Identification and Expression Analysis of Polyphenol Oxidase Gene Family Members in Response to Wound Stress in Lettuce (Lactuca sativa L.)
Abstract
1. Introduction
2. Results
2.1. Identification and Characteristics of PPO Genes in Lettuce
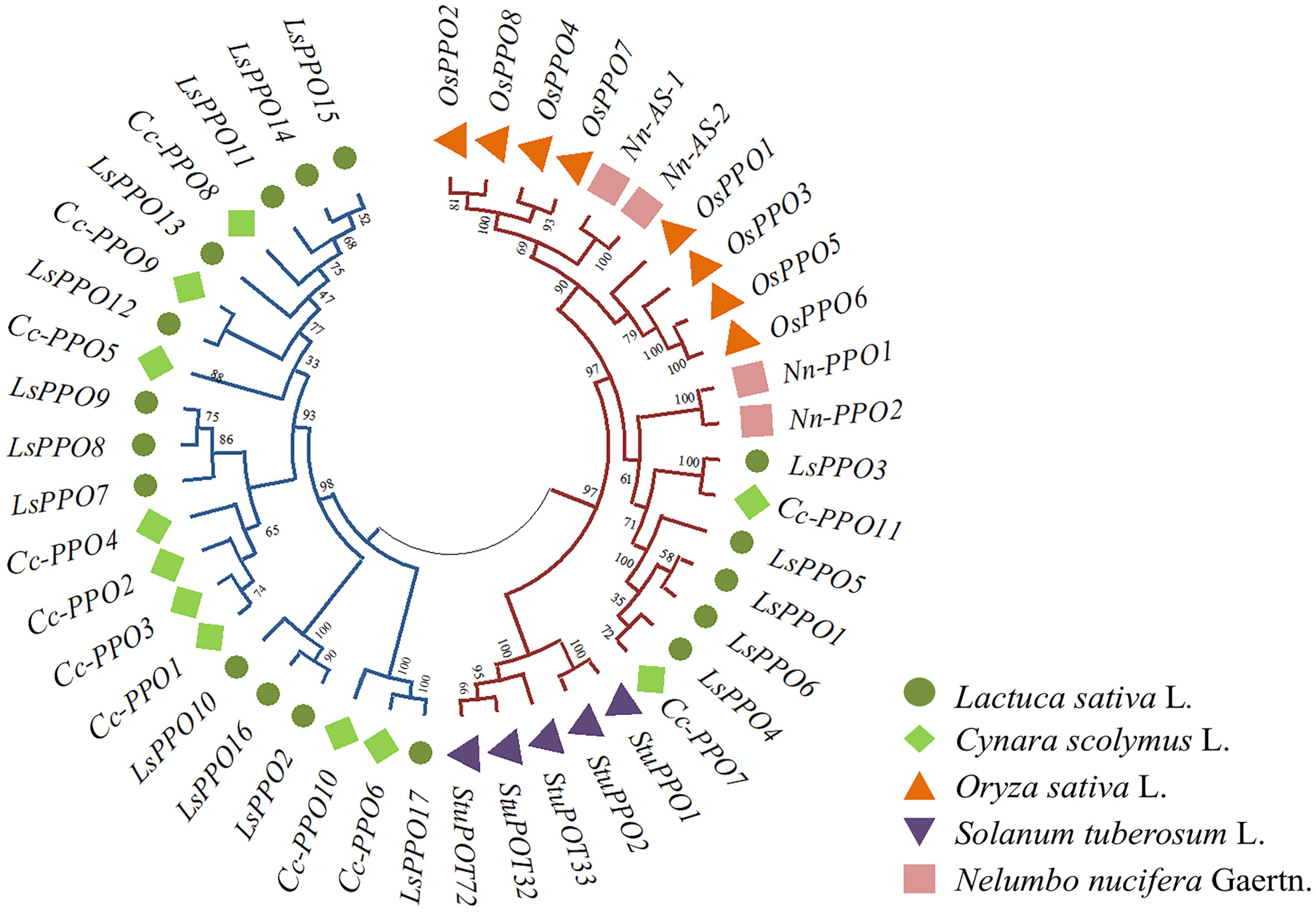
2.2. Evolutionary Analysis of LsPPOs Gene Family in Lettuce
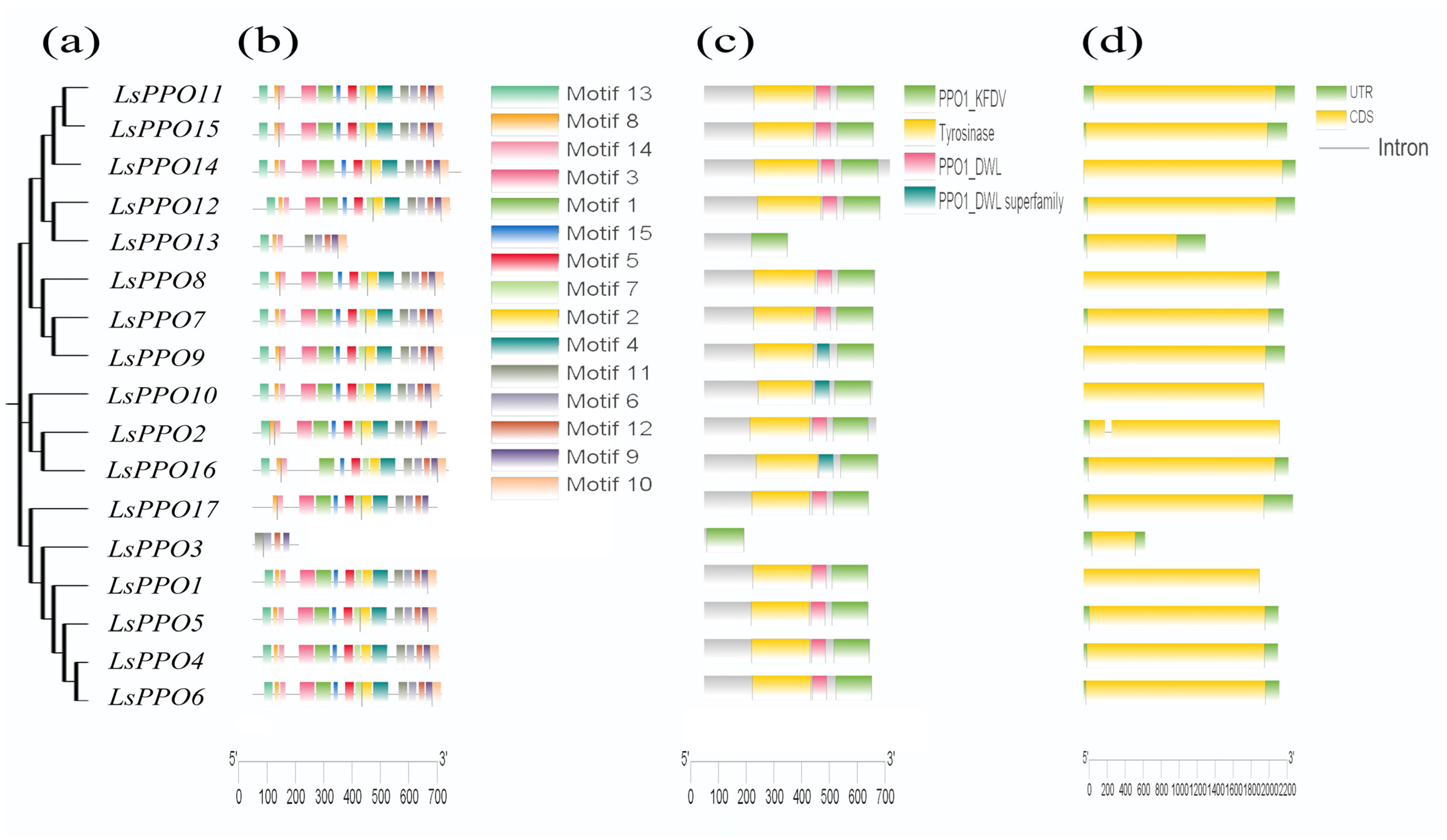
2.3. Predictive Analysis of Gene Structure and Protein Conserved Motifs of Lettuce LsPPOs Family
2.4. Analysis of PPO Gene Codon Bias in Lettuce
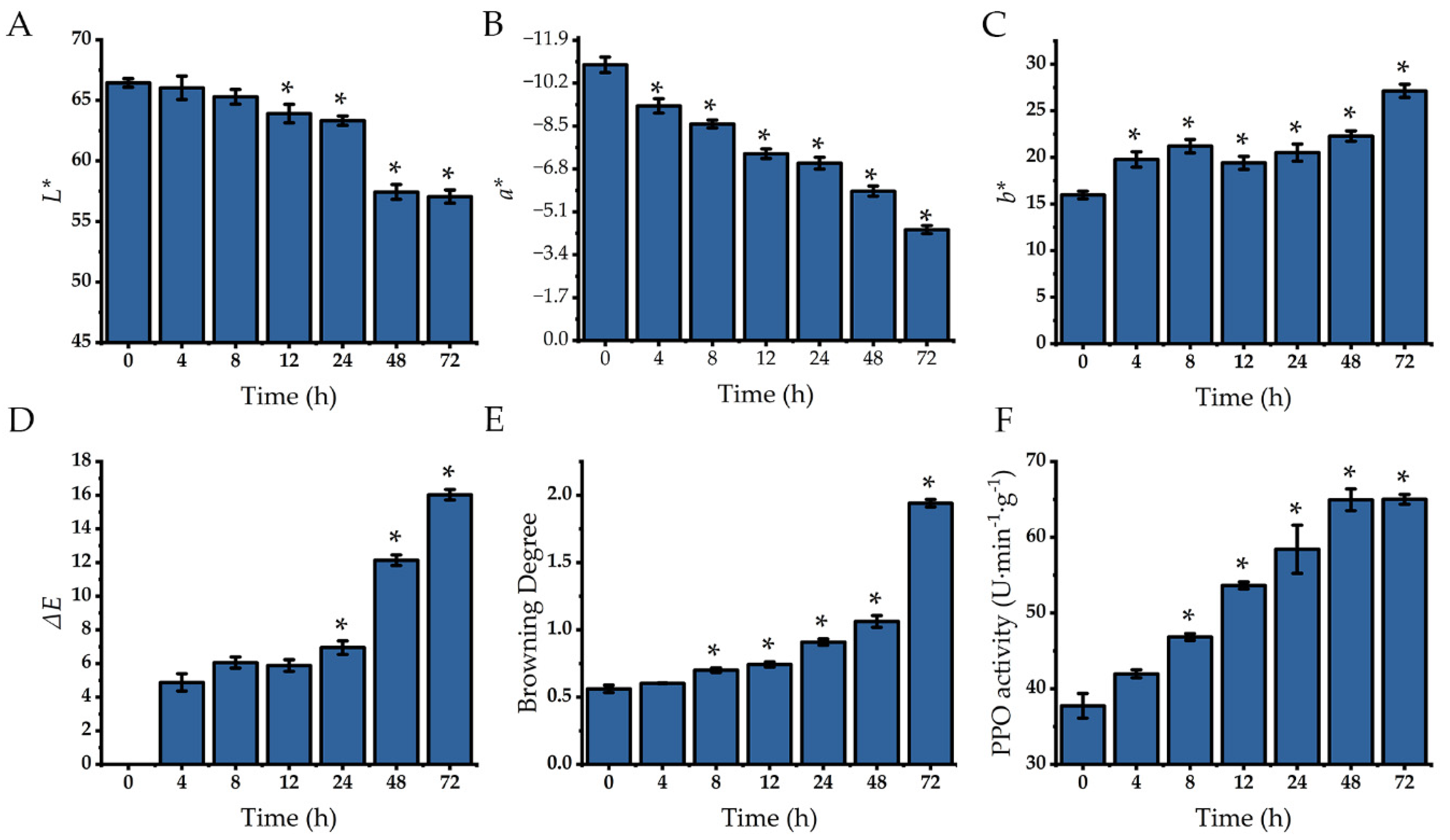
2.5. Changes in Lettuce Color and PPO Activity
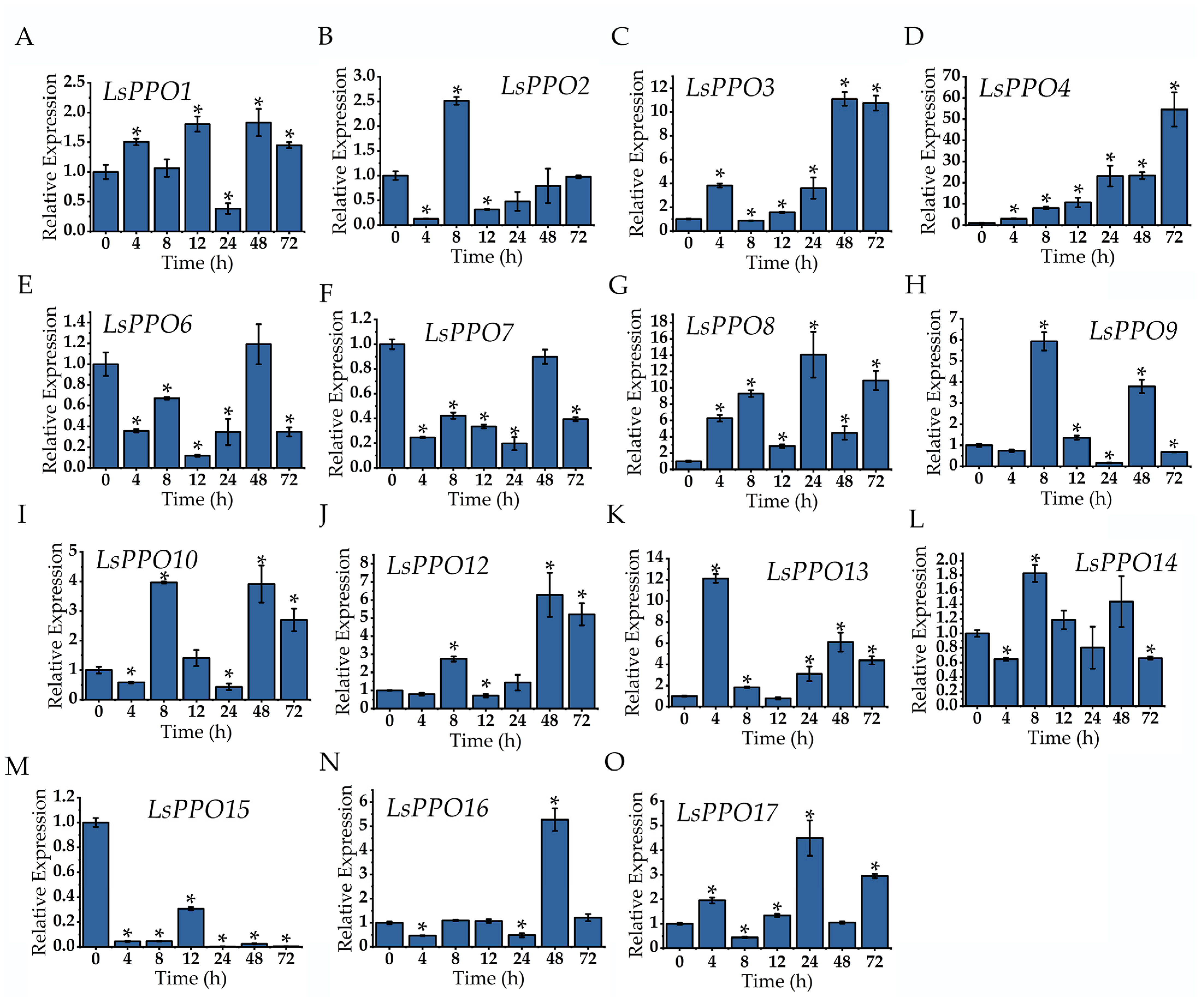
2.6. Expression Analysis of Lettuce LsPPOs
2.7. Correlation Analysis of PPO Activity and LsPPOs Expression Level with Lettuce Browning
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. LsPPOs Gene Family Identification and Physicochemical Analyses
4.3. Codon Usage Characteristics
4.4. Phylogenetic Analyses of the PPO Gene Family
4.5. Color
4.6. Browning Degree
4.7. PPO Activity
4.8. RNA Extraction and Quantitative Real-Time (qRT-PCR) Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Irazoqui, M.; Colazzo, M.; Fender, E.; Budelli, E.; Barrios, S.; Pérez, N.; Lema, P. Combined Peracetic Acid: Power Ultrasound Disinfection Process Enhances Bioactive Compounds and Preserves Quality Attributes of Fresh-Cut Lettuce (cv. Vera). Discov. Food 2024, 4, 14. [Google Scholar] [CrossRef]
- Wang, W.; Lin, Z.; Wang, W.; Shang, M.; Lv, H.; Zong, Q.; Li, J.; Liang, B.; Zhou, W. Elicitation with Hydrogen Peroxide Promotes Growth, Phenolic-Enrichment, Antioxidant Activity and Nutritional Values of Two Hydroponic Lettuce Genotypes. Food Chem. X 2023, 19, 100847. [Google Scholar] [CrossRef]
- Altunkaya, A. Effect of Whey Protein Concentrate on Phenolic Profile and Browning of Fresh-Cut Lettuce (Lactuca sativa). Food Chem. 2011, 128, 754–760. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Z.; Zeng, S.; Yuan, S.; Yue, X.; Tian, T.; Zhu, X.; Zheng, S.; Xu, X.; Zuo, J.; et al. Browning Mechanism in Stems of Fresh-cut Lettuce. Food Chem. 2023, 405, 134575. [Google Scholar] [CrossRef]
- Managa, M.G.; Tinyani, P.P.; Senyolo, G.M.; Soundy, P.; Sultanbawa, Y.; Sivakumar, D. Impact of Transportation, Storage, and Retail Shelf Conditions on Lettuce Quality and Phytonutrients Losses in the Supply Chain. Food Sci. Nutr. 2018, 6, 1527–1536. [Google Scholar] [CrossRef]
- Chen, Y.; Mao, J.; Zhang, L.; Zhu, C.; Qin, Q.; Li, N. Genome-Wide Identification and Expression Pattern Analysis of Polyphenol Oxidase Gene Family in Agaricus bisporus. Agronomy 2023, 13, 2534. [Google Scholar] [CrossRef]
- Mayer, A.M.; Harel, E. Polyphenol Oxidases in Plants. Phytochemistry 1979, 18, 193–215. [Google Scholar] [CrossRef]
- Tran, L.T.; Taylor, J.S.; Constabel, C.P. The Polyphenol Oxidase Gene Family in Land Plants: Lineage-Specific Duplication and Expansion. BMC Genom. 2012, 13, 395. [Google Scholar] [CrossRef]
- Bansal, R.; Srivastava, J.P. Antioxidative Defense System in Pigeonpea Roots under Waterlogging Stress. Acta Physiol. Plant 2012, 34, 515–522. [Google Scholar] [CrossRef]
- Nakayama, T.; Sato, T.; Fukui, Y.; Yonekura-Sakakibara, K.; Hayashi, H.; Tanaka, Y.; Kusumi, T.; Nishino, T. Specificity Analysis and Mechanism of Aurone Synthesis Catalyzed by Aureusidin Synthase, a Polyphenol Oxidase Homolog Responsible for £ower Coloration. FEBS Lett. 2001, 499, 107–111. [Google Scholar] [CrossRef]
- Thygesen, P.W.; Dry, I.B.; Robinson, S.P. Polyphenol Oxidase in Potato (A Multigene Family That Exhibits Differential Expression Patterns). Plant Physiol. 1995, 109, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Terefe, N.S.; Delon, A.; Buckow, R.; Versteeg, C. Blueberry Polyphenol Oxidase: Characterization and the Kinetics of Thermal and High Pressure Activation and Inactivation. Food Chem. 2015, 188, 193–200. [Google Scholar] [CrossRef]
- Zhu, H.S.; Kang, J.; Liu, J.T.; Chen, M.D.; Li, Y.P.; Wang, B.; Lin, B.Y.; Wen, Q.F. Cloning and Expression Analysis of Polyphenol Oxidase PPO Gene Family from Luffa cylindrical. J. Nucl. Agric. Sci. 2018, 32, 1502–1512. [Google Scholar]
- Chisari, M.; Barbagallo, R.N.; Spagna, G. Characterization and Role of Polyphenol Oxidase and Peroxidase in Browning of Fresh-Cut Melon. J. Agric. Food Chem. 2008, 56, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Bachem, C.W.B.; Speckmann, G.-J.; Van Der Linde, P.C.G.; Verheggen, F.T.M.; Hunt, M.D.; Steffens, J.C.; Zabeau, M. Antisense Expression of Polyphenol Oxidase Genes Inhibits Enzymatic Browning in Potato Tubers. Nat. Biotechnol. 1994, 12, 1101–1105. [Google Scholar] [CrossRef]
- Han, Q.-Y.; Liu, F.; Li, M.; Wang, K.-L.; Ni, Y.-Y. Comparison of Biochemical Properties of Membrane-Bound and Soluble Polyphenol Oxidase from Granny Smith Apple (Malus × domestica Borkh.). Food Chem. 2019, 289, 657–663. [Google Scholar] [CrossRef]
- Min, T.; Lu, K.; Chen, J.; Niu, L.; Lin, Q.; Yi, Y.; Hou, W.; Ai, Y.; Wang, H. Biochemical Mechanism of Fresh-Cut Lotus (Nelumbo nucifera Gaertn.) Root with Exogenous Melatonin Treatment by Multiomics Analysis. Foods 2023, 12, 44. [Google Scholar] [CrossRef]
- Zou, H.; Li, C.; Wei, X.; Xiao, Q.; Tian, X.; Zhu, L.; Ma, B.; Ma, F.; Li, M. Expression of the Polyphenol Oxidase Gene MdPPO7 Is Modulated by MdWRKY3 to Regulate Browning in Sliced Apple Fruit. Plant Physiol. 2024, 197, kiae614. [Google Scholar] [CrossRef]
- González, M.N. Comparative Potato Genome Editing: Agrobacterium Tumefaciens-Mediated Transformation and Protoplasts Transfection Delivery of CRISPR/Cas9 Components Directed to StPPO2 Gene. Plant Cell Tissue Organ Cult. 2021, 145, 291–305. [Google Scholar] [CrossRef]
- Maioli, A.; Gianoglio, S.; Moglia, A.; Acquadro, A.; Valentino, D.; Milani, A.M.; Prohens, J.; Orzaez, D.; Granell, A.; Lanteri, S.; et al. Simultaneous CRISPR/Cas9 Editing of Three PPO Genes Reduces Fruit Flesh Browning in Solanum melongena L. Front. Plant Sci. 2020, 11, 607161. [Google Scholar] [CrossRef]
- Waltz, E. Gene-Edited CRISPR Mushroom Escapes US Regulation. Nature 2016, 532, 293. [Google Scholar] [CrossRef] [PubMed]
- aan den Toorn, M.; Albrecht, C.; de Vries, S. On the Origin of SERKs: Bioinformatics Analysis of the Somatic Embryogenesis Receptor Kinases. Mol. Plant 2015, 8, 762–782. [Google Scholar] [CrossRef] [PubMed]
- Duan, W.; Song, X.; Liu, T.; Huang, Z.; Ren, J.; Hou, X.; Li, Y. Genome-Wide Analysis of the MADS-Box Gene Family in Brassica rapa (Chinese Cabbage). Mol. Genet. Genom. 2015, 290, 239–255. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.Y.; Li, Z.J.; Cheng, X.R.; Yang, T.B.; Yang, X.X.; Liu, L.; Dai, L.Y. Bioinformatics Analysis of SUT Gene Family in Sorghum bicolor. Genom. Appl. Biol. 2020, 39, 674–683. [Google Scholar] [CrossRef]
- Chi, M.; Bhagwat, B.; Lane, W.D.; Tang, G.; Su, Y.; Sun, R.; Oomah, B.D.; Wiersma, P.A.; Xiang, Y. Reduced Polyphenol Oxidase Gene Expression and Enzymatic Browning in Potato (Solanum tuberosum L.) with Artificial microRNAs. BMC Plant Biol. 2014, 14, 62. [Google Scholar] [CrossRef]
- Higuera-Rubio, J.M.; Ibarra-Laclette, E.; Reyes-López, M.A.; Sandoval-Castro, E.; Cruz-Mendívil, A.; Vega-García, M.O.; Calderón-Vázquez, C.L. Enzymatic Browning and Genome-Wide Polyphenol Oxidase Gene Identification in Three Contrasting Avocado Accessions. Plant Biotechnol. Rep. 2022, 16, 465–477. [Google Scholar] [CrossRef]
- Li, C.; Li, D.; Li, J.; Shao, F.; Lu, S. Characterization of the Polyphenol Oxidase Gene Family Reveals a Novel microRNA Involved in Posttranscriptional Regulation of PPOs in Salvia Miltiorrhiza. Sci. Rep. 2017, 7, 44622. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, C.; Cui, Q.; Fan, Y.; Zhang, J.; Rao, G. Genome-Wide Analysis of the Polyphenol Oxidase Gene Family in Olea Europaea Provides Insights into the Mechanism of Enzymatic Browning in Olive Fruit. Antioxidants 2023, 12, 1661. [Google Scholar] [CrossRef]
- Pompili, V.; Mazzocchi, E.; Moglia, A.; Acquadro, A.; Comino, C.; Rotino, G.L.; Lanteri, S. Structural and Expression Analysis of Polyphenol Oxidases Potentially Involved in Globe Artichoke (C. cardunculus var. scolymus L.) Tissue Browning. Sci. Rep. 2023, 13, 12288. [Google Scholar] [CrossRef]
- Pourcel, L.; Routaboul, J.-M.; Kerhoas, L.; Caboche, M.; Lepiniec, L.; Debeaujon, I. TRANSPARENT TESTA10 Encodes a Laccase-Like Enzyme Involved in Oxidative Polymerization of Flavonoids in Arabidopsis Seed Coat. Plant Cell 2005, 17, 2966–2980. [Google Scholar] [CrossRef]
- Zhao, P.; Wang, D.; Wang, R.; Kong, N.; Zhang, C.; Yang, C.; Wu, W.; Ma, H.; Chen, Q. Genome-Wide Analysis of the Potato Hsp20 Gene Family: Identification, Genomic Organization and Expression Profiles in Response to Heat Stress. BMC Genom. 2018, 19, 61. [Google Scholar] [CrossRef]
- Jeffares, D.C.; Penkett, C.J.; Bähler, J. Rapidly Regulated Genes Are Intron Poor. Trends Genet. 2008, 24, 375–378. [Google Scholar] [CrossRef]
- Zhao, Z.; Wang, F.; Deng, M.; Fan, G. Identification and Analysis of PPO Gene Family Members in Paulownia Fortunei. Plants 2024, 13, 2033. [Google Scholar] [CrossRef]
- Jiang, Y.; Duan, X.; Joyce, D.; Zhang, Z.; Li, J. Advances in Understanding of Enzymatic Browning in Harvested Litchi Fruit. Food Chem. 2004, 88, 443–446. [Google Scholar] [CrossRef]
- Li, T.T.; Li, Y.R.; Yan, M.L.; Zhang, H.X.; Wang, R.H.; Ke, W.D.; Guo, H.B. Identification and Analysis of PPO Gene Family Members in Lotus (Nelumbo nucifera). J. Agric. Biotechnol. 2022, 30, 38–49. [Google Scholar]
- Wang, J.; Liu, B.; Xiao, Q.; Li, H.; Sun, J. Cloning and Expression Analysis of Litchi (Litchi chinensis Sonn.) Polyphenol Oxidase Gene and Relationship with Postharvest Pericarp Browning. PLoS ONE 2014, 9, e93982. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Shi, Y.-J.; Zhao, Q.; Zhao, K.-J.; Cui, X.-L.; Chen, L.-H.; Yang, H.-B.; Zhang, F.; Mi, J.-X.; Huang, J.-L.; et al. Genome-Wide Investigation and Expression Profiling of Polyphenol Oxidase (PPO) Family Genes Uncover Likely Functions in Organ Development and Stress Responses in Populus Trichocarpa. BMC Genom. 2021, 22, 731. [Google Scholar] [CrossRef]
- Xiao, K.; Liu, X.; Zhang, A.; Zha, D.; Zhu, W.; Tan, F.; Huang, Q.; Zhou, Y.; Zhang, M.; Li, J.; et al. Genome-Wide Identification of Polyphenol Oxidase (PPO) Family Members in Eggplant (Solanum melongena L.) and Their Expression in Response to Low Temperature. Hortic. Environ. Biotechnol. 2022, 63, 747–758. [Google Scholar] [CrossRef]
- Teng, Z.; Luo, Y.; Bornhorst, E.R.; Zhou, B.; Simko, I.; Trouth, F. Identification of Romaine Lettuce (Lactuca sativa Var. Longifolia) Cultivars with Reduced Browning Discoloration for Fresh-Cut Processing. Postharvest. Biol. Technol. 2019, 156, 110931. [Google Scholar] [CrossRef]
- Yu, R.; Song, H.; Chen, Y.; Shi, N.; Shen, H.; Shi, P.; Shu, H.; Kong, X.; Yu, L.; Luo, H. Incorporation of Ascorbic Acid and L-Cysteine in Sodium Carboxymethyl Cellulose Coating Delays Color Deterioration and Extends the Shelf-Life of Fresh-Cut Asparagus Lettuce (Lactuca sativa Var. Angustata). Postharvest. Biol. Technol. 2023, 204, 112419. [Google Scholar] [CrossRef]
- Meade, J.C.; Shah, P.H.; Lushbaugh, W.B. Trichomonas Vaginalis:Analysis of Codon Usage. Exp. Parasitol. 1997, 87, 73–74. [Google Scholar] [CrossRef] [PubMed]
- Min, T.; Niu, L.-F.; Xie, J.; Yi, Y.; Wang, L.; Ai, Y.; Wang, H. Effects of Vacuum Packaging on NAC Gene Expression in Fresh-Cut Lotus Root. J. Am. Soc. Hortic. Sci. 2020, 145, 36–44. [Google Scholar] [CrossRef]
- Han, C.; Zhen, W.; Chen, Q.; Fu, M. UV-C Irradiation Inhibits Surface Discoloration and Delays Quality Degradation of Fresh-Cut Stem Lettuce. LWT 2021, 147, 111533. [Google Scholar] [CrossRef]


| Gene Name | Gene ID | CDS (bp) | Amino Acid | Molecular Weight (KDa) | Average Value of Total Hydrophilicity | Isoelectric Point | Instab-ility Index | Chr | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|
| LsPPO1 | XM_023878429.1 | 1773 | 590 | 66.43 | −0.487 | 5.9 | 44.69 | 2 | Chlo |
| LsPPO2 | XM_042898356.1 | 1913 | 618 | 70.12 | −0.584 | 6.05 | 41.07 | 3 | Chlo |
| LsPPO3 | XM_023907623.3 | 622 | 146 | 15.97 | −0.473 | 4.73 | 51.66 | 4 | Cyto |
| LsPPO4 | XM_023907626.3 | 1960 | 596 | 66.76 | −0.547 | 6.59 | 45.04 | 4 | Chlo |
| LsPPO5 | XM_023907627.3 | 1964 | 591 | 66.47 | −0.544 | 6.29 | 45.02 | 4 | Mito |
| LsPPO6 | XM_023907629.3 | 1974 | 603 | 67.94 | −0.595 | 6.59 | 49.56 | 4 | Chlo |
| LsPPO7 | XM_023909952.3 | 2018 | 609 | 68.52 | −0.451 | 6.27 | 43.13 | 7 | Pero |
| LsPPO8 | XM_023910066.2 | 1974 | 615 | 69.48 | −0.501 | 6.43 | 43.07 | 7 | Chlo |
| LsPPO9 | XM_023910589.3 | 2030 | 611 | 69.8 | −0.453 | 6.04 | 45.87 | 7 | Chlo |
| LsPPO10 | XM_023882917.2 | 1821 | 606 | 69.19 | −0.632 | 5.91 | 50.39 | 8 | Chlo |
| LsPPO11 | XM_023885882.3 | 2130 | 612 | 68.28 | −0.38 | 6 | 39.43 | 9 | Chlo |
| LsPPO12 | XM_023885884.3 | 2132 | 634 | 72.35 | −0.534 | 5.84 | 35.25 | 9 | Cyto |
| LsPPO13 | XM_023885885.3 | 1232 | 303 | 33.74 | −0.358 | 6.86 | 40.67 | 9 | Chlo |
| LsPPO14 | XM_023885921.3 | 2137 | 667 | 75.59 | −0.549 | 6.06 | 32.42 | 9 | Chlo |
| LsPPO15 | XM_023900746.3 | 2053 | 610 | 67.93 | −0.334 | 6.16 | 34.44 | 9 | Mito |
| LsPPO16 | XM_023900747.3 | 2066 | 627 | 71.57 | −0.735 | 8.32 | 44.73 | 9 | Chlo |
| LsPPO17 | XM_023908190.2 | 2112 | 591 | 65.99 | −0.514 | 7.2 | 45 | 9 | Chlo |
| Genes | Gene ID | Forward Primer (5′–3′) | Reverse Primer (5′–3′) |
|---|---|---|---|
| LsPPO1 | XM_023878429.1 | CTCCTGTATCCTACGACCA | GGAACCAGCACTTTTATCACC |
| LsPPO2 | XM_042898356.1 | ACTACTGCATGTTGCCCTCC | TTCATGTATCCTTCCGGGGC |
| LsPPO3 | XM_023907623.3 | GATGAGGATGAGGAGACAGCG | CGTTCCTGTGCTTATGTGGC |
| LsPPO4 | XM_023907626.3 | ATTGCGATGGGGCATACGAT | CCATCAGGTGCATCCCAGTT |
| LsPPO5 | XM_023907627.3 | TTCCACCACAGTCAAACCCC | ACCGAGTTGGACATATGCGG |
| LsPPO6 | XM_023907629.3 | AACAACAAGCAAGCTGTGGC | CCGTGGAACCAGCACTTTGA |
| LsPPO7 | XM_023909952.3 | CAGCGAAAAACCGAAGCCAA | TCTCGGATGCAGAGCTTGTG |
| LsPPO8 | XM_023910066.2 | AGGATGATGTCAGGGCAAGC | TAACATGTCGTCACCAGGGC |
| LsPPO9 | XM_023910589.3 | GGCCTTTACACCACCGTCA | CGCATGTGGAGAAGTTTGGC |
| LsPPO10 | XM_023882917.2 | CATCCGGAATGTCAATCACCT | GTTGCGCTTCATCTGCCTA |
| LsPPO11 | XM_023885882.3 | CCAATGACGCTGTAAGGGGT | GAAGGACTGGGTTGGTAGGC |
| LsPPO12 | XM_023885884.3 | GGAGCTCAGCAGAACGGATT | TTGTTGGTGGTGACTTCGGT |
| LsPPO13 | XM_023885885.3 | CCTCGGTGGTCTCTTTGTGG | AGCAGCACAAACCGAGTGAT |
| LsPPO14 | XM_023885921.3 | TTCCCATCGAAGCACCAGAC | CCGATGATGCAAACACCGGC |
| LsPPO15 | XM_023900746.3 | AGGCCAGCTACAAACAGGAC | ATGTGGCAACTGTGCGAAAC |
| LsPPO16 | XM_023900747.3 | TGGGCAACTTCTACTCTGCG | TCTGTCGGTTCTTTGCCTCC |
| LsPPO17 | XM_023908190.2 | ATCTTCAAGTGGATTTCCCGAT | CTCCCCAGGATTCTCTCGT |
| LsActin | XM_023878805.3 | AGCAACTGGGATGACATGGA | GGGTTGAGAGGTGCCTCAGT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, M.; Tang, Y.; Yang, Y.; Luo, J.; Gao, J. Identification and Expression Analysis of Polyphenol Oxidase Gene Family Members in Response to Wound Stress in Lettuce (Lactuca sativa L.). Plants 2025, 14, 972. https://doi.org/10.3390/plants14060972
Guo M, Tang Y, Yang Y, Luo J, Gao J. Identification and Expression Analysis of Polyphenol Oxidase Gene Family Members in Response to Wound Stress in Lettuce (Lactuca sativa L.). Plants. 2025; 14(6):972. https://doi.org/10.3390/plants14060972
Chicago/Turabian StyleGuo, Mei, Yueming Tang, Yiwen Yang, Jinghong Luo, and Jia Gao. 2025. "Identification and Expression Analysis of Polyphenol Oxidase Gene Family Members in Response to Wound Stress in Lettuce (Lactuca sativa L.)" Plants 14, no. 6: 972. https://doi.org/10.3390/plants14060972
APA StyleGuo, M., Tang, Y., Yang, Y., Luo, J., & Gao, J. (2025). Identification and Expression Analysis of Polyphenol Oxidase Gene Family Members in Response to Wound Stress in Lettuce (Lactuca sativa L.). Plants, 14(6), 972. https://doi.org/10.3390/plants14060972






