Assessing Functional Conservation Amongst FT- and TFL1-like Genes in Globe Artichoke
Abstract
1. Introduction
2. Results
2.1. Identification of Globe Artichoke PEBP Homologs
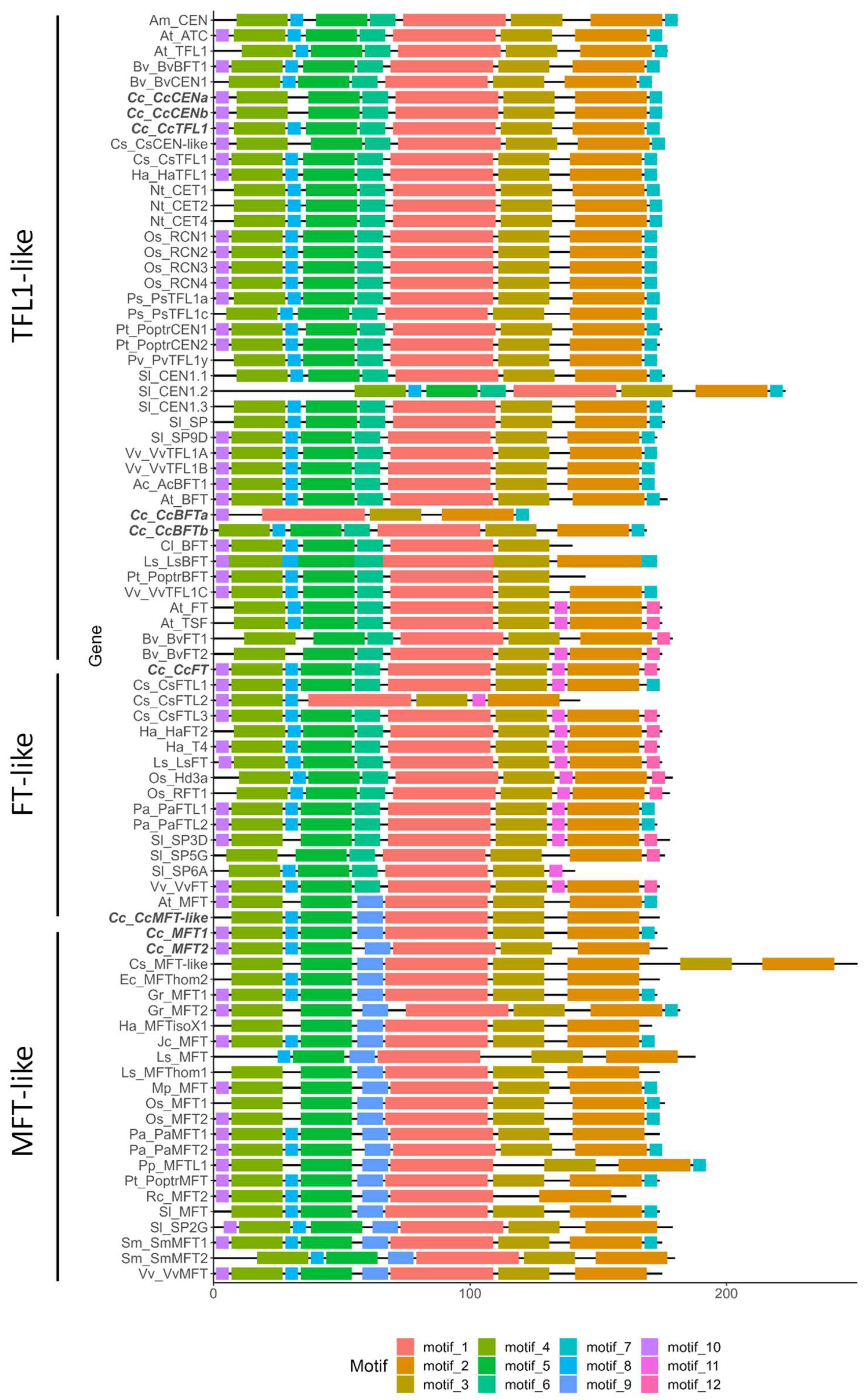
2.2. Protein Structure and Conserved Motifs in Globe Artichoke PEBP Family
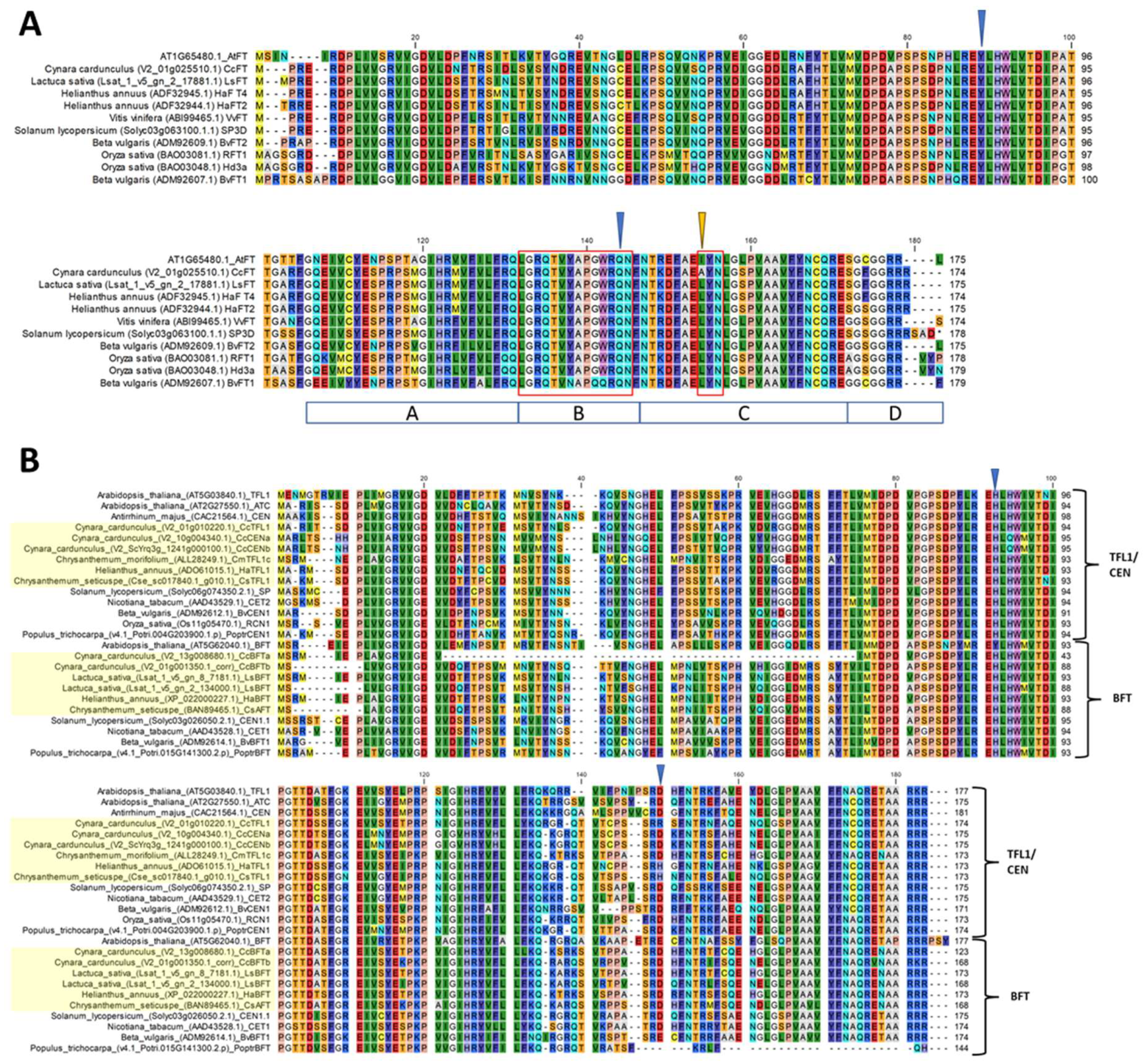
2.3. Sequence Variation in PEBP Homologs
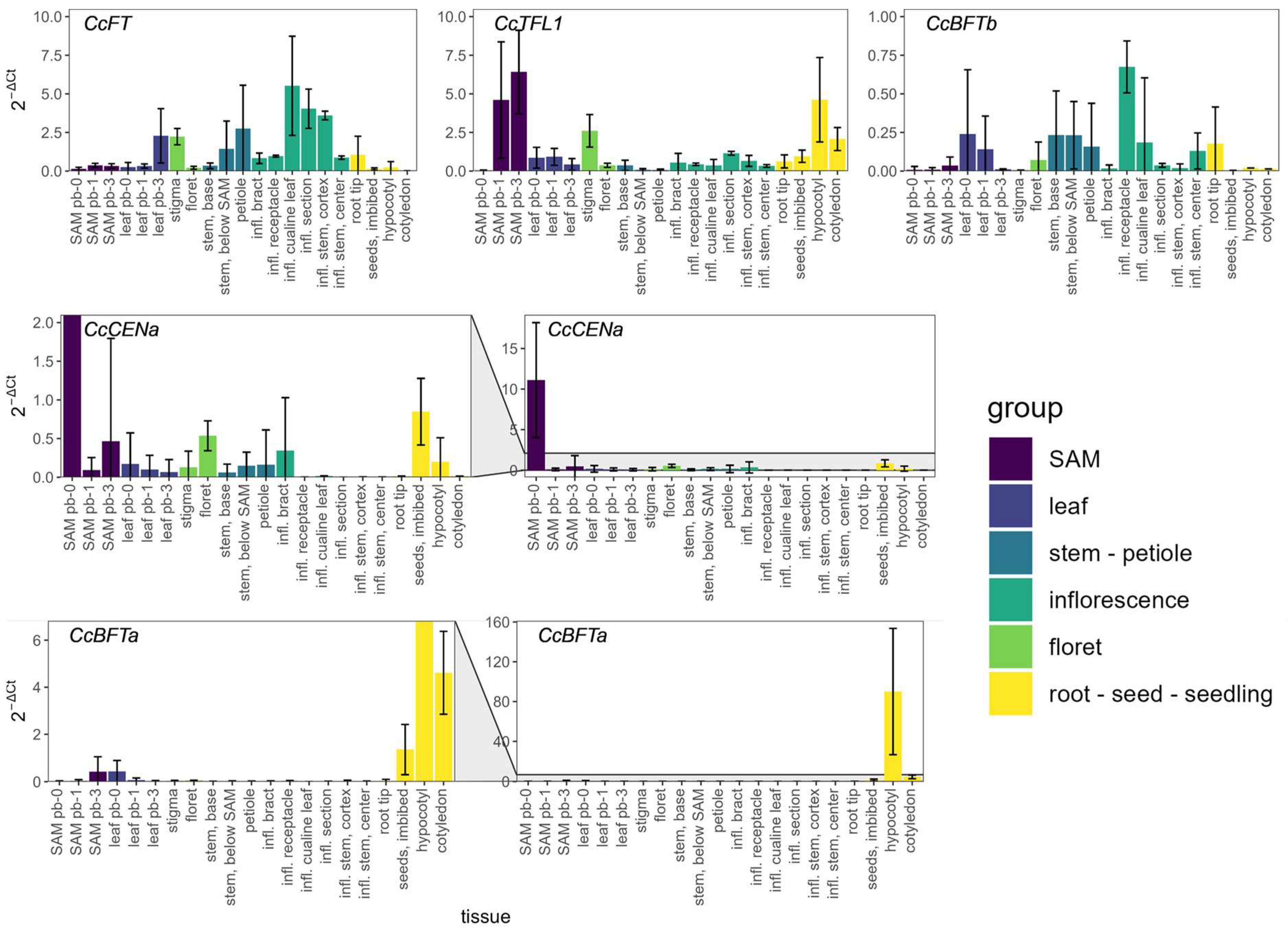
2.4. PEBP Gene Expression Profiles During Globe Artichoke Development
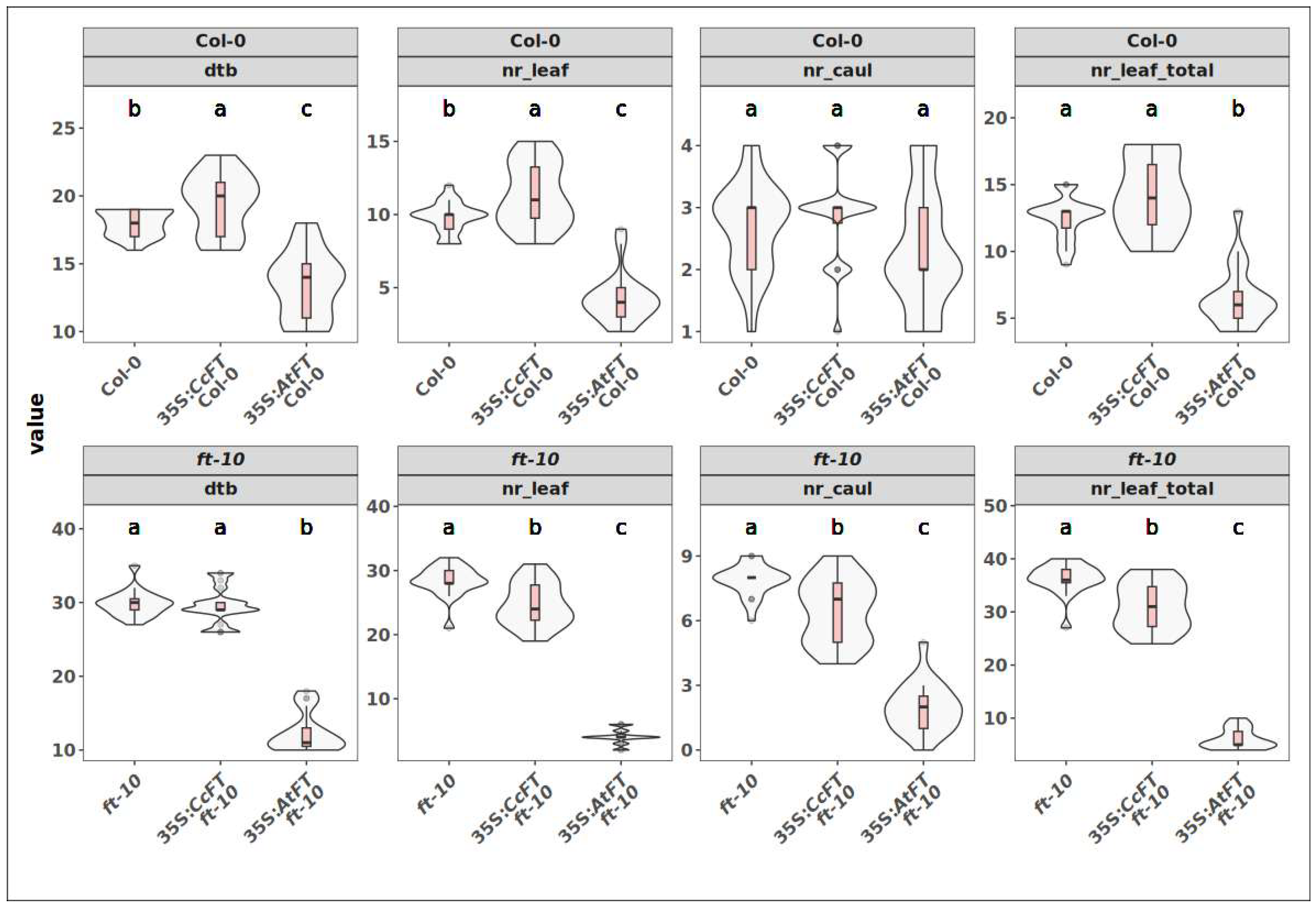
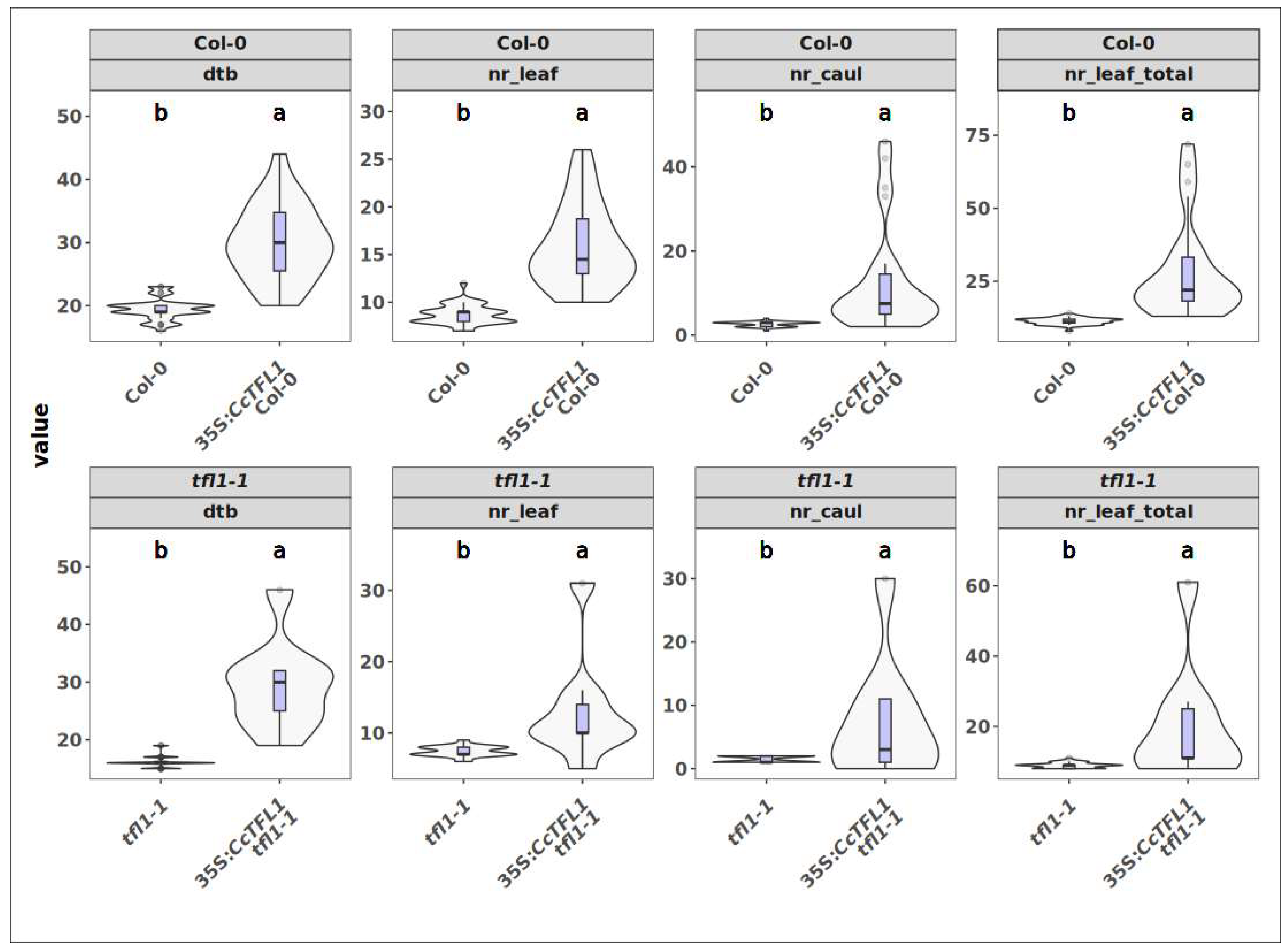
2.5. Assessment of Functional Conservation of CcFT and CcTFL1
3. Discussion
3.1. MFT-Likes from the Campanulid Subclade May Be Subjected to Higher Evolutionary Speed
3.2. Functional Conservation of CcFT Is Not Fully Certain
3.3. Homologs from the TFL1-Like Clade Are Conserved Between Asteraceae
4. Materials and Methods
4.1. Phylogenetic Study and Gene Characterization
4.2. Expression of PEBP Orthologues
4.3. Production of Entry Clones
4.4. Production of Expression Vectors
4.5. Transformation, Growing, and Observation of Transgenics
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| FT | FLOWERING LOCUS T |
| TFL1 | TERMINAL FLOWER 1 |
| dtb | Days to bolting |
| das | Days after sowing |
References
- Scaglione, D.; Reyes-Chin-Wo, S.; Acquadro, A.; Froenicke, L.; Portis, E.; Beitel, C.; Tirone, M.; Mauro, R.; Lo Monaco, A.; Mauromicale, G.; et al. The genome sequence of the outbreeding globe artichokes constructed de novo incorporating a phase-aware low-pass sequencing strategy of F1 progeny. Sci. Rep. 2016, 6, 19427. [Google Scholar] [CrossRef]
- FAOSTAT. FAOSTAT. Food and Agriculture Organization of the United Nations on 2022-01-28.2022. Available online: https://www.fao.org/faostat/ (accessed on 9 March 2022).
- Pignone, D.; Sonnante, G. Wild artichokes of south Italy: Did the story begin here? Genet. Resour. Crop. Evol. 2004, 51, 577–580. [Google Scholar] [CrossRef]
- Sonnante, G.; Pignone, D.; Hammer, K. The domestication of artichoke and cardoon: From Roman times to the genomic age. Ann. Bot. 2007, 100, 1095–1100. [Google Scholar] [CrossRef]
- Elia, A.; Miccolis, V. Relationships among 104 artichoke (Cynara scolymus L.) accessions using cluster analysis. Adv. Hortic. Sci. 1996, 10, 158–162. [Google Scholar]
- Basnizki, J.; Zohary, D. Breeding of Seed-Planted Artichoke. In Plant Breeding Reviews; Wiley: Hoboken, NJ, USA, 1994; pp. 253–269. [Google Scholar] [CrossRef]
- Macua, J. New Horizons for Artichoke Cultivation. In Proceedings of the 4th IS on Artichoke, Cardoon and Their Wild Relatives, Lorca, Spain, 28–31 March 2006; pp. 39–48. [Google Scholar] [CrossRef]
- Basnitzki, Y.; Zohary, D. A seed-planted cultivar of globe artichoke. HortScience 1987, 22, 678–679. [Google Scholar] [CrossRef]
- Macua, J.; Lahoz, I.; Bozal, J. Evolution in Seed-Propagated Artichoke Growing in Cold Zones of Spain. Acta Hortic. 2012, 331–336. [Google Scholar] [CrossRef]
- Calabrese, N.; De Palma, E.; Bianco, V. Yield and quality of seed propagated artichoke hybrid cultivars grown for four years. Acta Hortic. 2000, 135–142. [Google Scholar] [CrossRef]
- Calabrese, N.; De Palma, E.; Bianco, V. Yield and Quality of New Commercial Seed Grown Artichoke Hybrids. Acta Hortic. 2004, 77–82. [Google Scholar] [CrossRef]
- Maroto, J. Effects of gibberellic acid (GA3) applications on globe artichoke production. VI Int. Symp. Artichoke Cardoon Their Wild Relat. 2006, 137–142. [Google Scholar] [CrossRef]
- Berentsen, R.; Benlloch, R.; Visser, P.; Madueño, F.; Balanzà, V. A reduced vernalization requirement is a key component of the early-bolting trait in globe artichoke (Cynara cardunculus var. scolymus). iScience 2024, 27, 110829. [Google Scholar] [CrossRef]
- Soria, C.B.; Giner, A.; Aguilar, J.; Nájera, I. Productive and agronomic behaviour of new cultivars and lines of seed propagated artichoke. X Int. Symp. Artichoke Cardoon Their Wild Relat. 2019, 145–148. [Google Scholar] [CrossRef]
- Huijser, P.; Schmid, M. The control of developmental phase transitions in plants. Development 2011, 138, 4117–4129. [Google Scholar] [CrossRef] [PubMed]
- Amasino, R.M.; Michaels, S.D. The timing of flowering. Plant Physiol. 2010, 154, 516–520. [Google Scholar] [CrossRef] [PubMed]
- Andrés, F.; Coupland, G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012, 13, 627–639. [Google Scholar] [CrossRef]
- Karlgren, A.; Gyllenstrand, N.; Källman, T.; Sundström, J.F.; Moore, D.; Lascoux, M.; Lagercrantz, U. Evolution of the PEBP Gene Family in Plants: Functional Diversification in Seed Plant Evolution. Plant Physiol. 2011, 156, 1967–1977. [Google Scholar] [CrossRef]
- Colleoni, P.E.; van Es, S.W.; Winkelmolen, T.; Immink, R.G.H.; van Esse, G.W. Flowering time genes branching out. J. Exp. Bot. 2024, 75, 4195–4209. [Google Scholar] [CrossRef]
- Schoentgen, F.; Jollès, P. From structure to function: Possible biological roles of a new widespread protein family binding hydrophobic ligands and displaying a nucleotide binding site. FEBS Lett. 1995, 369, 22–26. [Google Scholar] [CrossRef]
- Banfield, M.J.; Barker, J.J.; Perry, A.C.; Brady, R.L. Function from structure? The crystal structure of human phosphatidylethanolamine-binding protein suggests a role in membrane signal transduction. Structure 1998, 6, 1245–1254. [Google Scholar] [CrossRef]
- Bradley, D.; Carpenter, R.; Copsey, L.; Vincent, C.; Rothstein, S.; Coen, E. Control of inflorescence architecture in Antirrhinum. Nature 1996, 379, 791–797. [Google Scholar] [CrossRef]
- Bradley, D.; Ratcliffe, O.; Vincent, C.; Carpenter, R.; Coen, E. Inflorescence Commitment and Architecture in Arabidopsis. Science 1997, 275, 80–83. [Google Scholar] [CrossRef]
- Kardailsky, I.; Shukla, V.K.; Ahn, J.H.; Dagenais, N.; Christensen, S.K.; Nguyen, J.T.; Chory, J.; Harrison, M.J.; Weigel, D. Activation Tagging of the Floral Inducer FT. Science 1999, 286, 1962–1965. [Google Scholar] [CrossRef] [PubMed]
- Wickland, D.P.; Hanzawa, Y. The FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family: Functional evolution and molecular mechanisms. Mol. Plant 2015, 8, 983–997. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, C.; Li, Y.; Yu, H. Mobile TERMINAL FLOWER1 determines seed size in Arabidopsis. Nat. Plants 2020, 6, 1146–1157. [Google Scholar] [CrossRef] [PubMed]
- Pin, P.A.; Nilsson, O. The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ. 2012, 35, 1742–1755. [Google Scholar] [CrossRef]
- González-Suárez, P.; Walker, C.H.; Bennett, T. FLOWERING LOCUS T mediates photo-thermal timing of inflorescence meristem arrest in Arabidopsis thaliana. Plant Physiol. 2023, 192, 2276–2289. [Google Scholar] [CrossRef]
- Bigas, J.N.; Fiers, M.; van der Wal, F.; Willems, L.A.J.; Willemsen, V.; Nijveen, H.; Angenent, G.C.; Immink, R.G.H. The PEBP genes FLOWERING LOCUS T and TERMINAL FLOWER 1 modulate seed dormancy and size. J. Exp. Bot. 2025, 76, 1049–1067. [Google Scholar] [CrossRef]
- Hedman, H.; Källman, T.; Lagercrantz, U. Early evolution of the MFT-like gene family in plants. Plant Mol. Biol. 2009, 70, 359–369. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Yang, K.Z.; Wei, X.X.; Wang, X.Q. Revisiting the phosphatidylethanolamine-binding protein (PEBP) gene family reveals cryptic FLOWERING LOCUS T gene homologs in gymnosperms and sheds new light on functional evolution. New Phytol. 2016, 212, 730–744. [Google Scholar] [CrossRef]
- Yoo, S.Y.; Kardailsky, I.; Lee, J.S.; Weigel, D.; Ahn, J.H. Acceleration of flowering by overexpression of MFT (Mother of FT and TFL1). Mol. Cells 2004, 17, 95–101. [Google Scholar] [CrossRef]
- Xi, W.; Liu, C.; Hou, X.; Yu, H. MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 2010, 22, 1733–1748. [Google Scholar] [CrossRef]
- Li, Q.; Fan, C.; Zhang, X.; Wang, X.; Wu, F.; Hu, R.; Fu, Y. Identification of a soybean MOTHER OF FT AND TFL1 homolog involved in regulation of seed germination. PLoS ONE 2014, 9, e99642. [Google Scholar] [CrossRef] [PubMed]
- Chardon, F.; Damerval, C. Phylogenomic analysis of the PEBP gene family in cereals. J. Mol. Evol. 2005, 61, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Gyllenstrand, N.; Clapham, D.; Kallman, T.; Lagercrantz, U. A Norway spruce FLOWERING LOCUS T homolog is implicated in control of growth rhythm in conifers. Plant Physiol. 2007, 144, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Pelayo, C.; Ambrose, B.A.; Vasco, A.; Alzate, J.F.; Pabón-Mora, N. Tracking Ancestral Flowering Integrators: Evolution and Expression of PEBP Genes in Lycophytes and Ferns. Int. J. Plant Sci. 2022, 183, 251–267. [Google Scholar] [CrossRef]
- Bennett, T.; Dixon, L.E. Asymmetric expansions of FT and TFL1 lineages characterize differential evolution of the EuPEBP family in the major angiosperm lineages. BMC Biol. 2021, 19, 181. [Google Scholar] [CrossRef]
- Huang, N.C.; Jane, W.N.; Chen, J.; Yu, T.S. Arabidopsis thaliana CENTRORADIALIS homologue (ATC) acts systemically to inhibit floral initiation in Arabidopsis. Plant J. 2012, 72, 175–184. [Google Scholar] [CrossRef]
- Ryu, J.Y.; Lee, H.-J.; Seo, P.J.; Jung, J.-H.; Ahn, J.H.; Park, C.-M. The Arabidopsis floral repressor BFT delays flowering by competing with FT for FD binding under high salinity. Mol. Plant 2014, 7, 377–387. [Google Scholar] [CrossRef]
- Goretti, D.; Silvestre, M.; Collani, S.; Langenecker, T.; Méndez, C.; Madueño, F.; Schmid, M. TERMINAL FLOWER1 functions as a mobile transcriptional cofactor in the shoot apical meristem. Plant Physiol. 2020, 182, 2081–2095. [Google Scholar] [CrossRef]
- Zhu, Y.; Klasfeld, S.; Jeong, C.W.; Jin, R.; Goto, K.; Yamaguchi, N.; Wagner, D. TERMINAL FLOWER 1-FD complex target genes and competition with FLOWERING LOCUS T. Nat. Commun. 2020, 11, 5118. [Google Scholar] [CrossRef]
- Zhu, Y.; Klasfeld, S.; Wagner, D. Molecular regulation of plant developmental transitions and plant architecture via PEPB family proteins: An update on mechanism of action. J. Exp. Bot. 2021, 72, 2301–2311. [Google Scholar] [CrossRef]
- Ratcliffe, O.J.; Bradley, D.J.; Coen, E.S. Separation of shoot and floral identity in Arabidopsis. Development 1999, 126, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Shannon, S.; Meeks-Wagner, D.R. A mutation in the Arabidopsis TFL1 gene affects inflorescence meristem development. Plant Cell 1991, 3, 877. [Google Scholar] [CrossRef] [PubMed]
- Conti, L.; Bradley, D. TERMINAL FLOWER1 Is a Mobile Signal Controlling Arabidopsis Architecture. Plant Cell 2007, 19, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, Y.; Narumi, T.; Oda, A.; Nakano, Y.; Sumitomo, K.; Fukai, S.; Hisamatsu, T. The gated induction system of a systemic floral inhibitor, antiflorigen, determines obligate short-day flowering in chrysanthemums. Proc. Natl. Acad. Sci. USA 2013, 110, 17137–17142. [Google Scholar] [CrossRef]
- Gao, Y.; Gao, Y.; Wu, Z.; Bu, X.; Fan, M.; Zhang, Q. Characterization of TEMINAL FLOWER1 homologs CmTFL1c gene from Chrysanthemum morifolium. Plant Mol. Biol. 2019, 99, 587–601. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, Y.; Juntheikki, I.; Mouhu, K.; Broholm, S.K.; Rijpkema, A.S.; Kins, L.; Lan, T.; Albert, V.A.; Teeri, T.H.; et al. Dissecting functions of SEPALLATA-like MADS box genes in patterning of the pseudanthial inflorescence of Gerbera hybrida. New Phytol. 2017, 216, 939–954. [Google Scholar] [CrossRef]
- Yoo, S.J.; Chung, K.S.; Jung, S.H.; Yoo, S.Y.; Lee, J.S.; Ahn, J.H. BROTHER OF FT AND TFL1(BFT) has TFL1-like activity and functions redundantly with TFL1 in inflorescence meristem development in Arabidopsis. Plant J. 2010, 63, 241–253. [Google Scholar] [CrossRef]
- Ryu, J.Y.; Park, C.-M.; Seo, P.J. The floral repressor BROTHER OF FT AND TFL1 (BFT) modulates flowering initiation under high salinity in Arabidopsis. Mol. Cells 2011, 32, 295–304. [Google Scholar] [CrossRef]
- Voogd, C.; Brian, L.A.; Wang, T.; Allan, A.C.; Varkonyi-Gasic, E. Three FT and multiple CEN and BFT genes regulate maturity, flowering, and vegetative phenology in kiwifruit. J. Exp. Bot. 2017, 68, 1539–1553. [Google Scholar] [CrossRef]
- Herath, D.; Voogd, C.; Mayo-Smith, M.; Yang, B.; Allan, A.C.; Putterill, J.; Varkonyi-Gasic, E. CRISPR-Cas9-mediated mutagenesis of kiwifruit BFT genes results in an evergrowing but not early flowering phenotype. Plant Biotechnol. J. 2022, 20, 2064–2076. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Kaya, H.; Goto, K.; Iwabuchi, M.; Araki, T. A pair of related genes with antagonistic roles in mediating flowering signals. Science 1999, 286, 1960–1962. [Google Scholar] [CrossRef] [PubMed]
- Turck, F.; Fornara, F.; Coupland, G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 2008, 59, 573–594. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.C. Constans activates suppressor of overexpression of constans 1 through Flowering Locus T to promote flowering in Arabidopsis. Plant Physiol. 2005, 139, 770–778. [Google Scholar] [CrossRef] [PubMed]
- Hanzawa, Y.; Money, T.; Bradley, D. A single amino acid converts a repressor to an activator of flowering. Proc. Natl. Acad. Sci. USA 2005, 102, 7748–7753. [Google Scholar] [CrossRef]
- Ahn, J.H.; Miller, D.; Winter, V.J.; Banfield, M.J.; Lee, J.H.; Yoo, S.Y.; Henz, S.R.; Brady, R.L.; Weigel, D. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 2006, 25, 605–614. [Google Scholar] [CrossRef]
- Ho, W.W.H.; Weigel, D. Structural features determining flower-promoting activity of Arabidopsis FLOWERING LOCUS T. Plant Cell 2014, 26, 552–564. [Google Scholar] [CrossRef]
- Pin, P.A.; Benlloch, R.; Bonnet, D.; Wremerth-Weich, E.; Kraft, T.; Gielen, J.J.L.; Nilsson, O. An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 2010, 330, 1397–1400. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Kobayashi, Y.; Goto, K.; Abe, M.; Araki, T. TWIN SISTER OF FT (TSF) acts as a floral pathway inte-grator redundantly with FT. Plant Cell Physiol. 2005, 46, 1175–1189. [Google Scholar] [CrossRef]
- Blackman, B.K.; Rasmussen, D.A.; Strasburg, J.L.; Raduski, A.R.; Burke, J.M.; Knapp, S.J.; Michaels, S.D.; Rieseberg, L.H. Contributions of Flowering Time Genes to Sunflower Domestication and Improvement. Genetics 2011, 187, 271–287. [Google Scholar] [CrossRef]
- Fukuda, M.; Matsuo, S.; Kikuchi, K.; Kawazu, Y.; Fujiyama, R.; Honda, I. Isolation and functional characterization of the FLOWERING LOCUS T homolog, the LsFT gene, in lettuce. J. Plant Physiol. 2011, 168, 1602–1607. [Google Scholar] [CrossRef]
- Acquadro, A.; Portis, E.; Valentino, D.; Barchi, L.; Lanteri, S. “Mind the gap”: Hi-C technology boosts contiguity of the globe artichoke genome in low-recombination regions. G3 Genes Genomes Genet. 2020, 10, 3557–3564. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, Z.; Bao, Z.; Li, H.; Lyu, Y.; Zan, Y.; Wu, Y.; Cheng, L.; Fang, Y.; Wu, K.; et al. Graph pangenome captures missing heritability and empowers tomato breeding. Nature 2022, 606, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Carmona, M.J.; Calonje, M.; Martínez-Zapater, J.M. The FT/TFL1 gene family in grapevine. Plant Mol. Biol. 2007, 63, 637–650. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, H.; Sumitomo, K.; Hisamatsu, T.; Nagano, S.; Shirasawa, K.; Higuchi, Y.; Kusaba, M.; Koshioka, M.; Nakano, Y.; Yagi, M.; et al. De novo whole-genome assembly in Chrysanthemum seticuspe, a model species of Chrysanthemums, and its application to genetic and gene discovery analysis. DNA Res. 2019, 26, 195–203. [Google Scholar] [CrossRef]
- Blackman, B.K.; Strasburg, J.L.; Raduski, A.R.; Michaels, S.D.; Rieseberg, L.H. The role of recently derived FT paralogs in sunflower domestication. Curr. Biol. 2010, 20, 629–635. [Google Scholar] [CrossRef]
- Oda, A.; Narumi, T.; Li, T.; Kando, T.; Higuchi, Y.; Sumitomo, K.; Fukai, S.; Hisamatsu, T. CsFTL3, a chrysanthemum FLOWERING LOCUS T-like gene, is a key regulator of photoperiodic flowering in chrysanthemums. J. Exp. Bot. 2012, 63, 1461–1477. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, D.; Ou, C.; Hao, W.; Zhao, Z.; Zhuang, F. Genome-wide identification and characterization profile of phosphatidy ethanolamine-binding protein family genes in carrot. Front. Genet. 2022, 13, 1047890. [Google Scholar] [CrossRef]
- Acquadro, A.; Barchi, L.; Portis, E.; Mangino, G.; Valentino, D.; Mauromicale, G.; Lanteri, S. Genome reconstruction in Cynara cardunculus taxa gains access to chromosome-scale DNA variation. Sci. Rep. 2017, 7, 5617. [Google Scholar] [CrossRef]
- Fukuda, M.; Yanai, Y.; Nakano, Y.; Sasaki, H.; Uragami, A.; Okada, K. Isolation and gene expression analysis of flowering-related genes in lettuce (Lactuca sativa L.). Hortic. J. 2017, 86, 340–348. [Google Scholar] [CrossRef]
- Chen, Z.; Han, Y.; Ning, K.; Ding, Y.; Zhao, W.; Yan, S.; Luo, C.; Jiang, X.; Ge, D.; Liu, R.; et al. Inflorescence development and the role of LsFT in regulating bolting in lettuce (Lactuca sativa L.). Front. Plant Sci. 2018, 8, 2248. [Google Scholar] [CrossRef]
- Nakano, Y.; Higuchi, Y.; Sumitomo, K.; Hisamatsu, T. Flowering retardation by high temperature in chrysanthemums: Involvement of FLOWERING LOCUS T-like 3 gene repression. J. Exp. Bot. 2013, 64, 909–920. [Google Scholar] [CrossRef] [PubMed]
- Hecht, V.; Laurie, R.E.; Vander Schoor, J.K.; Ridge, S.; Knowles, C.L.; Liew, L.C.; Sussmilch, F.C.; Murfet, I.C.; Macknight, R.C.; Weller, J.L. The Pea GIGAS Gene Is a FLOWERING LOCUS THomolog Necessary for Graft-Transmissible Specification of Flowering but Not for Responsiveness to Photoperiod. Plant Cell 2011, 23, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Muszynski, M.G.; Danilevskaya, O.N. The FT-Like ZCN8 Gene Functions as a Floral Activator and Is Involved in Photoperiod Sensitivity in Maize. Plant Cell 2011, 23, 942–960. [Google Scholar] [CrossRef]
- Kikuchi, R.; Kawahigashi, H.; Ando, T.; Tonooka, T.; Handa, H. Molecular and functional characterization of PEBP genes in barley reveal the diversification of their roles in flowering. Plant Physiol. 2009, 149, 1341–1353. [Google Scholar] [CrossRef]
- Soyk, S.; Müller, N.A.; Park, S.J.; Schmalenbach, I.; Jiang, K.; Hayama, R.; Zhang, L.; Van Eck, J.; Jiménez-Gómez, J.M.; Lippman, Z.B. Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat. Genet. 2017, 49, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Kalia, D.; Jose-Santhi, J.; Kumar, R.; Singh, R.K. Analysis of PEBP genes in saffron identifies a FLOWERING LOCUS T homologue involved in flowering regulation. J. Plant Growth Regul. 2023, 42, 2486–2505. [Google Scholar] [CrossRef]
- Fu, J.; Wang, L.; Wang, Y.; Yang, L.; Yang, Y.; Dai, S. Photoperiodic control of FT-like gene ClFT initiates flowering in Chrysanthemum lavandulifolium. Plant Physiol. Biochem. 2014, 74, 230–238. [Google Scholar] [CrossRef]
- Berardini, T.Z.; Reiser, L.; Li, D.; Mezheritsky, Y.; Muller, R.; Strait, E.; Huala, E. The Arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 2015, 53, 474–485. [Google Scholar] [CrossRef]
- University of Torino. Globe Artichoke Genome Database. 2012. Available online: http://www.artichokegenome.unito.it/ (accessed on 15 March 2022).
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Bodenhofer, U.; Bonatesta, E.; Horejš-Kainrath, C.; Hochreiter, S. msa: An R package for multiple sequence alignment. Bioinformatics 2015, 31, 3997–3999. [Google Scholar] [CrossRef] [PubMed]
- Schliep, K.P. phangorn: Phylogenetic analysis in R. Bioinformatics 2011, 27, 592–593. [Google Scholar] [CrossRef] [PubMed]
- Whelan, S.; Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef]
- Gascuel, O. BIONJ: An improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 1997, 14, 685–695. [Google Scholar] [CrossRef]
- Posada, D.; Crandall, K.A. MODELTEST: Testing the model of DNA substitution. Bioinformatics 1998, 14, 817–818. [Google Scholar] [CrossRef]
- Yu, G. Using ggtree to Visualize Data on Tree-Like Structures. Curr. Protoc. Bioinform. 2020, 69, e96. [Google Scholar] [CrossRef]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, Stanford, CA, USA, 14–17 August 1994; pp. 28–36. [Google Scholar]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Li, X.; Ma, L.; Mei, X.; Liu, Y.; Huang, H. ggmotif: An R Package for the extraction and visualization of motifs from MEME software. PLoS ONE 2022, 17, e0276979. [Google Scholar] [CrossRef]
- Ochogavía, A.C.; Novello, M.A.; Picardi, L.A.; Nestares, G.M. Identification of suitable reference genes by quan-titative real-time PCR for gene expression normalization in sunflower. Plant Omics 2017, 10, 210–218. [Google Scholar] [CrossRef]
- Bevis, B.J.; Glick, B.S. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nat. Biotechnol. 2002, 20, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Brukhin, V.; Clapham, D.; Elfstrand, M.; von Arnold, S. Basta tolerance as a selectable and screening marker for transgenic plants of Norway spruce. Plant Cell Rep. 2000, 19, 899–903. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Mislata, A.; Fernández-Nohales, P.; Doménech, M.J.; Hanzawa, Y.; Bradley, D.; Madueño, F. Separate elements of the TERMINAL FLOWER 1 cis-regulatory region integrate pathways to control flowering time and shoot meristem identity. Development 2016, 143, 3315–3327. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Length, R. Emmeans: Estimated Marginal Means, aka Least-Squares Means. R package version 1.11.0-003. 2025. Available online: https://github.com/rvlenth/emmeans (accessed on 14 April 2025).



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berentsen, R.; Domenech, M.J.; Visser, P.; Madueño, F.; Balanzà, V.; Benlloch, R. Assessing Functional Conservation Amongst FT- and TFL1-like Genes in Globe Artichoke. Plants 2025, 14, 1364. https://doi.org/10.3390/plants14091364
Berentsen R, Domenech MJ, Visser P, Madueño F, Balanzà V, Benlloch R. Assessing Functional Conservation Amongst FT- and TFL1-like Genes in Globe Artichoke. Plants. 2025; 14(9):1364. https://doi.org/10.3390/plants14091364
Chicago/Turabian StyleBerentsen, Rick, María José Domenech, Peter Visser, Francisco Madueño, Vicente Balanzà, and Reyes Benlloch. 2025. "Assessing Functional Conservation Amongst FT- and TFL1-like Genes in Globe Artichoke" Plants 14, no. 9: 1364. https://doi.org/10.3390/plants14091364
APA StyleBerentsen, R., Domenech, M. J., Visser, P., Madueño, F., Balanzà, V., & Benlloch, R. (2025). Assessing Functional Conservation Amongst FT- and TFL1-like Genes in Globe Artichoke. Plants, 14(9), 1364. https://doi.org/10.3390/plants14091364







