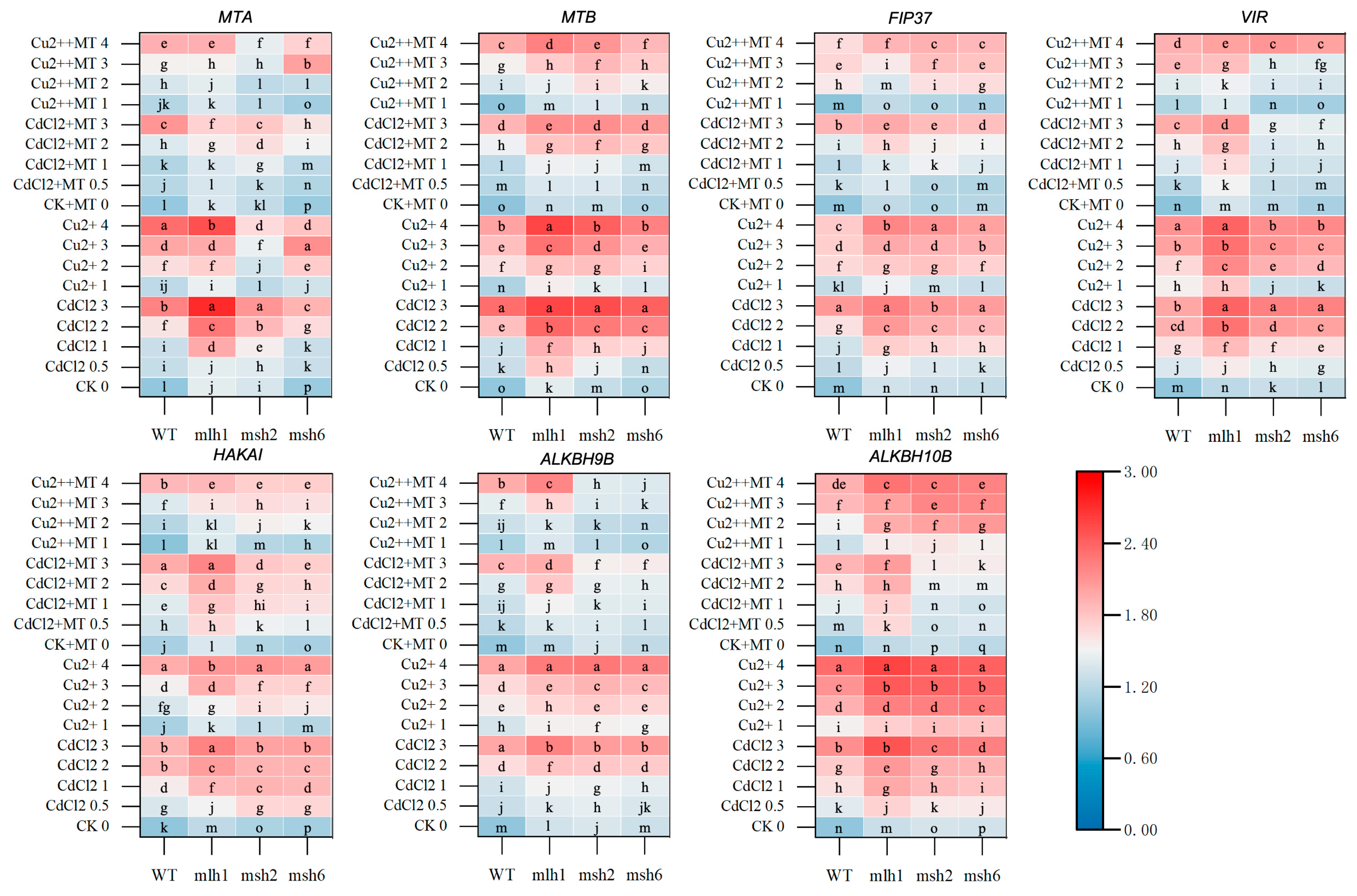
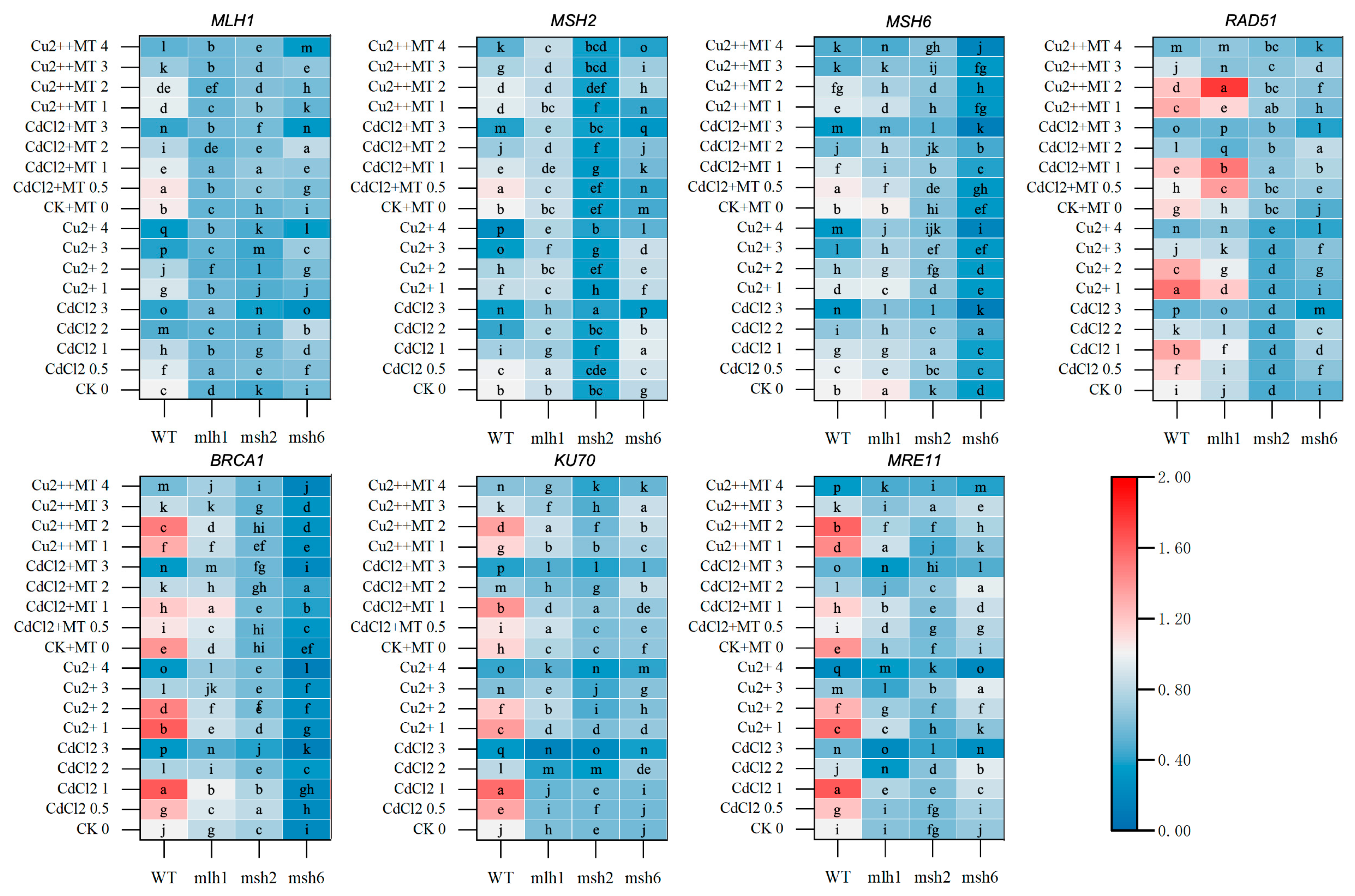
3. Results
3.1. Cd- and Cu-Induced Arabidopsis Growth Repression and Its Recovery Mediated by MT
3.2. Restored Effect of MT on Cd- and Cu-Induced m6A Hypermethylation
3.3. The m6A-Related Gene Expression Mediated by MMR Genes and MT Responsible for m6A Levels
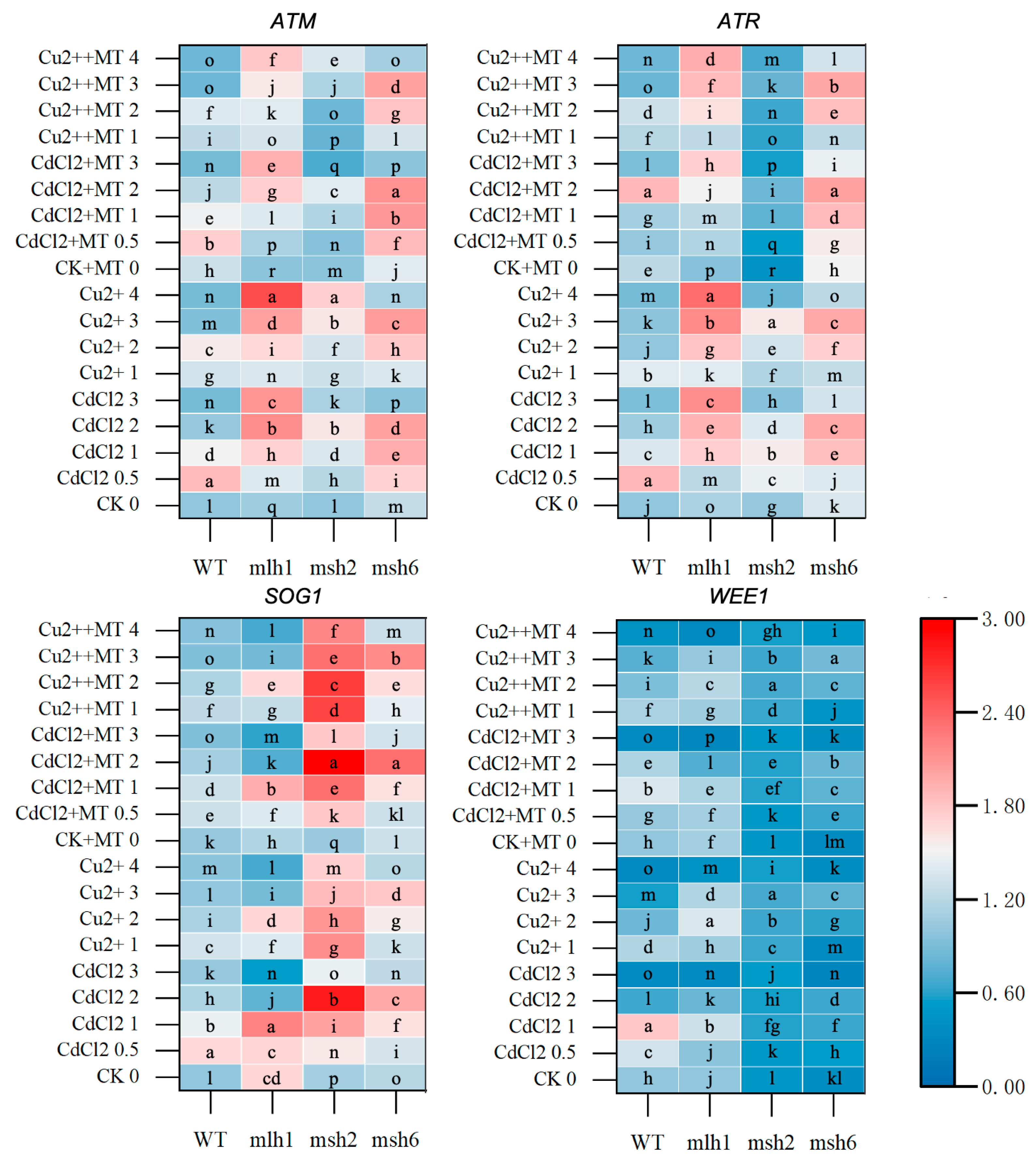
3.4. DNA-Repair-Gene Expression Mediated by MT in the Cd- and Cu-Induced DDRs
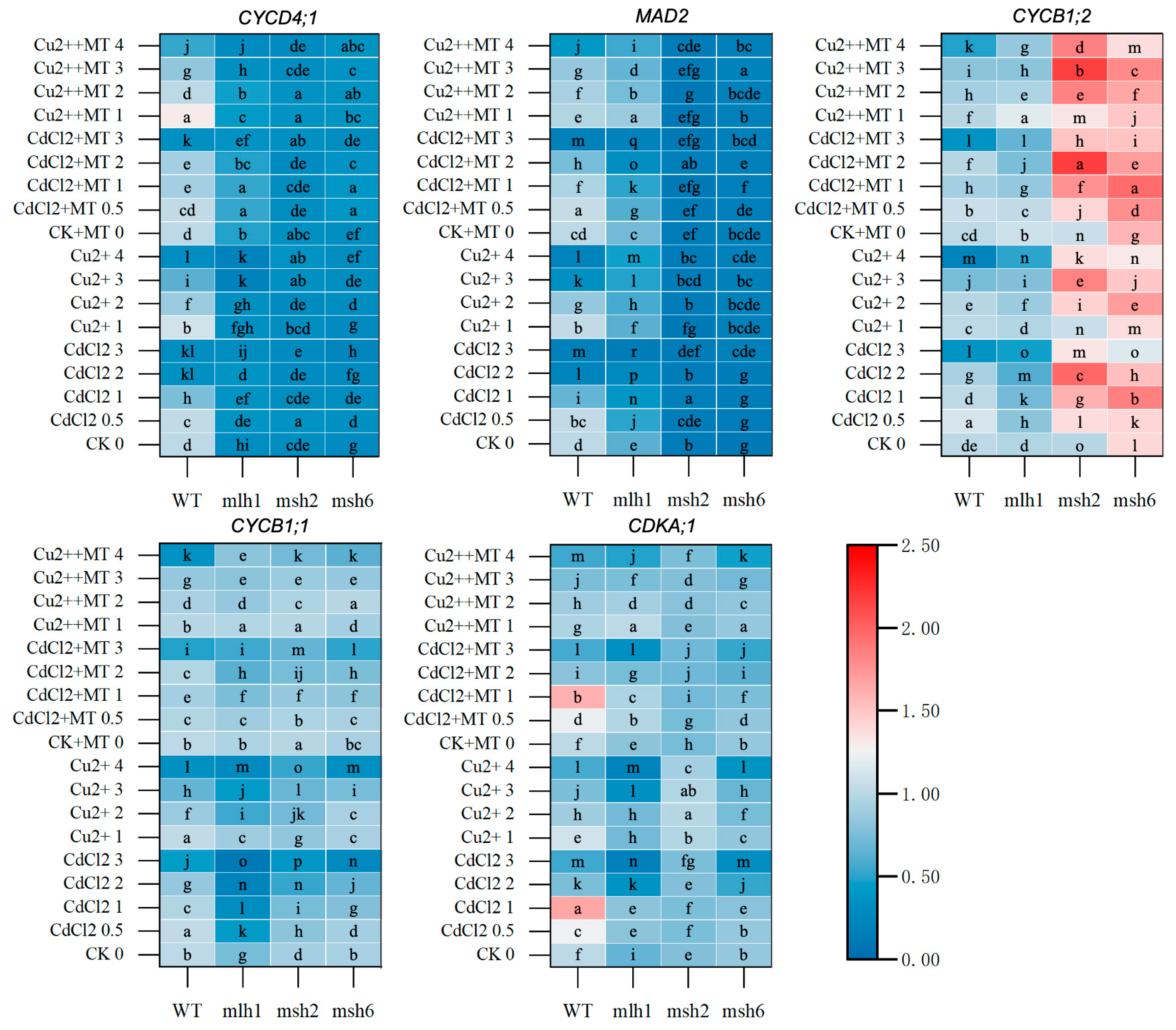
3.5. The DDR and Cell-Cycle-Related Gene Expression Mediated by MT Under Cd and Cu Stress
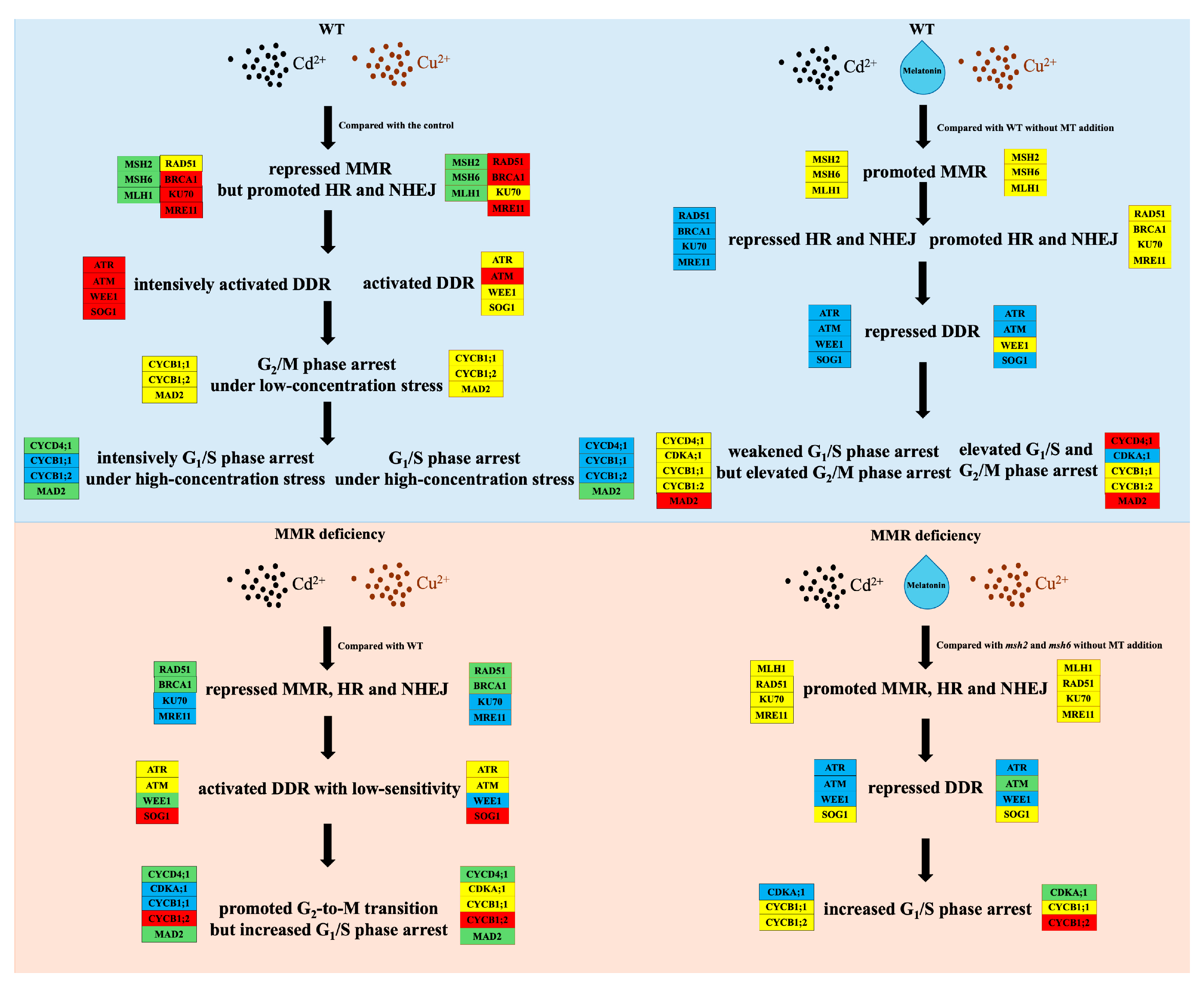
4. Discussions
4.1. MT Promotes Root Growth by Enhancing the DDR
4.2. MT Reduced m6A Hypermethylation Mediated by the DDR
4.3. MT Restored Cd-Induced Growth Repression via the MMR-Mediated DDR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Abbreviations | Full forms |
| DDR | DNA damage response |
| MT | Melatonin |
| MMR | Mismatch repair |
| HMs | Heavy metals |
| RPA | Replication protein A |
| MRN | MRE11-RAD50-NBS1 |
| ICLs | Interstrand cross-links |
| DSBs | Double-strand breaks |
| m6A | N6-methyladenosine |
| CDS | Coding sequence |
| 3′UTR | 3′ untranslated region |
| CPDs | Pyrimidine dimers |
| ROS | Reactive oxygen species |
| MS | Murashige and Skoog |
| SD | Standard deviation |
| HR | Homologous recombination |
| NHEJ | Non-homologous end joining |
References
- ATSDR. Agency for Toxic Substances and Disease Registry (ATSDR). Available online: https://www.atsdr.cdc.gov/programs/substance-priority-list.html?CDC_AAref_Val/ (accessed on 14 April 2025).
- Sljivic Husejnovic, M.; Bergant, M.; Jankovic, S.; Zizek, S.; Smajlovic, A.; Softic, A.; Music, O.; Antonijevic, B. Assessment of Pb, Cd and Hg soil contamination and its potential to cause cytotoxic and genotoxic effects in human cell lines (CaCo-2 and HaCaT). Environ. Geochem. Health 2018, 40, 1557–1572. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; He, L.; Song, J.; Cui, W.; Zhang, Y.; Jia, C.; Francis, D.; Rogers, H.J.; Sun, L.; Tai, P.; et al. Cadmium-induced genomic instability in Arabidopsis: Molecular toxicological biomarkers for early diagnosis of cadmium stress. Chemosphere 2016, 150, 258–265. [Google Scholar] [CrossRef]
- Zou, L.; Elledge, S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003, 300, 1542–1548. [Google Scholar] [CrossRef]
- Hu, Z.; Cools, T.; De Veylder, L. Mechanisms Used by Plants to Cope with DNA Damage. Annu. Rev. Plant Biol. 2016, 67, 439–462. [Google Scholar] [CrossRef]
- Aymard, F.; Bugler, B.; Schmidt, C.K.; Guillou, E.; Caron, P.; Briois, S.; Iacovoni, J.S.; Daburon, V.; Miller, K.M.; Jackson, S.P.; et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 2014, 21, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Pearlman, A.H.; Hsieh, P. DNA mismatch repair and the DNA damage response. DNA Repair. 2016, 38, 94–101. [Google Scholar] [CrossRef]
- Campregher, C.; Luciani, M.G.; Gasche, C. Activated neutrophils induce an hMSH2-dependent G2/M checkpoint arrest and replication errors at a (CA)13-repeat in colon epithelial cells. Gut 2008, 57, 780–787. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Wang, H.; Zhuang, D.; Zhu, H.; Du, Y.; Cheng, Z.; Cui, W.; Rogers, H.J.; Zhang, Q.; Jia, C. Roles of MSH2 and MSH6 in cadmium-induced G2/M checkpoint arrest in Arabidopsis roots. Chemosphere 2018, 201, 586. [Google Scholar] [CrossRef]
- Zhao, Q.; Wang, H.; Du, Y.; Rogers, H.; Wu, Z.; Jia, S.; Yao, X.; Xie, F.; Liu, W. MSH2 and MSH6 in Mismatch Repair System Account for Soybean (Glycine max (L.) Merr.) Tolerance to Cadmium Toxicity by Determining DNA Damage Response. J. Agric. Food Chem. 2020, 68, 1974–1985. [Google Scholar] [CrossRef]
- Wang, H.; Cao, Q.; Zhao, Q.; Arfan, M.; Liu, W. Mechanisms used by DNA MMR system to cope with Cadmium-induced DNA damage in plants. Chemosphere 2020, 246, 125614. [Google Scholar] [CrossRef]
- Salem, M.E.; Bodor, J.N.; Puccini, A.; Xiu, J.; Goldberg, R.M.; Grothey, A.; Korn, W.M.; Shields, A.F.; Worrilow, W.M.; Kim, E.S.; et al. Relationship between MLH1, PMS2, MSH2 and MSH6 gene-specific alterations and tumor mutational burden in 1057 microsatellite instability-high solid tumors. Int. J. Cancer 2020, 147, 2948–2956. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.-M.; Kang, Y.; Park, J.; Sung, Y.; Kim, D.; Seo, Y.; Lee, E.A.; Ra, J.S.; Amarsanaa, E.; Park, Y.-U. MSH2-MSH3 promotes DNA end resection during homologous recombination and blocks polymerase theta-mediated end-joining through interaction with SMARCAD1 and EXO1. Nucleic Acids Res. 2023, 51, 5584–5602. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Wei, J.; He, C. Where, when, and how: Context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 2019, 74, 640–650. [Google Scholar] [CrossRef]
- Luo, G.Z.; MacQueen, A.; Zheng, G.; Duan, H.; Dore, L.C.; Lu, Z.; Liu, J.; Chen, K.; Jia, G.; Bergelson, J.; et al. Unique features of the m6A methylome in Arabidopsis thaliana. Nat. Commun. 2014, 5, 5630. [Google Scholar] [CrossRef] [PubMed]
- Duan, H.C.; Wei, L.H.; Zhang, C.; Wang, Y.; Chen, L.; Lu, Z.; Chen, P.R.; He, C.; Jia, G. ALKBH10B is An RNA N6-Methyladenosine Demethylase Affecting Arabidopsis Floral Transition. Plant Cell 2017, 29, 2995–3011. [Google Scholar] [CrossRef]
- Robbens, S.; Rouze, P.; Cock, J.M.; Spring, J.; Worden, A.Z.; Van de Peer, Y. The FTO gene, implicated in human obesity, is found only in vertebrates and marine algae. J. Mol. Evol. 2008, 66, 80–84. [Google Scholar] [CrossRef]
- Martinez-Perez, M.; Aparicio, F.; Lopez-Gresa, M.P.; Belles, J.M.; Sanchez-Navarro, J.A.; Pallas, V. Arabidopsis m(6)A demethylase activity modulates viral infection of a plant virus and the m(6)A abundance in its genomic RNAs. Proc. Natl. Acad. Sci. USA 2017, 114, 10755–10760. [Google Scholar] [CrossRef]
- Xiang, Y.; Laurent, B.; Hsu, C.H.; Nachtergaele, S.; Lu, Z.; Sheng, W.; Xu, C.; Chen, H.; Ouyang, J.; Wang, S.; et al. RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature 2017, 543, 573–576. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Zhou, K.; Wu, T.; Zhao, B.S.; Sun, M.; Chen, Z.; Deng, X.; Xiao, G.; Auer, F.; et al. Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature 2019, 567, 414–419. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Su, T.; Fu, L.; Kuang, L.; Chen, D.; Zhang, G.; Shen, Q.; Wu, D. Transcriptome-wide m6A methylation profile reveals regulatory networks in roots of barley under cadmium stress. J. Hazard. Mater. 2022, 423, 127140. [Google Scholar] [CrossRef]
- Han, X.; Wang, J.; Zhang, Y.; Kong, Y.; Dong, H.; Feng, X.; Li, T.; Zhou, C.; Yu, J.; Xin, D.; et al. Changes in the m6A RNA methylome accompany the promotion of soybean root growth by rhizobia under cadmium stress. J. Hazard. Mater. 2023, 441, 129843. [Google Scholar] [CrossRef]
- Li, W.; Tan, M.; Wang, H.; Wang, Z.; Pang, Y.; Yang, R.; Zhong, S.; Pan, X.; Chen, S.; Wang, Q.; et al. METTL3-mediated m6A mRNA modification was involved in cadmium-induced liver injury. Environ. Pollut. 2023, 331, 121887. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Cao, H.; Chen, D.; Kuang, L.; Wu, D. Transcriptome-wide analysis of m6A methylation reveals genetic responses to cadmium stress at germination stage in rice. Environ. Exp. Bot. 2023, 205, 105130. [Google Scholar] [CrossRef]
- Yin, H.; Hong, H.; Yin, P.; Lu, W.; Niu, S.; Chen, X.; Xia, Y.; Jiang, P.; Huang, Z. Increased levels of N6-methyladenosine in peripheral blood RNA: A perspective diagnostic biomarker and therapeutic target for non-small cell lung cancer. Clin. Chem. Lab. Med. 2023, 61, 473–484. [Google Scholar] [CrossRef]
- Chen, H.; Zhao, X.; Yang, W.; Zhang, Q.; Hao, R.; Jiang, S.; Han, H.; Yu, Z.; Xing, S.; Feng, C.; et al. RNA N6-methyladenosine modification-based biomarkers for absorbed ionizing radiation dose estimation. Nat. Commun. 2023, 14, 6912. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Lu, Y.; Du, A. m6A-related lncRNAs as potential biomarkers and the lncRNA ELFN1-AS1/miR-182-5p/BCL-2 regulatory axis in diffuse large B-cell lymphoma. J. Cell. Mol. Med. 2024, 28, e18046. [Google Scholar] [CrossRef] [PubMed]
- Bai, Z.; Xia, Q.; Xu, W.; Wu, Z.; He, X.; Zhang, X.; Wang, Z.; Luo, M.; Sun, H.; Liu, S. N6-Methylandenosine-related lncRNAs as potential biomarkers for predicting prognosis and the immunotherapy response in pancreatic cancer. Cell. Mol. Life Sci. 2025, 82, 48. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L.; et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018, 20, 285–295. [Google Scholar] [CrossRef]
- Du, H.; Zhao, Y.; He, J.; Zhang, Y.; Xi, H.; Liu, M.; Ma, J.; Wu, L. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 2016, 7, 12626. [Google Scholar] [CrossRef]
- Xiong, Y.W.; Tan, L.L.; Zhang, J.; Zhu, H.L.; Zheng, X.M.; Chang, W.; Gao, L.; Wei, T.; Xu, D.X.; Wang, H. Combination of high-fat diet and cadmium impairs testicular spermatogenesis in an m6A-YTHDF2-dependent manner. Environ. Pollut. 2022, 313, 120112. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Yasui, M.; Sassa, A.; You, X.; Wan, J.; Cao, Y.; Xi, J.; Zhang, X.; Honma, M.; Luan, Y. FTO regulates the DNA damage response via effects on cell-cycle progression. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2023, 887, 503608. [Google Scholar] [CrossRef] [PubMed]
- Lerner, A.B.; Case, J.D.; Takahashi, Y.; Lee, T.H.; Mori, W. ISOLATION OF MELATONIN, THE PINEAL GLAND FACTOR THAT LIGHTENS MELANOCYTES1. J. Am. Chem. Soc. 1958, 80, 2587. [Google Scholar] [CrossRef]
- Maestroni, G.J. The immunoneuroendocrine role of melatonin. J. Pineal Res. 1993, 14, 1–10. [Google Scholar] [CrossRef]
- Bonilla, E.; Valero, N.; Chacín-Bonilla, L.; Medina-Leendertz, S. Melatonin and viral infections. J. Pineal Res. 2004, 36, 73–79. [Google Scholar] [CrossRef]
- Hosseinzadeh, A.; Kamrava, S.K.; Joghataei, M.T.; Darabi, R.; Shakeri-Zadeh, A.; Shahriari, M.; Reiter, R.J.; Ghaznavi, H.; Mehrzadi, S. Apoptosis signaling pathways in osteoarthritis and possible protective role of melatonin. J. Pineal Res. 2016, 61, 411–425. [Google Scholar] [CrossRef]
- Zisapel, N. New perspectives on the role of melatonin in human sleep, circadian rhythms and their regulation. Br. J. Pharmacol. 2018, 175, 3190–3199. [Google Scholar] [CrossRef]
- Guan, Q.; Wang, Z.; Cao, J.; Dong, Y.; Chen, Y. Mechanisms of Melatonin in Obesity: A Review. Int. J. Mol. Sci. 2022, 23, 218. [Google Scholar] [CrossRef]
- Hattori, A.; Migitaka, H.; Iigo, M.; Itoh, M.; Yamamoto, K.; Ohtani-Kaneko, R.; Hara, M.; Suzuki, T.; Reiter, R.J. Identification of melatonin in plants and its effects on plasma melatonin levels and binding to melatonin receptors in vertebrates. Biochem. Mol. Biol. Int. 1995, 35, 627–634. [Google Scholar]
- Dubbels, R.; Reiter, R.J.; Klenke, E.; Goebel, A.; Schnakenberg, E.; Ehlers, C.; Schiwara, H.W.; Schloot, W. Melatonin in edible plants identified by radioimmunoassay and by high performance liquid chromatography-mass spectrometry. J. Pineal Res. 1995, 18, 28–31. [Google Scholar] [CrossRef] [PubMed]
- Arnao, M.B.; Hernández-Ruiz, J. Melatonin: Plant growth regulator and/or biostimulator during stress? Trends Plant Sci. 2014, 19, 789–797. [Google Scholar] [CrossRef] [PubMed]
- Arnao, M.B.; Hernández-Ruiz, J. Melatonin: A new plant hormone and/or a plant master regulator? Trends Plant Sci. 2019, 24, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Kong, M.; Sheng, T.; Liang, J.; Ali, Q.; Gu, Q.; Wu, H.; Chen, J.; Liu, J.; Gao, X. Melatonin and Its Homologs Induce Immune Responses via Receptors trP47363-trP13076 in Nicotiana benthamiana. Front. Plant Sci. 2021, 12, 691835. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Rodríguez-Ruiz, M.; Muñoz-Vargas, M.A.; González-Gordo, S.; Reiter, R.J.; Palma, J.M. Interactions of melatonin, reactive oxygen species, and nitric oxide during fruit ripening: An update and prospective view. J. Exp. Bot. 2022, 73, 5947–5960. [Google Scholar] [CrossRef]
- Wei, J.; Li, D.X.; Zhang, J.R.; Shan, C.; Rengel, Z.; Song, Z.B.; Chen, Q. Phytomelatonin receptor PMTR1-mediated signaling regulates stomatal closure in Arabidopsis thaliana. J. Pineal Res. 2018, 65, e12500. [Google Scholar] [CrossRef]
- Chen, Q.; Arnao, M.B. Phytomelatonin: An emerging new hormone in plants. J. Exp. Bot. 2022, 73, 5773–5778. [Google Scholar] [CrossRef]
- Arnao, M.B.; Hernández-Ruiz, J. Functions of melatonin in plants: A review. J. Pineal Res. 2015, 59, 133–150. [Google Scholar] [CrossRef]
- Argueta, J.; Solís-Chagoyán, H.; Estrada-Reyes, R.; Constantino-Jonapa, L.A.; Oikawa-Sala, J.; Velázquez-Moctezuma, J.; Benítez-King, G. Further Evidence of the Melatonin Calmodulin Interaction: Effect on CaMKII Activity. Int. J. Mol. Sci. 2022, 23, 2479. [Google Scholar] [CrossRef]
- Wang, Z.; Mu, Y.; Zhang, L.; Liu, Z.; Liu, D.; Jin, Z.; Pei, Y. Hydrogen sulfide mediated the melatonin induced stoma closure by regulating the K+ channel in Arabidopsis thaliana. Environ. Exp. Bot. 2023, 205, 105125. [Google Scholar] [CrossRef]
- Zhao, G.; Zhao, Y.; Yu, X.; Kiprotich, F.; Han, H.; Guan, R.; Wang, R.; Shen, W. Nitric Oxide Is Required for Melatonin-Enhanced Tolerance against Salinity Stress in Rapeseed (Brassica napus L.) Seedlings. Int. J. Mol. Sci. 2018, 19, 1912. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Gao, H.; Lu, M.; Hao, C.; Pu, Z.; Guo, M.; Hou, D.; Chen, L.-Y.; Huang, X. Melatonin-Nitric Oxide Crosstalk and Their Roles in the Redox Network in Plants. Int. J. Mol. Sci. 2019, 20, 6200. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.S.S.; Ahmed, S.; Ikram, A.u.; Hannan, F.; Yasin, M.U.; Wang, J.; Zhao, B.; Islam, F.; Chen, J. Phytomelatonin: A key regulator of redox and phytohormones signaling against biotic/abiotic stresses. Redox Biol. 2023, 64, 102805. [Google Scholar]
- Altaf, M.A.; Hao, Y.; Shu, H.; Mumtaz, M.A.; Cheng, S.; Alyemeni, M.N.; Ahmad, P.; Wang, Z. Melatonin enhanced the heavy metal-stress tolerance of pepper by mitigating the oxidative damage and reducing the heavy metal accumulation. J. Hazard. Mater. 2023, 454, 131468. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, F.; Tang, M.; Wang, Y.; Dong, J.; Ying, J.; Chen, Y.; Hu, B.; Li, C.; Liu, L. Melatonin confers cadmium tolerance by modulating critical heavy metal chelators and transporters in radish plants. J. Pineal Res. 2020, 69, e12659. [Google Scholar] [CrossRef]
- Shehzadi, K.; Maqsood, M.F.; Kanwal, R.; Shahbaz, M.; Naqve, M.; Zulfiqar, U.; Jamil, M.; Khalid, N.; Ali, M.F.; Soufan, W. Enhancing cadmium stress resilience in chickpea (Cicer arietinum L.) via exogenous melatonin application. Int. J. Phytoremediat. 2025, 27, 794–809. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Raychaudhuri, S.S. Melatonin improves the lead tolerance in Plantago ovata by modulating ROS homeostasis, phytohormone status and expression of stress-responsive genes. Plant Cell Rep. 2025, 44, 39. [Google Scholar] [CrossRef]
- Gao, S.; Ma, W.; Lyu, X.; Cao, X.; Yao, Y. Melatonin may increase disease resistance and flavonoid biosynthesis through effects on DNA methylation and gene expression in grape berries. BMC Plant Biol. 2020, 20, 231. [Google Scholar] [CrossRef]
- Shan, S.; Wang, Z.; Pu, H.; Duan, W.; Song, H.; Li, J.; Zhang, Z.; Xu, X. DNA methylation mediated by melatonin was involved in ethylene signal transmission and ripening of tomato fruit. Sci. Hortic. 2022, 291, 110566. [Google Scholar] [CrossRef]
- Tang, M.; Xu, L.; Wang, Y.; Dong, J.; Zhang, X.; Wang, K.; Ying, J.; Li, C.; Liu, L. Melatonin-induced DNA demethylation of metal transporters and antioxidant genes alleviates lead stress in radish plants. Hortic. Res. 2021, 8, 124. [Google Scholar] [CrossRef]
- Davoodvandi, A.; Nikfar, B.; Reiter, R.J.; Asemi, Z. Melatonin and cancer suppression: Insights into its effects on DNA methylation. Cell. Mol. Biol. Lett. 2022, 27, 73. [Google Scholar] [CrossRef]
- Sharma, R.; Ottenhof, T.; Rzeczkowska, P.A.; Niles, L.P. Epigenetic targets for melatonin: Induction of histone H3 hyperacetylation and gene expression in C17. 2 neural stem cells. J. Pineal Res. 2008, 45, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Bargunam, S.; Roy, R.; Shetty, D.; H, A.S.; V S, S.; Babu, V.S. Melatonin-governed growth and metabolome divergence: Circadian and stress responses in key plant species. Plant Physiol. Biochem. 2025, 221, 109635. [Google Scholar] [CrossRef]
- Yang, L.; Liu, X.; Song, L.; Su, G.; Di, A.; Bai, C.; Wei, Z.; Li, G. Melatonin restores the pluripotency of long-term-cultured embryonic stem cells through melatonin receptor-dependent m6A RNA regulation. J. Pineal Res. 2020, 69, e12669. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Li, T.; Yang, M.; Su, L.; Zhu, Z.; Zhao, S.; Zeng, W.; Zheng, Y. Melatonin Attenuates Chromium (VI)-Induced Spermatogonial Stem Cell/Progenitor Mitophagy by Restoration of METTL3-Mediated RNA N6-Methyladenosine Modification. Front. Cell Dev. Biol. 2021, 9, 684398. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Qin, K.; Zhao, L.; Ash, R.D.; McDonough, W.F.; Zhao, R.Y. ATM-mediated transcriptional elevation of prion in response to copper-induced oxidative stress. J. Biol. Chem. 2009, 284, 4582–4593. [Google Scholar] [CrossRef]
- Baumgarten, S.; Bryant, J.M.; Sinha, A.; Reyser, T.; Preiser, P.R.; Dedon, P.C.; Scherf, A. Transcriptome-wide dynamics of extensive m6A mRNA methylation during Plasmodium falciparum blood-stage development. Nat. Microbiol. 2019, 4, 2246–2259. [Google Scholar] [CrossRef]
- Yang, S.; Lian, G. ROS and diseases: Role in metabolism and energy supply. Mol. Cell. Biochem. 2020, 467, 1–12. [Google Scholar] [CrossRef]
- Sharma, B.; Govindan, G.; Li, Y.; Sunkar, R.; Gregory, B.D. RNA N6-Methyladenosine Affects Copper-Induced Oxidative Stress Response in Arabidopsis thaliana. Non-Coding RNA 2024, 10, 8. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.-X.; Wang, Z.-H.; Sun, Y.-D.; Wang, L.-L.; Li, M.; Liu, Y.-T.; Zhang, H.-M.; Jing, P.-W.; Shi, Q.-F.; Yu, Y.-H. Molecular mechanism of plant response to copper stress: A review. Environ. Exp. Bot. 2023, 218, 105590. [Google Scholar] [CrossRef]
- Jin, J.; Ma, M.; Shi, S.; Wang, J.; Xiao, P.; Yu, H.F.; Zhang, C.; Guo, Q.; Yu, Z.; Lou, Z.; et al. Copper enhances genotoxic drug resistance via ATOX1 activated DNA damage repair. Cancer Lett. 2022, 536, 215651. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Jing, H.K.; Zhang, Y.; Chen, S.Y.; Wang, H.Y.; Cao, Y.; Zhang, Z.; Lu, Y.H.; Zheng, Q.S.; Shen, R.F. Melatonin reduces cadmium accumulation via mediating the nitric oxide accumulation and increasing the cell wall fixation capacity of cadmium in rice. J. Hazard. Mater. 2023, 445, 130529. [Google Scholar] [CrossRef] [PubMed]
- Xing, Q.; Hasan, M.K.; Li, Z.; Yang, T.; Jin, W.; Qi, Z.; Yang, P.; Wang, G.; Ahammed, G.J.; Zhou, J. Melatonin-induced plant adaptation to cadmium stress involves enhanced phytochelatin synthesis and nutrient homeostasis in Solanum lycopersicum L. J. Hazard. Mater. 2023, 456, 131670. [Google Scholar] [CrossRef]
- Li, G.-Z.; Wang, Y.-Y.; Liu, J.; Liu, H.-T.; Liu, H.-P.; Kang, G.-Z. Exogenous melatonin mitigates cadmium toxicity through ascorbic acid and glutathione pathway in wheat. Ecotoxicol. Environ. Saf. 2022, 237, 113533. [Google Scholar] [CrossRef]
- Qiu, C.-W.; Richmond, M.; Ma, Y.; Zhang, S.; Liu, W.; Feng, X.; Ahmed, I.M.; Wu, F. Melatonin enhances cadmium tolerance in rice via long non-coding RNA-mediated modulation of cell wall and photosynthesis. J. Hazard. Mater. 2024, 465, 133251. [Google Scholar] [CrossRef]
- Sharma, P.; Thakur, N.; Mann, N.A.; Umar, A. Melatonin as plant growth regulator in sustainable agriculture. Sci. Hortic. 2024, 323, 112421. [Google Scholar] [CrossRef]
- Rakosy-Tican, E.; Lörincz-Besenyei, E.; Molnár, I.; Thieme, R.; Hartung, F.; Sprink, T.; Antonova, O.; Famelaer, I.; Angenon, G.; Aurori, A. New Phenotypes in Potato Co-Induced by Mismatch Repair Deficiency and Somatic Hybridization. Front. Plant Sci. 2019, 10, 3. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, Z.; Wang, H.; Wang, X.; Ludlow, R.A.; Liu, Z.; Zhang, M.; Cao, Q.; Liu, W.; Zhao, Q. Melatonin Mitigates Cd-Induced Growth Repression and RNA m6A Hypermethylation by Triggering MMR-Mediated DNA Damage Response. Plants 2025, 14, 1398. https://doi.org/10.3390/plants14091398
Tang Z, Wang H, Wang X, Ludlow RA, Liu Z, Zhang M, Cao Q, Liu W, Zhao Q. Melatonin Mitigates Cd-Induced Growth Repression and RNA m6A Hypermethylation by Triggering MMR-Mediated DNA Damage Response. Plants. 2025; 14(9):1398. https://doi.org/10.3390/plants14091398
Chicago/Turabian StyleTang, Zihan, Hetong Wang, Xianpeng Wang, Richard A. Ludlow, Zhouli Liu, Min Zhang, Qijiang Cao, Wan Liu, and Qiang Zhao. 2025. "Melatonin Mitigates Cd-Induced Growth Repression and RNA m6A Hypermethylation by Triggering MMR-Mediated DNA Damage Response" Plants 14, no. 9: 1398. https://doi.org/10.3390/plants14091398
APA StyleTang, Z., Wang, H., Wang, X., Ludlow, R. A., Liu, Z., Zhang, M., Cao, Q., Liu, W., & Zhao, Q. (2025). Melatonin Mitigates Cd-Induced Growth Repression and RNA m6A Hypermethylation by Triggering MMR-Mediated DNA Damage Response. Plants, 14(9), 1398. https://doi.org/10.3390/plants14091398






