Transcript-Wide Identification and Characterization of the BBX Gene Family in Trichosanthes kirilowii and Its Potential Roles in Development and Abiotic Stress
Abstract
1. Introduction
2. Results
2.1. Identification of TkBBX Genes
2.2. Phylogenetic and Classification Analysis of TkBBX Genes
2.3. Conserved Motif and Selective Pressure Analysis of TkBBX Genes
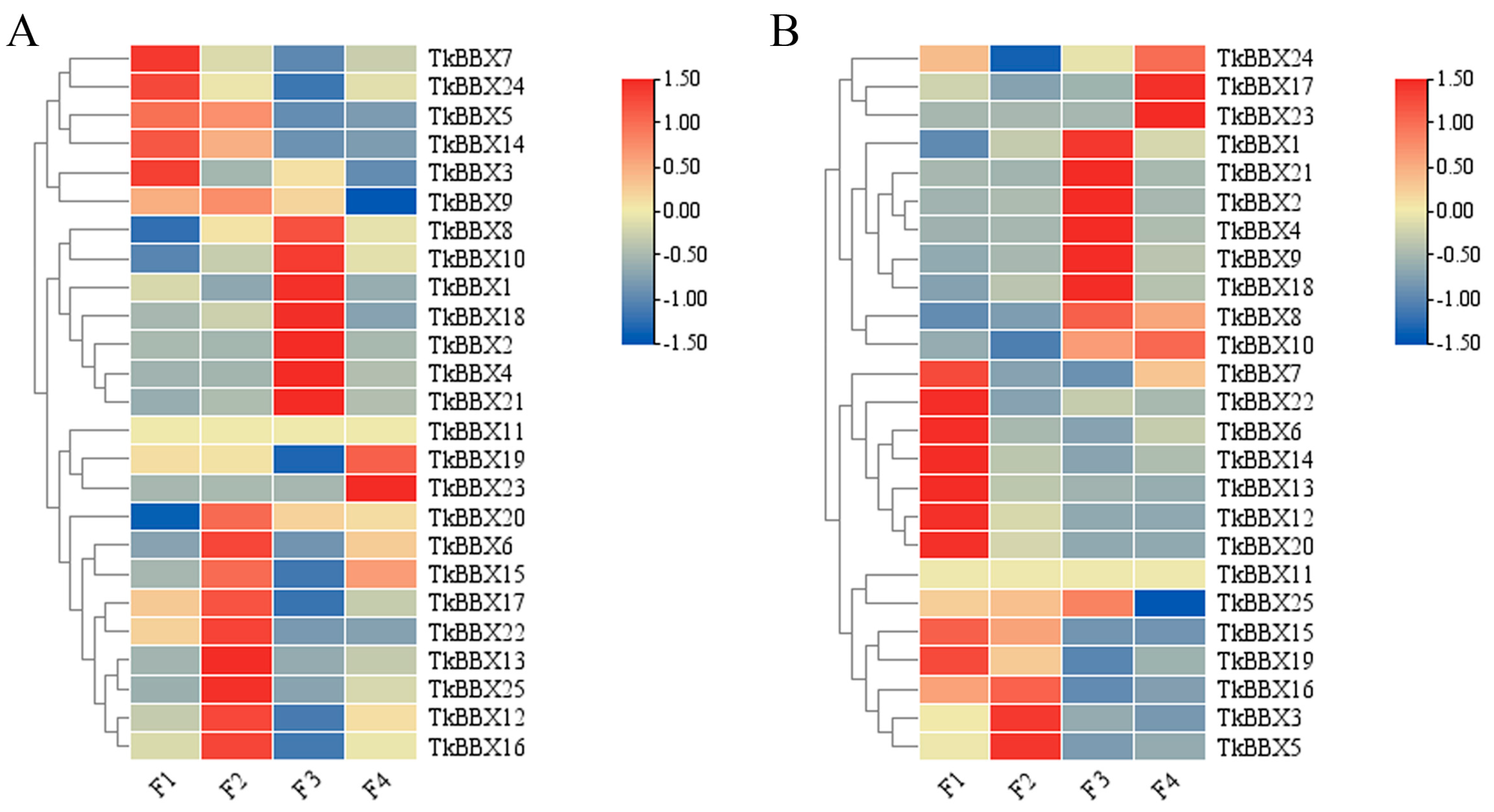
2.4. Expression Analyses of TkBBX Genes During Flowering Stage
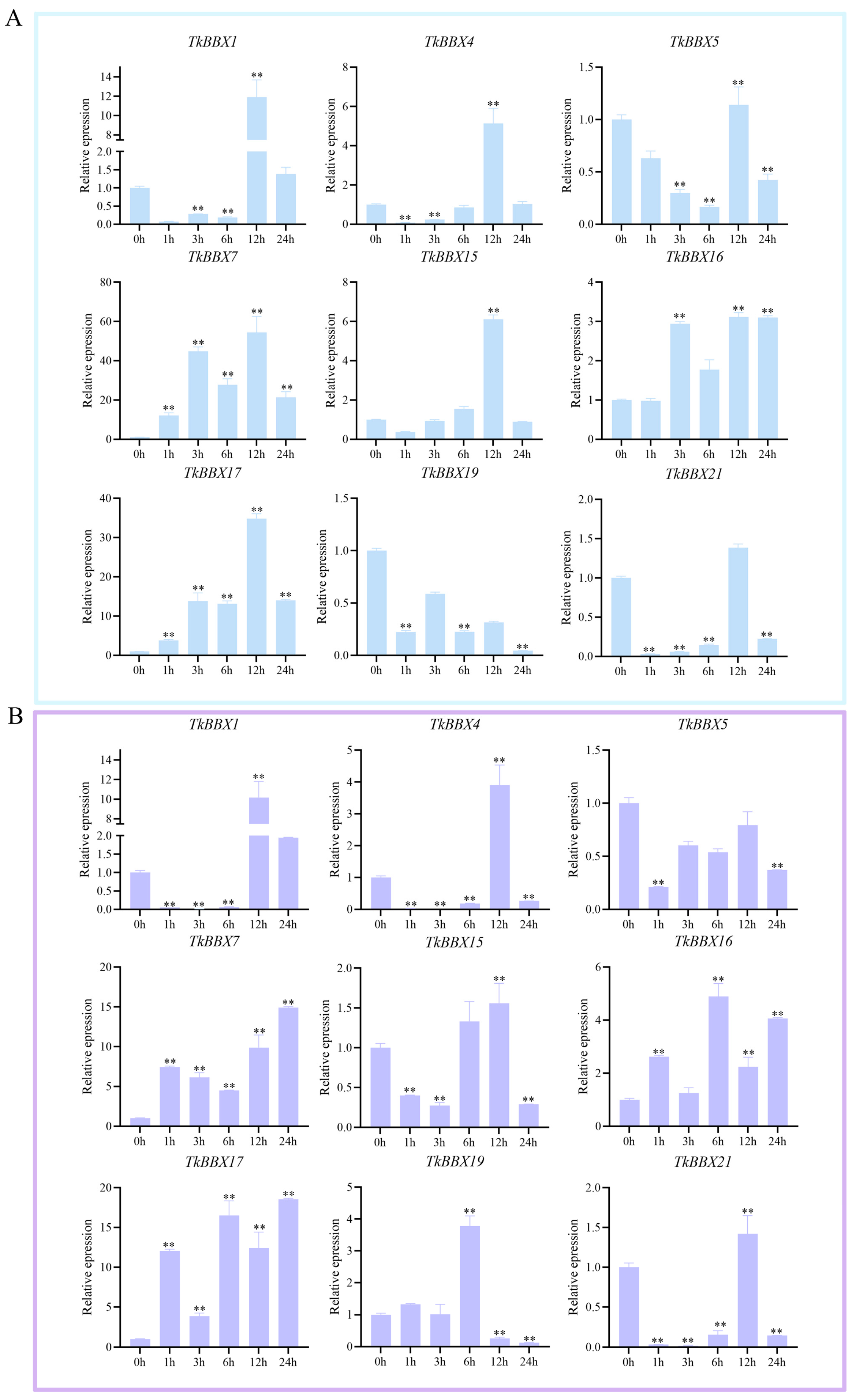
2.5. TkBBX Expression Pattern Responds to Salt and Drought Stresses
2.6. Subcellular Localization of TkBBX Genes
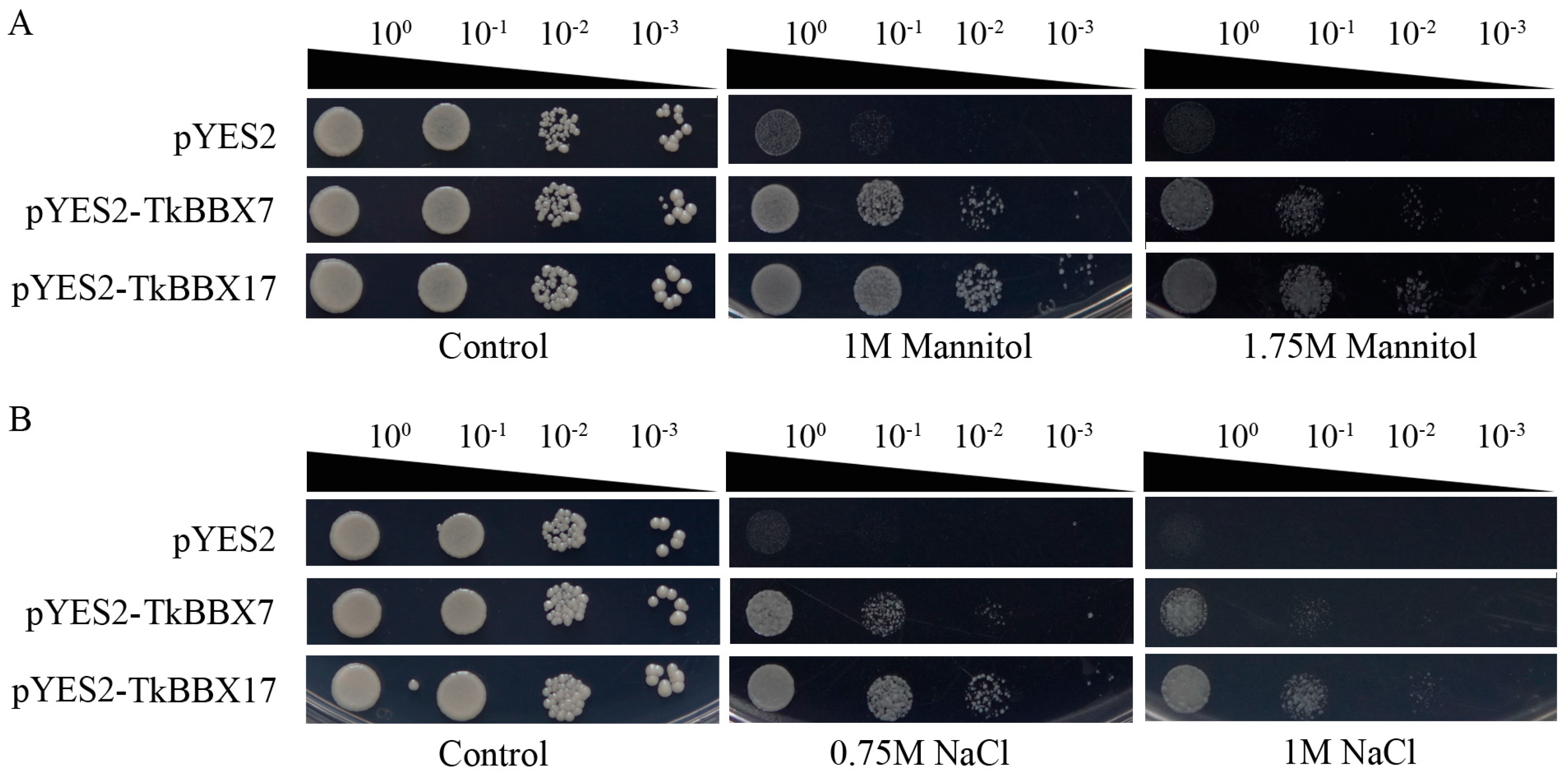
2.7. The Tolerance of TkBBXs to Salt and Drought Stresses in Yeast
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Stress Treatments
4.2. Identification of the BBX Gene Family in T. kirilowii
4.3. Phylogenetic and Conserved Motif Analysis of TkBBX Gene Family
4.4. Expression Pattern Analysis of TkBBX Genes in Different Flower Periods
4.5. RNA Extraction and qRT-PCR Analysis
4.6. Subcellular Localization Assay
4.7. Functional Verification of TkBBX7 and TkBBX17 in Yeast
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gangappa, S.N.; Botto, J.F. The BBX family of plant transcription factors. Trends Plant Sci. 2014, 19, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Qiao, Z.; Li, Y.; Yang, Z.; Wang, C.; Zhang, Y.; Liu, L.; Wang, B. RING Zinc Finger Proteins in Plant Abiotic Stress Tolerance. Front. Plant Sci. 2022, 13, 877011. [Google Scholar] [CrossRef]
- Liu, Y.; Khan, A.R.; Gan, Y. C2H2 Zinc Finger Proteins Response to Abiotic Stress in Plants. Int. J. Mol. Sci. 2022, 23, 2730. [Google Scholar] [CrossRef]
- Cheng, X.; Lei, S.; Li, J.; Tian, B.; Li, C.; Cao, J.; Lu, J.; Ma, C.; Chang, C.; Zhang, H. In silico analysis of the wheat BBX gene family and identification of candidate genes for seed dormancy and germination. BMC Plant Biol. 2024, 24, 334. [Google Scholar] [CrossRef]
- Cao, J.; Yuan, J.; Zhang, Y.; Chen, C.; Zhang, B.; Shi, X.; Niu, R.; Lin, F. Multi-layered roles of BBX proteins in plant growth and development. Stress Biol. 2023, 3, 1. [Google Scholar] [CrossRef]
- Putterill, J.; Robson, F.; Lee, K.; Simon, R.; Coupland, G. The CONSTANS gene of arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 1995, 80, 847–857. [Google Scholar] [CrossRef]
- Hassidim, M.; Harir, Y.; Yakir, E.; Kron, I.; Green, R.M. Over-expression of CONSTANS-LIKE 5 can induce flowering in short-day grown Arabidopsis. Planta 2009, 230, 481–491. [Google Scholar] [CrossRef]
- Li, F.; Sun, J.; Wang, D.; Bai, S.; Clarke, A.K.; Holm, M. The B-box family gene STO (BBX24) in Arabidopsis thaliana regulates flowering time in different pathways. PLoS ONE 2014, 9, e87544. [Google Scholar] [CrossRef]
- Cheng, X.F.; Wang, Z.Y. Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J. 2005, 43, 758–768. [Google Scholar] [CrossRef]
- Tripathi, P.; Carvallo, M.; Hamilton, E.E.; Preuss, S.; Kay, S.A. Arabidopsis B-BOX32 interacts with CONSTANS-LIKE3 to regulate flowering. Proc. Natl. Acad. Sci. USA 2017, 114, 172–177. [Google Scholar] [CrossRef]
- Wang, L.; Sun, J.; Ren, L.; Zhou, M.; Han, X.; Ding, L.; Zhang, F.; Guan, Z.; Fang, W.; Chen, S.; et al. CmBBX8 accelerates flowering by targeting CmFTL1 directly in summer chrysanthemum. Plant Biotechnol. J. 2020, 18, 1562–1572. [Google Scholar] [CrossRef] [PubMed]
- Bai, B.; Zhao, J.; Li, Y.; Zhang, F.; Zhou, J.; Chen, F.; Xie, X. OsBBX14 delays heading date by repressing florigen gene expression under long and short-day conditions in rice. Plant Sci. 2016, 247, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Dong, S.; Sun, D.; Liu, W.; Gu, F.; Liu, Y.; Guo, T.; Wang, H.; Wang, J.; Chen, Z. CONSTANS-Like 9 (OsCOL9) Interacts with Receptor for Activated C-Kinase 1(OsRACK1) to Regulate Blast Resistance through Salicylic Acid and Ethylene Signaling Pathways. PLoS ONE 2016, 11, e0166249. [Google Scholar] [CrossRef]
- Park, H.J.; Kim, W.Y.; Pardo, J.M.; Yun, D.J. Molecular Interactions Between Flowering Time and Abiotic Stress Pathways. Int. Rev. Cell Mol. Biol. 2016, 327, 371–412. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Hettiarachchi, C.; Johansson, H.; Holm, M. SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 2007, 19, 3242–3255. [Google Scholar] [CrossRef]
- Datta, S.; Johansson, H.; Hettiarachchi, C.; Irigoyen, M.L.; Desai, M.; Rubio, V.; Holm, M. LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 2008, 20, 2324–2338. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Hettiarachchi, G.H.; Deng, X.W.; Holm, M. Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 2006, 18, 70–84. [Google Scholar] [CrossRef]
- Chang, C.S.; Li, Y.H.; Chen, L.T.; Chen, W.C.; Hsieh, W.P.; Shin, J.; Jane, W.N.; Chou, S.J.; Choi, G.; Hu, J.M.; et al. LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J. 2008, 54, 205–219. [Google Scholar] [CrossRef]
- Fan, X.Y.; Sun, Y.; Cao, D.M.; Bai, M.Y.; Luo, X.M.; Yang, H.J.; Wei, C.Q.; Zhu, S.W.; Sun, Y.; Chong, K.; et al. BZS1, a B-box protein, promotes photomorphogenesis downstream of both brassinosteroid and light signaling pathways. Mol. Plant 2012, 5, 591–600. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Crocco, C.D.; Johansson, H.; Datta, S.; Hettiarachchi, C.; Holm, M.; Botto, J.F. The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 2013, 25, 1243–1257. [Google Scholar] [CrossRef]
- Holtan, H.E.; Bandong, S.; Marion, C.M.; Adam, L.; Tiwari, S.; Shen, Y.; Maloof, J.N.; Maszle, D.R.; Ohto, M.A.; Preuss, S.; et al. BBX32, an Arabidopsis B-Box protein, functions in light signaling by suppressing HY5-regulated gene expression and interacting with STH2/BBX21. Plant Physiol. 2011, 156, 2109–2123. [Google Scholar] [CrossRef]
- Indorf, M.; Cordero, J.; Neuhaus, G.; Rodríguez-Franco, M. Salt tolerance (SO), a stress-related protein, has a major role in light signalling. Plant J. 2007, 51, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, T.; Ito, S.; Nakamichi, N.; Niwa, Y.; Murakami, M.; Yamashino, T.; Mizuno, T. The common function of a novel subfamily of B-Box zinc finger proteins with reference to circadian-associated events in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2008, 72, 1539–1549. [Google Scholar] [CrossRef] [PubMed]
- Khanna, R.; Shen, Y.; Toledo-Ortiz, G.; Kikis, E.A.; Johannesson, H.; Hwang, Y.S.; Quail, P.H. Functional profiling reveals that only a small number of phytochrome-regulated early-response genes in Arabidopsis are necessary for optimal deetiolation. Plant Cell 2006, 18, 2157–2171. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Wang, X.F.; Zhang, X.W.; Bi, S.Q.; You, C.X.; Hao, Y.J. MdBBX22 regulates UV-B-induced anthocyanin biosynthesis through regulating the function of MdHY5 and is targeted by MdBT2 for 26S proteasome-mediated degradation. Plant Biotechnol. J. 2019, 17, 2231–2233. [Google Scholar] [CrossRef]
- Fang, H.; Dong, Y.; Yue, X.; Hu, J.; Jiang, S.; Xu, H.; Wang, Y.; Su, M.; Zhang, J.; Zhang, Z.; et al. The B-box zinc finger protein MdBBX20 integrates anthocyanin accumulation in response to ultraviolet radiation and low temperature. Plant Cell Environ. 2019, 42, 2090–2104. [Google Scholar] [CrossRef]
- Crocco, C.D.; Holm, M.; Yanovsky, M.J.; Botto, J.F. Function of B-BOX under shade. Plant Signal Behav. 2011, 6, 101–104. [Google Scholar] [CrossRef]
- Crocco, C.D.; Holm, M.; Yanovsky, M.J.; Botto, J.F. AtBBX21 and COP1 genetically interact in the regulation of shade avoidance. Plant J. 2010, 64, 551–562. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.F.; Espley, R.V.; Lin-Wang, K.; Bi, S.Q.; You, C.X.; Hao, Y.J. An Apple B-Box Protein MdBBX37 Modulates Anthocyanin Biosynthesis and Hypocotyl Elongation Synergistically with MdMYBs and MdHY5. Plant Cell Physiol. 2020, 61, 130–143. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, X.W.; Liu, X.; Zheng, P.F.; Su, L.; Wang, G.L.; Wang, X.F.; Li, Y.Y.; You, C.X.; An, J.P. Phytochrome interacting factor MdPIF7 modulates anthocyanin biosynthesis and hypocotyl growth in apple. Plant Physiol. 2022, 188, 2342–2363. [Google Scholar] [CrossRef]
- Xuefen, D.; Wei, X.; Wang, B.; Xiaolin, Z.; Xian, W.; Jincheng, L. Genome-wide identification and expression pattern analysis of quinoa BBX family. PeerJ 2022, 10, e14463. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Saito, T.; Honda, C.; Hatsuyama, Y.; Ito, A.; Moriguchi, T. An apple B-box protein, MdCOL11, is involved in UV-B- and temperature-induced anthocyanin biosynthesis. Planta 2014, 240, 1051–1062. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Tao, R.; Tang, Y.; Yin, L.; Ma, Y.; Ni, J.; Yan, X.; Yang, Q.; Wu, Z.; Zeng, Y.; et al. BBX16, a B-box protein, positively regulates light-induced anthocyanin accumulation by activating MYB10 in red pear. Plant Biotechnol J. 2019, 17, 1985–1997. [Google Scholar] [CrossRef]
- Nagaoka, S.; Takano, T. Salt tolerance-related protein STO binds to a Myb transcription factor homologue and confers salt tolerance in Arabidopsis. J. Exp. Bot. 2003, 54, 2231–2237. [Google Scholar] [CrossRef]
- Liu, X.; Li, R.; Dai, Y.; Yuan, L.; Sun, Q.; Zhang, S.; Wang, X. A B-box zinc finger protein, MdBBX10, enhanced salt and drought stresses tolerance in Arabidopsis. Plant Mol. Biol. 2019, 99, 437–447. [Google Scholar] [CrossRef]
- Huang, S.; Chen, C.; Xu, M.; Wang, G.; Xu, L.A.; Wu, Y. Overexpression of Ginkgo BBX25 enhances salt tolerance in Transgenic Populus. Plant Physiol. Biochem. 2021, 167, 946–954. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Wang, X.; Cao, Y.; Zhang, H.; Hua, R.; Liu, H.; Sui, S. CpBBX19, a B-Box Transcription Factor Gene of Chimonanthus praecox, Improves Salt and Drought Tolerance in Arabidopsis. Genes 2021, 12, 1456. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, H.; Ping, Q.; Zhang, Z.; Guan, Z.; Fang, W.; Chen, S.; Chen, F.; Jiang, J.; Zhang, F. The heterologous expression of CmBBX22 delays leaf senescence and improves drought tolerance in Arabidopsis. Plant Cell Rep. 2019, 38, 15–24. [Google Scholar] [CrossRef]
- Wang, Q.; Tu, X.; Zhang, J.; Chen, X.; Rao, L. Heat stress-induced BBX18 negatively regulates the thermotolerance in Arabidopsis. Mol. Biol. Rep. 2013, 40, 2679–2688. [Google Scholar] [CrossRef]
- Ouyang, Y.; Pan, X.; Wei, Y.; Wang, J.; Xu, X.; He, Y.; Zhang, X.; Li, Z.; Zhang, H. Genome-wide identification and characterization of the BBX gene family in pineapple reveals that candidate genes are involved in floral induction and flowering. Genomics 2022, 114, 110397. [Google Scholar] [CrossRef]
- Yu, L.; Lyu, Z.; Liu, H.; Zhang, G.; He, C.; Zhang, J. Insights into the evolutionary origin and expansion of the BBX gene family. Plant Biotechnol Rep. 2022, 16, 205–214. [Google Scholar] [CrossRef]
- Song, Z.; Bian, Y.; Liu, J.; Sun, Y.; Xu, D. B-box proteins: Pivotal players in light-mediated development in plants. J. Integr. Plant Biol. 2020, 62, 1293–1309. [Google Scholar] [CrossRef]
- Talar, U.; Kiełbowicz-Matuk, A. Beyond Arabidopsis: BBX Regulators in Crop Plants. Int. J. Mol. Sci. 2021, 22, 2906. [Google Scholar] [CrossRef] [PubMed]
- Susila, H.; Nasim, Z.; Gawarecka, K.; Jung, J.Y.; Jin, S.; Youn, G.; Ahn, J.H. Chloroplasts prevent precocious flowering through a GOLDEN2-LIKE-B-BOX DOMAIN PROTEIN module. Plant Commun. 2023, 4, 100515. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Xu, J.; Yuan, C.; Chen, Q.; Liu, Q.; Wang, X.; Qin, C. BBX17 Interacts with CO and Negatively Regulates Flowering Time in Arabidopsis thaliana. Plant Cell Physiol. 2022, 63, 401–409. [Google Scholar] [CrossRef]
- Böhlenius, H.; Huang, T.; Charbonnel-Campaa, L.; Brunner, A.M.; Jansson, S.; Strauss, S.H.; Nilsson, O. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 2006, 312, 1040–1043. [Google Scholar] [CrossRef]
- Shi, G.; Ai, K.; Yan, X.; Zhou, Z.; Cai, F.; Bao, M.; Zhang, J. Genome-Wide Analysis of the BBX Genes in Platanus × acerifolia and Their Relationship with Flowering and/or Dormancy. Int. J. Mol. Sci. 2023, 24, 8576. [Google Scholar] [CrossRef]
- Li, S.; Guo, S.; Gao, X.; Wang, X.; Liu, Y.; Wang, J.; Li, X.; Zhang, J.; Fu, B. Genome-wide identification of B-box zinc finger (BBX) gene family in Medicago sativa and their roles in abiotic stress responses. BMC Genom. 2024, 25, 110. [Google Scholar] [CrossRef]
- Shan, B.; Bao, G.; Shi, T.; Zhai, L.; Bian, S.; Li, X. Genome-wide identification of BBX gene family and their expression patterns under salt stress in soybean. BMC Genom. 2022, 23, 820. [Google Scholar] [CrossRef]
- Papiernik, S.K.; Grieve, C.M.; Lesch, S.M.; Yates, S.R. Effects of salinity, imazethapyr, and chlorimuron application on soybean growth and yield. Commun. Soil Sci. Plant Anal. 2005, 36, 951–967. [Google Scholar]
- Dai, Y.Q.; Lu, Y.; Zhou, Z.; Wang, X.Y.; Ge, H.J.; Sun, Q.H. B-box containing protein 1 from Malus domestica (MdBBX1) is involved in the abiotic stress response. Peerj. 2022, 10, e12852. [Google Scholar] [CrossRef] [PubMed]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adamscollier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, 585–587. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Xiong, R.; Chu, Z.; Peng, X.; Cui, G.; Li, W.; Dong, L. Transcript-wide identification and expression pattern analysis to comprehend the roles of AP2/ERF genes under development and abiotic stress in Trichosanthes kirilowii. BMC Plant Biol. 2023, 23, 354. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101. [Google Scholar] [CrossRef]
- Li, D.; He, Y.; Li, S.; Shi, S.; Li, L.; Liu, Y.; Chen, H. Genome-wide characterization and expression analysis of AP2/ERF genes in eggplant (Solanum melongena L.). Plant Physiol. Biochem. 2021, 167, 492–503. [Google Scholar] [CrossRef]
- Mitteer, D.R.; Greer, B.D. Using GraphPad Prism’s Heat Maps for Efficient, Fine-Grained Analyses of Single-Case Data. Behav. Anal. Pract. 2022, 15, 505–514. [Google Scholar] [CrossRef]



| Name | ORF Length (bp) | Protein | Subcellular Localization | ||
|---|---|---|---|---|---|
| Length (aa) | PI | MW (Da) | |||
| TkBBX1 | 1251 | 416 | 5.34 | 45,315.65 | nucleus |
| TkBBX2 | 591 | 196 | 5.42 | 21,528.00 | nucleus |
| TkBBX3 | 1071 | 356 | 4.74 | 40,194.50 | nucleus |
| TkBBX4 | 1227 | 408 | 5.48 | 44,870.00 | nucleus |
| TkBBX5 | 1197 | 398 | 6.22 | 44,811.11 | nucleus |
| TkBBX6 | 933 | 310 | 5.30 | 34,189.24 | nucleus |
| TkBBX7 | 834 | 277 | 4.24 | 30,376.77 | nucleus |
| TkBBX8 | 1251 | 416 | 5.27 | 45,256.58 | nucleus |
| TkBBX9 | 954 | 317 | 4.79 | 35,435.72 | nucleus |
| TkBBX10 | 1101 | 366 | 4.82 | 39,790.15 | nucleus |
| TkBBX11 | 687 | 228 | 4.86 | 25,319.24 | nucleus |
| TkBBX12 | 954 | 317 | 8.22 | 34,707.13 | nucleus |
| TkBBX13 | 480 | 159 | 6.86 | 17,469.76 | nucleus |
| TkBBX14 | 312 | 103 | 10.42 | 11,615.83 | nucleus |
| TkBBX15 | 888 | 295 | 5.36 | 32,186.03 | nucleus |
| TkBBX16 | 708 | 235 | 5.02 | 25,915.49 | nucleus |
| TkBBX17 | 846 | 281 | 4.20 | 31,019.26 | nucleus |
| TkBBX18 | 651 | 216 | 6.37 | 23,578.75 | nucleus |
| TkBBX19 | 951 | 316 | 7.51 | 34,439.71 | nucleus |
| TkBBX20 | 456 | 151 | 5.70 | 16,623.66 | nucleus |
| TkBBX21 | 1227 | 408 | 5.31 | 44,798.88 | nucleus |
| TkBBX22 | 228 | 75 | 5.13 | 8068.35 | nucleus |
| TkBBX23 | 669 | 222 | 5.60 | 24,372.74 | nucleus |
| TkBBX24 | 462 | 153 | 6.74 | 17,176.36 | nucleus |
| TkBBX25 | 1323 | 440 | 6.12 | 49,928.00 | nucleus |
| Paralogous Pairs | Ka | Ks | Ka/Ks |
|---|---|---|---|
| TkBBX1/TkBBX18 | 0.062811 | 0.084951 | 0.73938 |
| TkBBX2/TkBBX4 | 0.013396 | 0.022376 | 0.598669 |
| TkBBX3/TkBBX25 | 0.027515 | 0.055871 | 0.492471 |
| TkBBX5/TkBBX25 | 0.349804354 | 3.031300959 | 0.115397434 |
| TkBBX8/TkBBX18 | 0.064963628 | 0.077480506 | 0.838451254 |
| TkBBX7/TkBBX17 | 0.007762036 | 0.010979153 | 0.706979455 |
| TkBBX9/TkBBX11 | 0.008541693 | 0.009793393 | 0.872189379 |
| TkBBX10/TkBBX21 | 0.00471746 | 0.012224379 | 0.385905915 |
| TkBBX13/TkBBX20 | 0.017279382 | 0.06141537 | 0.281352732 |
| TkBBX22/TkBBX24 | 0.064695258 | 0.18555613 | 0.348656001 |
| Orthologous pairs | Ka | Ks | Ka/Ks |
| TkBBX8/AtBBX7 | 0.257420338 | 2.43798484 | 0.105587342 |
| TkBBX11/AtBBX15 | 0.3645691 | ||
| TkBBX12/AtBBX6 | 0.373713424 | 3.080075312 | 0.12133256 |
| TkBBX18/AtBBX25 | 0.223789243 | ||
| TkBBX18/AtBBX8 | 0.356460289 | ||
| TkBBX20/AtBBX18 | 0.256192316 | ||
| TkBBX23/AtBBX20 | 0.324254708 | ||
| TkBBX24/AtBBX19 | 0.168224215 | ||
| TkBBX25/AtBBX14 | 0.419126264 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Xiong, R.; Chu, Z.; Peng, X.; Cui, G.; Dong, L. Transcript-Wide Identification and Characterization of the BBX Gene Family in Trichosanthes kirilowii and Its Potential Roles in Development and Abiotic Stress. Plants 2025, 14, 975. https://doi.org/10.3390/plants14060975
Li W, Xiong R, Chu Z, Peng X, Cui G, Dong L. Transcript-Wide Identification and Characterization of the BBX Gene Family in Trichosanthes kirilowii and Its Potential Roles in Development and Abiotic Stress. Plants. 2025; 14(6):975. https://doi.org/10.3390/plants14060975
Chicago/Turabian StyleLi, Weiwen, Rui Xiong, Zhuannan Chu, Xingxing Peng, Guangsheng Cui, and Ling Dong. 2025. "Transcript-Wide Identification and Characterization of the BBX Gene Family in Trichosanthes kirilowii and Its Potential Roles in Development and Abiotic Stress" Plants 14, no. 6: 975. https://doi.org/10.3390/plants14060975
APA StyleLi, W., Xiong, R., Chu, Z., Peng, X., Cui, G., & Dong, L. (2025). Transcript-Wide Identification and Characterization of the BBX Gene Family in Trichosanthes kirilowii and Its Potential Roles in Development and Abiotic Stress. Plants, 14(6), 975. https://doi.org/10.3390/plants14060975





