Small-Sample Authenticity Identification and Variety Classification of Anoectochilus roxburghii (Wall.) Lindl. Using Hyperspectral Imaging and Machine Learning
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Hyperspectral Image System
2.3. Data Preprocessing
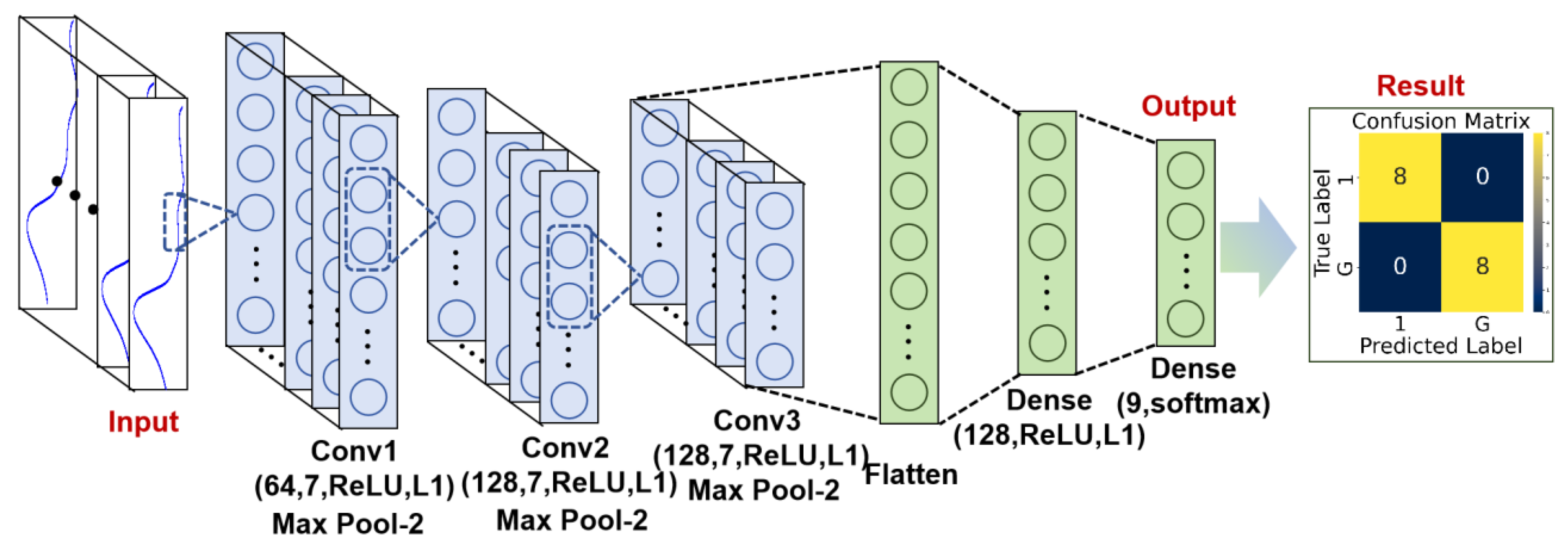
2.4. Spectral Data Processing
3. Results and Discussion
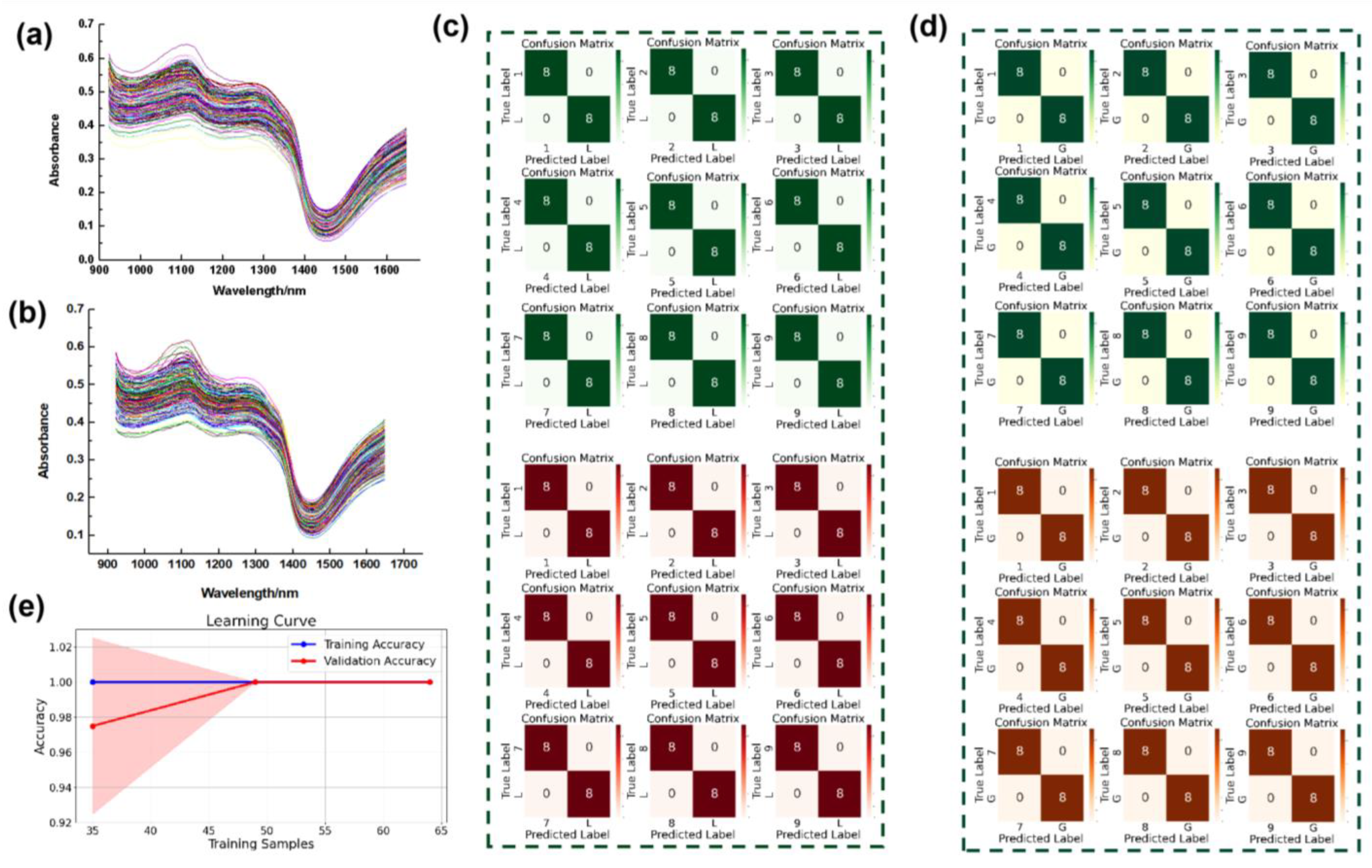
3.1. Authenticity Identification
3.2. Traditional Machine Learning Models for Goldthread Classification
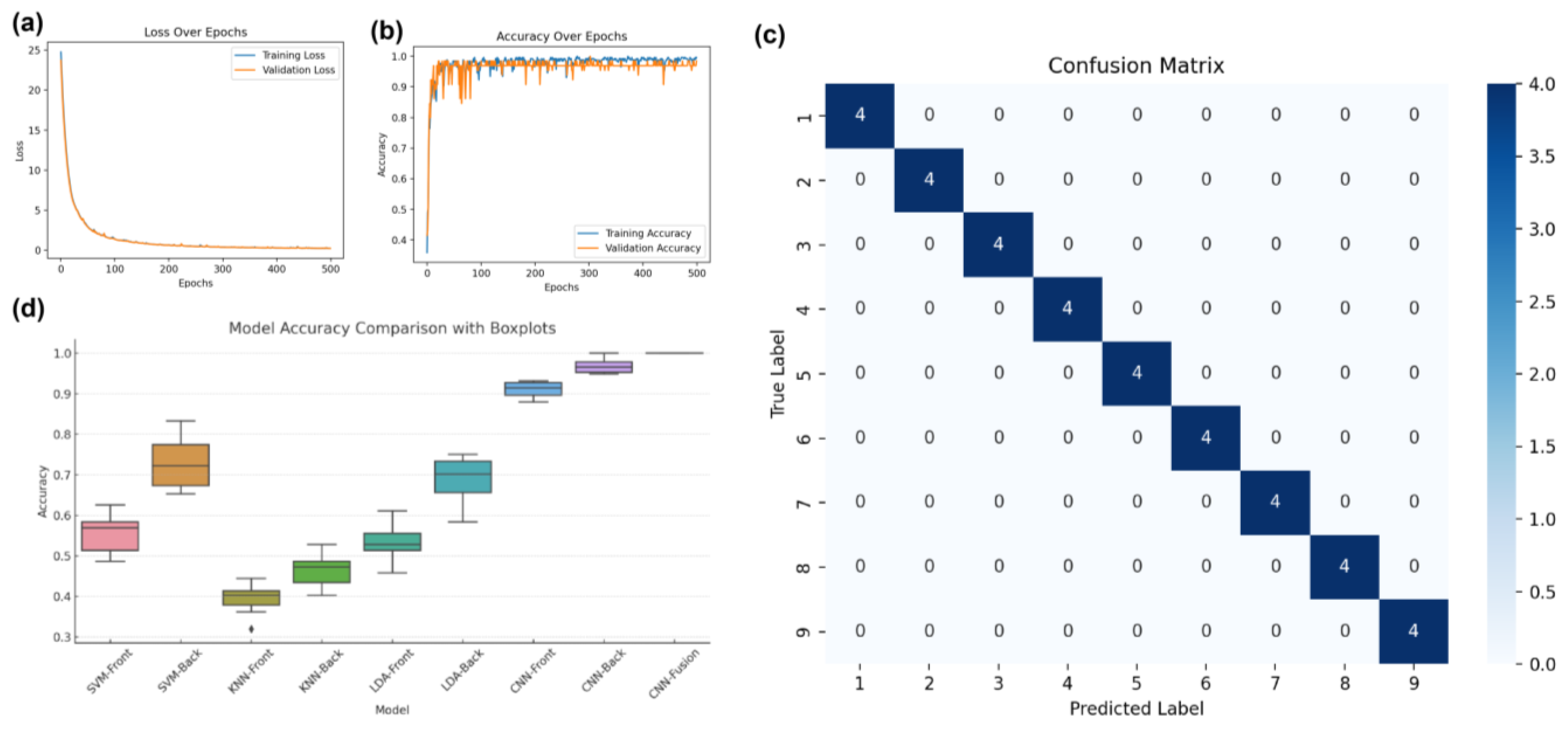
3.3. Multi-View Spectral Fusion Model
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, C.Q.; Chen, X.Y.; Liu, Y.H.; Dai, D.J. First report of fusarium oxysporum associated with stem rot on seedlings of Jinxianlian (Anoectochilus roxburghii) in China. Plant Dis. 2022, 106, 1991. [Google Scholar] [CrossRef]
- Chen, X.Y.; Zhang, C.Q.; Zhou, X.J.; Zhu, L.Y.; He, X.C. First report of gray mold on Jinxianlian (Anoectochilus roxburghii) caused by Botrytis cinerea in China. Plant Dis. 2020, 104, 1861. [Google Scholar] [CrossRef]
- Xing, B.; Wan, S.; Su, L.; Riaz, M.W.; Li, L.; Ju, Y.; Shao, Q. Two polyamines-responsive WRKY transcription factors from Anoectochilus roxburghii play opposite functions on flower development. Plant Sci. 2023, 327, 111566. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, X.; Yan, X.; Xu, Z.; Shao, Q.; Wu, X.; Wang, H. Induction, proliferation, regeneration and kinsenoside and flavonoid content analysis of the Anoectochilus roxburghii (Wall.) Lindl protocorm-like body. Plants 2022, 11, 2465. [Google Scholar] [CrossRef]
- Jin, Q.R.; Mao, J.W.; Zhu, F. The effects of Anoectochilus roxburghii polysaccharides on the innate immunity and disease resistance of Procambarus clarkii. Aquaculture 2022, 555, 738210. [Google Scholar] [CrossRef]
- Han, T.; Xu, E.; Yao, L.; Zheng, B.; Younis, A.; Shao, Q. Regulation of flowering time using temperature, photoperiod and spermidine treatments in Anoectochilus roxburghii. Physiol. Mol. Biol. Plants 2020, 26, 247–260. [Google Scholar] [CrossRef]
- Klein-Junior, L.C.; de Souza, M.R.; Viaene, J.; Bresolin, T.M.B.; de Gasper, A.L.; Henriques, A.T.; Heyden, Y.V. Quality Control of Herbal Medicines: From Traditional Techniques to State-of-the-art Approaches. Planta. Med. 2021, 87, 964–988. [Google Scholar] [CrossRef]
- Song, C.; Liu, Y.; Song, A.; Dong, G.; Zhao, H.; Sun, W.; Chen, S. The Chrysanthemum nankingense genome provides insights into the evolution and diversification of chrysanthemum flowers and medicinal traits. Mol. Plant 2018, 11, 1482–1491. [Google Scholar] [CrossRef]
- He, S.; Wang, D.; Zhang, Y.; Yang, S.; Li, X.; Wei, D.; Qin, J. Chemical components and biological activities of the essential oil from traditional medicinal food, Euryale ferox Salisb., seeds. J. Essent. Oil Bear. Plants. 2019, 22, 73–81. [Google Scholar] [CrossRef]
- Li, Q.; Zhu, T.; Zhang, R.; Bu, Q.; Yin, J.; Zhang, L.; Chen, W. Molecular cloning and functional analysis of hyoscyamine 6β-hydroxylase (H6H) in the poisonous and medicinal plant Datura innoxia mill. Plant Physiol. Biochem. 2020, 153, 11–19. [Google Scholar] [CrossRef]
- Tong, Y.; Xue, J.; Li, Q.; Zhang, L. A generalist regulator: MYB transcription factors regulate the biosynthesis of active compounds in medicinal plants. J. Exp. Bot. 2024, 75, 4729–4744. [Google Scholar] [CrossRef] [PubMed]
- Kiani, S.; van Ruth, S.M.; Minaei, S. Hyperspectral imaging, a non-destructive technique in medicinal and aromatic plant products industry: Current status and potential future applications. Comput. Electron. Agric. 2018, 152, 9–18. [Google Scholar] [CrossRef]
- Li, S.; Wang, Z.; Shao, Q.; Fang, H.; Zhu, J.; Wu, X.; Zheng, B. Rapid detection of adulteration in Anoectochilus roxburghii by near-infrared spectroscopy coupled with chemometric methods. J. Food Sci. Technol. 2018, 55, 3518–3525. [Google Scholar] [CrossRef]
- Chai, Q.; Zeng, J.; Lin, D.; Li, X.; Huang, J.; Wang, W. Improved 1D convolutional neural network adapted to near-infraredspectroscopy for rapid discrimination of Anoectochilus roxburghii and its counterfeits. J. Pharm. Biomed. Anal. 2021, 199, 114035. [Google Scholar] [CrossRef]
- Li, Y.; Cai, B. Study on Classification of Anoectochilus roxburghii Strains by Hand-Held Near Infrared Spectrometer. LNEE 2022, 805, 319–326. [Google Scholar]
- Naeem, S.; Ali, A.; Chesneau, C.; Tahir, M.H.; Jamal, F.; Sherwani, R.A.K.; Ul Hassan, M. The Classification of Medicinal Plant Leaves Based on Multispectral and Texture Feature Using Machine Learning Approach. Agronomy 2021, 11, 263. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, X.; Chen, C.; Zhou, L.; Zhao, Y.; Chen, J.; Du, H. Coupling the PROSAIL Model and Machine Learning Approach for Canopy Parameter Estimation of Moso Bamboo Forests from UAV Hyperspectral Data. Forests 2024, 15, 946. [Google Scholar] [CrossRef]
- García-Vera, Y.E.; Polochè-Arango, A.; Mendivelso-Fajardo, C.A.; Gutiérrez-Bernal, F.J. Hyperspectral image analysis and machine learning techniques for crop disease detection and identification: A review. Sustainability 2024, 16, 6064. [Google Scholar] [CrossRef]
- Dai, Y.; Gao, X.; Liu, Z. Accuracy Improvement of Mn Element in Aluminum Alloy by the Combination of LASSO-LSSVM and Laser-Induced Breakdown Spectroscopy. Spectrosc. Spectral Anal. 2024, 44, 977–982. [Google Scholar]
- Hu, G.; Liu, Y.; Chu, X.; Liu, Z. Fourier ptychographic layer-based imaging of hazy environments. Results Phys. 2024, 56, 107216. [Google Scholar] [CrossRef]
- Ma, Q.; Liu, Z.; Sun, T.; Gao, X.; Dai, Y. Small-sample stacking model for qualitative analysis of aluminum alloys based on femtosecond laser-induced breakdown spectroscopy. Opt. Express. 2023, 31, 27633–27653. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Liu, Z.; Zhang, T.; Zhao, S.; Gao, X.; Sun, T.; Dai, Y. Multielement simultaneous quantitative analysis of trace elements in stainless steel via full spectrum laser-induced breakdown spectroscopy. Talanta 2024, 272, 125745. [Google Scholar] [CrossRef]
- Liu, Z.; Ma, Q.; Zhang, T.; Zhao, S.; Gao, X.; Sun, T.; Dai, Y. Quantitative modeling and uncertainty estimation for small-sample LIBS using Gaussian negative log-likelihood and monte carlo dropout methods. Opt. Laser Technol. 2025, 181, 111720. [Google Scholar] [CrossRef]
- Dai, Y.; Ma, Q.; Zhang, T.; Zhao, S.; Zhou, L.; Gao, X.; Liu, Z. Classification of aluminum alloy using laser-induced breakdown spectroscopy combined with discriminative restricted Boltzmann machine. Chemom. Intell. Lab. Syst. 2025, 258, 105342. [Google Scholar] [CrossRef]
- Song, Y.; Cao, S.; Chu, X.; Zhou, Y.; Xu, Y.; Sun, T.; Zhou, G.; Liu, X. Non-destructive detection of moisture and fatty acid content in rice using hyperspectral imaging and chemometrics. J. Food Compos. Anal. 2023, 121, 105397. [Google Scholar] [CrossRef]
- Bischl, B.; Binder, M.; Lang, M.; Pielok, T.; Richter, J.; Coors, S.; Lindauer, M. Hyperparameter optimization: Foundations, algorithms, best practices, and open challenges. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2023, 13, e1484. [Google Scholar] [CrossRef]
- Gong, Q.; Yu, J.; Guo, Z.; Fu, K.; Xu, Y.; Zou, H.; Han, Z. Accumulation mechanism of metabolite markers identified by machine learning between Qingyuan and Xiushui counties in Polygonatum cyrtonema Hua. BMC Plant Biol. 2024, 24, 173. [Google Scholar] [CrossRef] [PubMed]
- Nie, F.; Hao, Z.; Wang, R. Multi-Class Support Vector Machine with Maximizing Minimum Margin. In Proceedings of the AAAI’24/IAAI’24/EAAI’24, Vancouver, BC, Canada, 20–27 February 2024; pp. 14466–14473. [Google Scholar]
- Salcedo-Sanz, S.; Rojo-Álvarez, L.; Martínez-Ramón, M.; Camps-Valls, G. Support vector machines in engineering: An overview. Data Min. Knowl. Disc. 2014, 4, 234–267. [Google Scholar] [CrossRef]
- Zhu, Q.; Zhang, P.; Wang, Z.; Ye, X. A New Loss Function for CNN Classifier Based on Predefined Evenly-Distributed Class Centroids. IEEE Access 2020, 8, 10888–10895. [Google Scholar] [CrossRef]

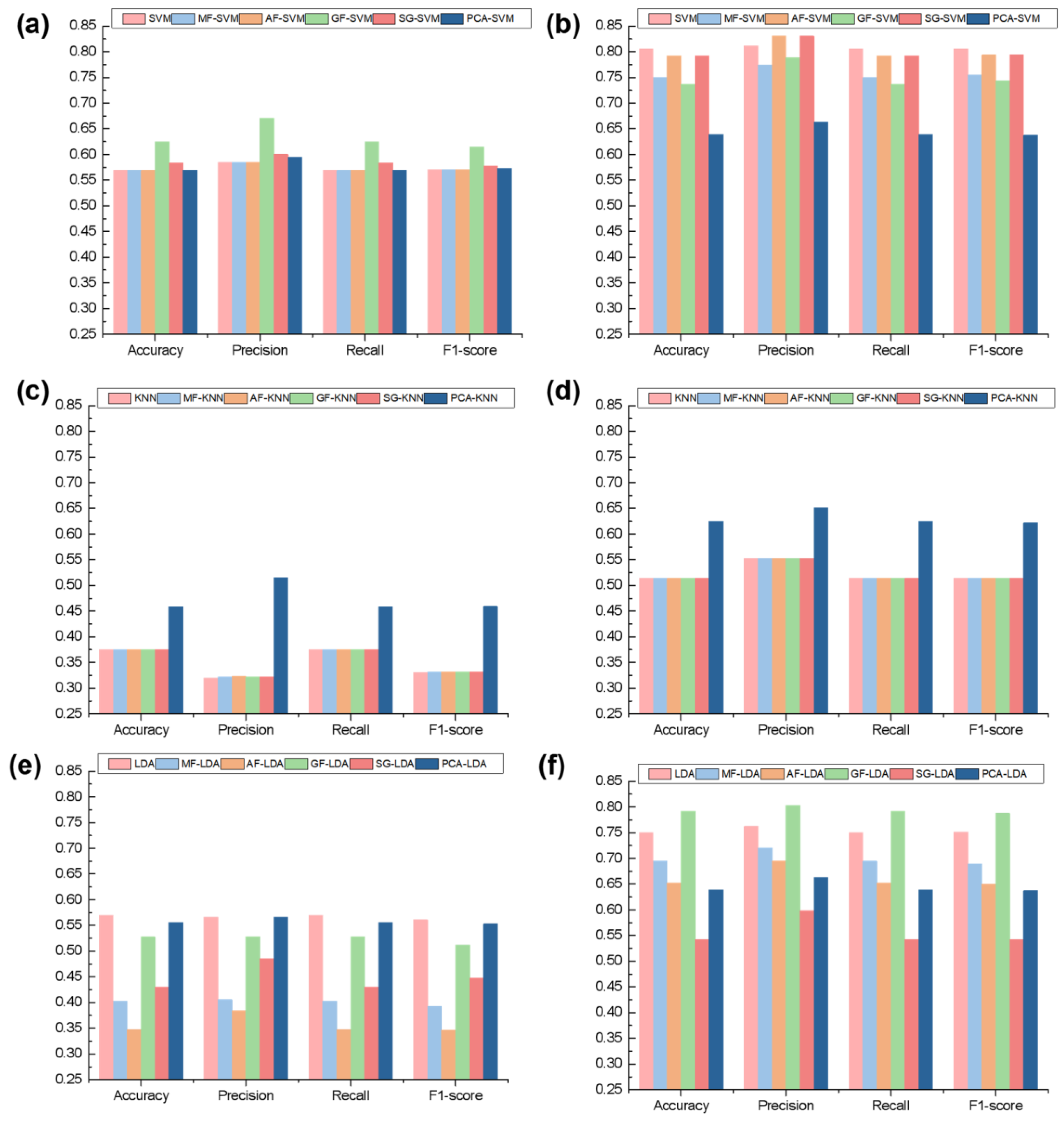
| Model | Accuracy | Precision | Recall | F1-Score | Train Accuracy | 95% CI (Train Accuracy) | |
|---|---|---|---|---|---|---|---|
| SVM | front | 0.5694 | 0.5844 | 0.5694 | 0.5704 | 0.5243 ± 0.0474 | [0.4584, 0.5901] |
| back | 0.8056 | 0.8113 | 0.8056 | 0.8055 | 0.6665 ± 0.0276 | [0.6281, 0.7048] | |
| MF-SVM | front | 0.5694 | 0.5844 | 0.5694 | 0.5704 | 0.5383 ± 0.0404 | [0.4881, 0.5885] |
| back | 0.7500 | 0.7738 | 0.7500 | 0.7545 | 0.6629 ± 0.0350 | [0.6195, 0.7063] | |
| AF-SVM | front | 0.5694 | 0.5844 | 0.5694 | 0.5704 | 0.5382 ± 0.0388 | [0.4843, 0.5920] |
| back | 0.7917 | 0.8304 | 0.7917 | 0.7936 | 0.6699 ± 0.0380 | [0.6171, 0.7227] | |
| GF-SVM | front | 0.6250 | 0.6713 | 0.6250 | 0.6147 | 0.5416 ± 0.0263 | [0.5089, 0.5743] |
| back | 0.7361 | 0.7882 | 0.7361 | 0.7428 | 0.6700 ± 0.0213 | [0.6436, 0.6964] | |
| SG-SVM | front | 0.5833 | 0.6001 | 0.5833 | 0.5782 | 0.5415 ± 0.0343 | [0.5115, 0.5715] |
| back | 0.7917 | 0.8304 | 0.7917 | 0.7936 | 0.6699 ± 0.0380 | [0.6366, 0.7033] | |
| PCA-SVM | front | 0.5694 | 0.5953 | 0.5694 | 0.5731 | 0.5070 ± 0.0344 | [0.4643, 0.5497] |
| back | 0.6389 | 0.6624 | 0.6389 | 0.6371 | 0.6495 ± 0.0275 | [0.6153, 0.6837] | |
| KNN | front | 0.3750 | 0.3199 | 0.3750 | 0.3302 | 0.4056 ± 0.0893 | [0.2946, 0.5165] |
| back | 0.5139 | 0.5528 | 0.5139 | 0.5144 | 0.4750 ± 0.0753 | [0.3816, 0.5684] | |
| MF-KNN | front | 0.3750 | 0.3215 | 0.3750 | 0.3310 | 0.4056 ± 0.0972 | [0.2849, 0.5262] |
| back | 0.5139 | 0.5528 | 0.5139 | 0.5144 | 0.4806 ± 0.0759 | [0.3864, 0.5748] | |
| AF-KNN | front | 0.3750 | 0.3235 | 0.3750 | 0.3319 | 0.4586 ± 0.0359 | [0.4140, 0.5032] |
| back | 0.5139 | 0.5528 | 0.5139 | 0.5144 | 0.4615 ± 0.0653 | [0.3805, 0.5426] | |
| GF-KNN | front | 0.3750 | 0.3215 | 0.3750 | 0.3310 | 0.4056 ± 0.0972 | [0.2849, 0.5262] |
| back | 0.5139 | 0.5528 | 0.5139 | 0.5144 | 0.4806 ± 0.0759 | [0.3864, 0.5748] | |
| SG-KNN | front | 0.3750 | 0.3215 | 0.3750 | 0.3310 | 0.4517 ± 0.0380 | [0.4184, 0.4850] |
| back | 0.5139 | 0.5528 | 0.5139 | 0.5144 | 0.4615 ± 0.0653 | [0.4043, 0.5187] | |
| PCA-KNN | front | 0.4583 | 0.5152 | 0.4583 | 0.4585 | 0.4064 ± 0.0655 | [0.3489, 0.4638] |
| back | 0.6250 | 0.6515 | 0.6250 | 0.6224 | 0.5694 ± 0.0359 | [0.5379, 0.6008] | |
| LDA | front | 0.5694 | 0.5665 | 0.5694 | 0.5609 | 0.5139 ± 0.1185 | [0.3667, 0.6610] |
| back | 0.7500 | 0.7625 | 0.7500 | 0.7517 | 0.6833 ± 0.0671 | [0.6000, 0.7667] | |
| MF-LDA | front | 0.4028 | 0.4059 | 0.4028 | 0.3923 | 0.4410 ± 0.0817 | [0.3694, 0.5125] |
| back | 0.6944 | 0.7205 | 0.6944 | 0.6893 | 0.7044 ± 0.0734 | [0.6401, 0.7688] | |
| AF-LDA | front | 0.3472 | 0.3847 | 0.3472 | 0.3464 | 0.4417 ± 0.0397 | [0.3924, 0.4909] |
| back | 0.6528 | 0.6943 | 0.6528 | 0.6500 | 0.6194 ± 0.0509 | [0.5562, 0.6827] | |
| GF-LDA | front | 0.5278 | 0.5284 | 0.5278 | 0.5125 | 0.4621 ± 0.1120 | [0.3231, 0.6012] |
| back | 0.7917 | 0.8037 | 0.7917 | 0.7878 | 0.6909 ± 0.0589 | [0.6178, 0.7640] | |
| SG-LDA | front | 0.4306 | 0.4858 | 0.4306 | 0.4470 | 0.4204 ± 0.0864 | [0.3005, 0.5404] |
| back | 0.5417 | 0.5983 | 0.5417 | 0.5424 | 0.6245 ± 0.0620 | [0.5385, 0.7105] | |
| PCA-LDA | front | 0.5556 | 0.5663 | 0.5556 | 0.5532 | 0.5106 ± 0.0654 | [0.4532, 0.5680] |
| back | 0.6389 | 0.6629 | 0.6389 | 0.6372 | 0.6075 ± 0.0435 | [0.5694, 0.6456] | |
| CNN | front | 0.9028 | 0.9387 | 0.9028 | 0.9039 | 0.9531 ± 0.0064 | [0.9452, 0.9610] |
| back | 0.9722 | 0.9773 | 0.9722 | 0.9690 | 0.9939 ± 0.0052 | [0.9874, 1.0000] | |
| CNN | fusion | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000±0 | [1.0000, 1.0000] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Ding, H.; Zhang, T.; Wang, Z.; Wang, H.; Zhou, L.; Dai, Y.; Liu, Z. Small-Sample Authenticity Identification and Variety Classification of Anoectochilus roxburghii (Wall.) Lindl. Using Hyperspectral Imaging and Machine Learning. Plants 2025, 14, 1177. https://doi.org/10.3390/plants14081177
Xu Y, Ding H, Zhang T, Wang Z, Wang H, Zhou L, Dai Y, Liu Z. Small-Sample Authenticity Identification and Variety Classification of Anoectochilus roxburghii (Wall.) Lindl. Using Hyperspectral Imaging and Machine Learning. Plants. 2025; 14(8):1177. https://doi.org/10.3390/plants14081177
Chicago/Turabian StyleXu, Yiqing, Haoyuan Ding, Tingsong Zhang, Zhangting Wang, Hongzhen Wang, Lu Zhou, Yujia Dai, and Ziyuan Liu. 2025. "Small-Sample Authenticity Identification and Variety Classification of Anoectochilus roxburghii (Wall.) Lindl. Using Hyperspectral Imaging and Machine Learning" Plants 14, no. 8: 1177. https://doi.org/10.3390/plants14081177
APA StyleXu, Y., Ding, H., Zhang, T., Wang, Z., Wang, H., Zhou, L., Dai, Y., & Liu, Z. (2025). Small-Sample Authenticity Identification and Variety Classification of Anoectochilus roxburghii (Wall.) Lindl. Using Hyperspectral Imaging and Machine Learning. Plants, 14(8), 1177. https://doi.org/10.3390/plants14081177






