Development of Protoplast-Based Gene Editing System for Areca Palm
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
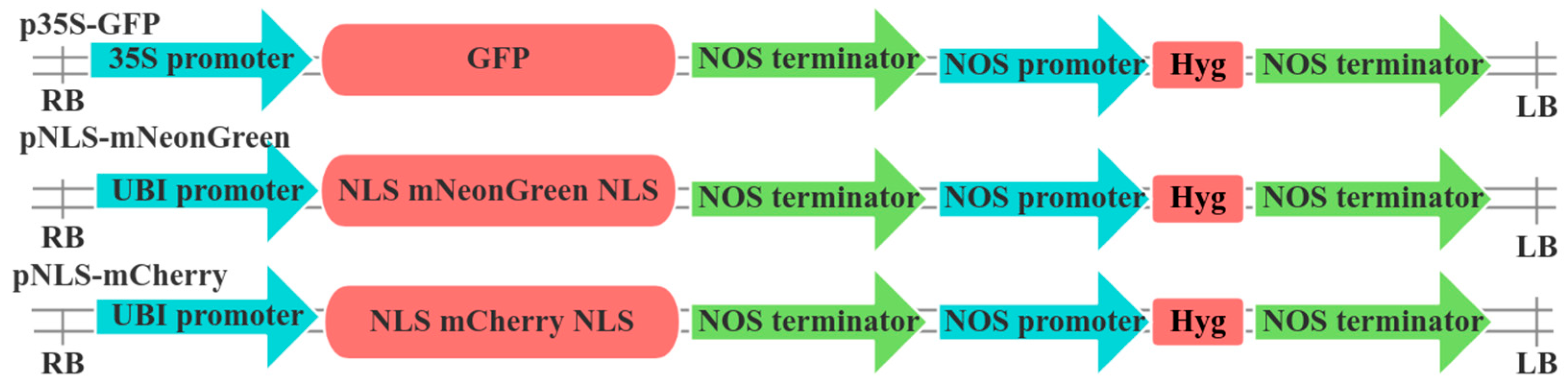
2.2. Vector Construction
2.3. Protoplast Isolation
2.4. Protoplast Yield and Viability Assay
2.5. Protoplast Transformation
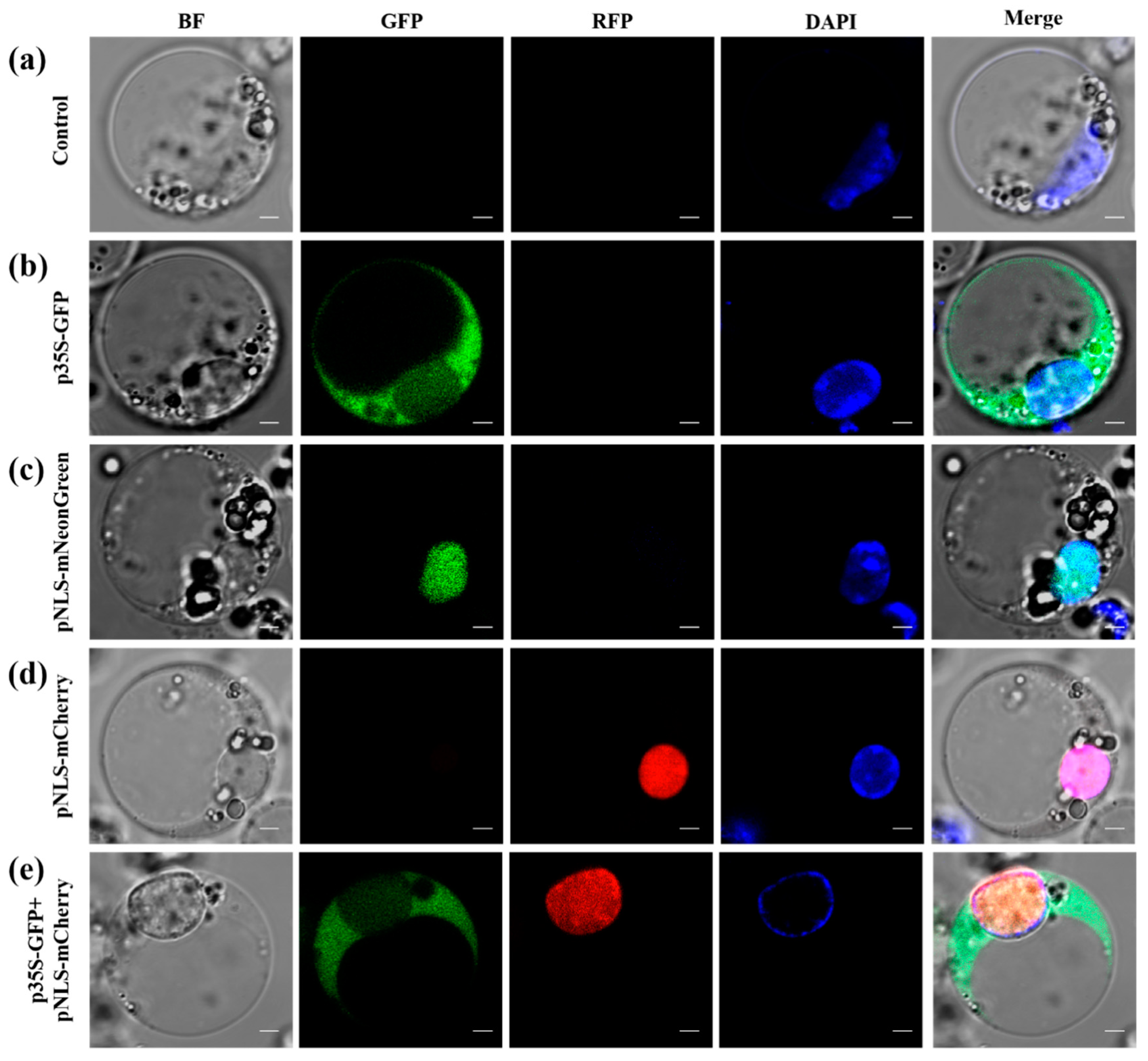
2.6. Subcellular Localization
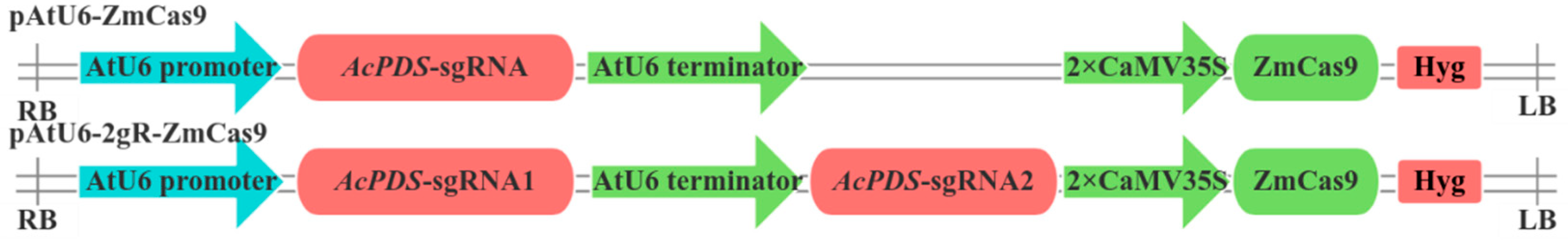
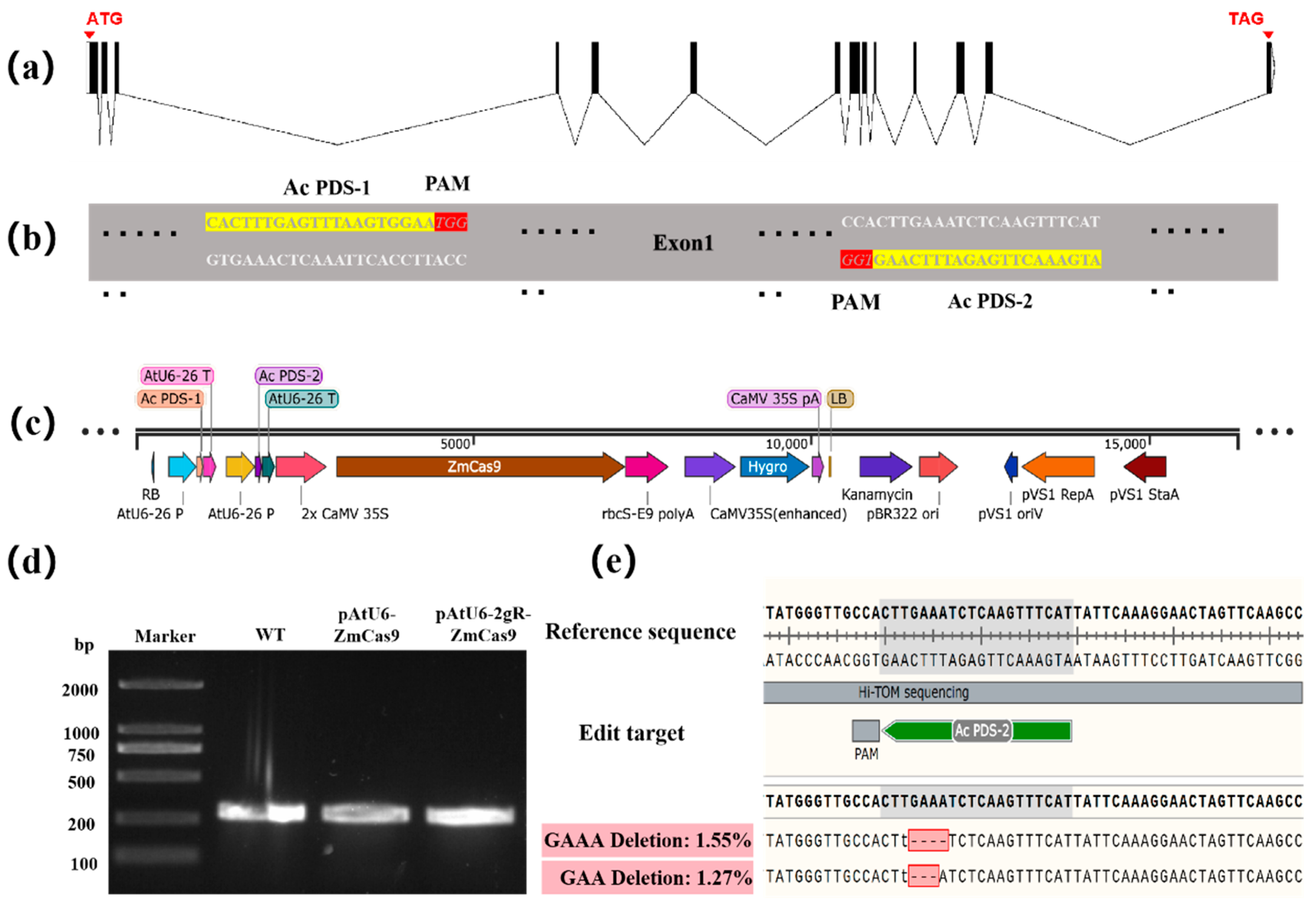
2.7. Gene Editing Targeting AcPDS Gene Using the CRISPR-Cas9 System
2.8. Statistical Analysis
3. Results
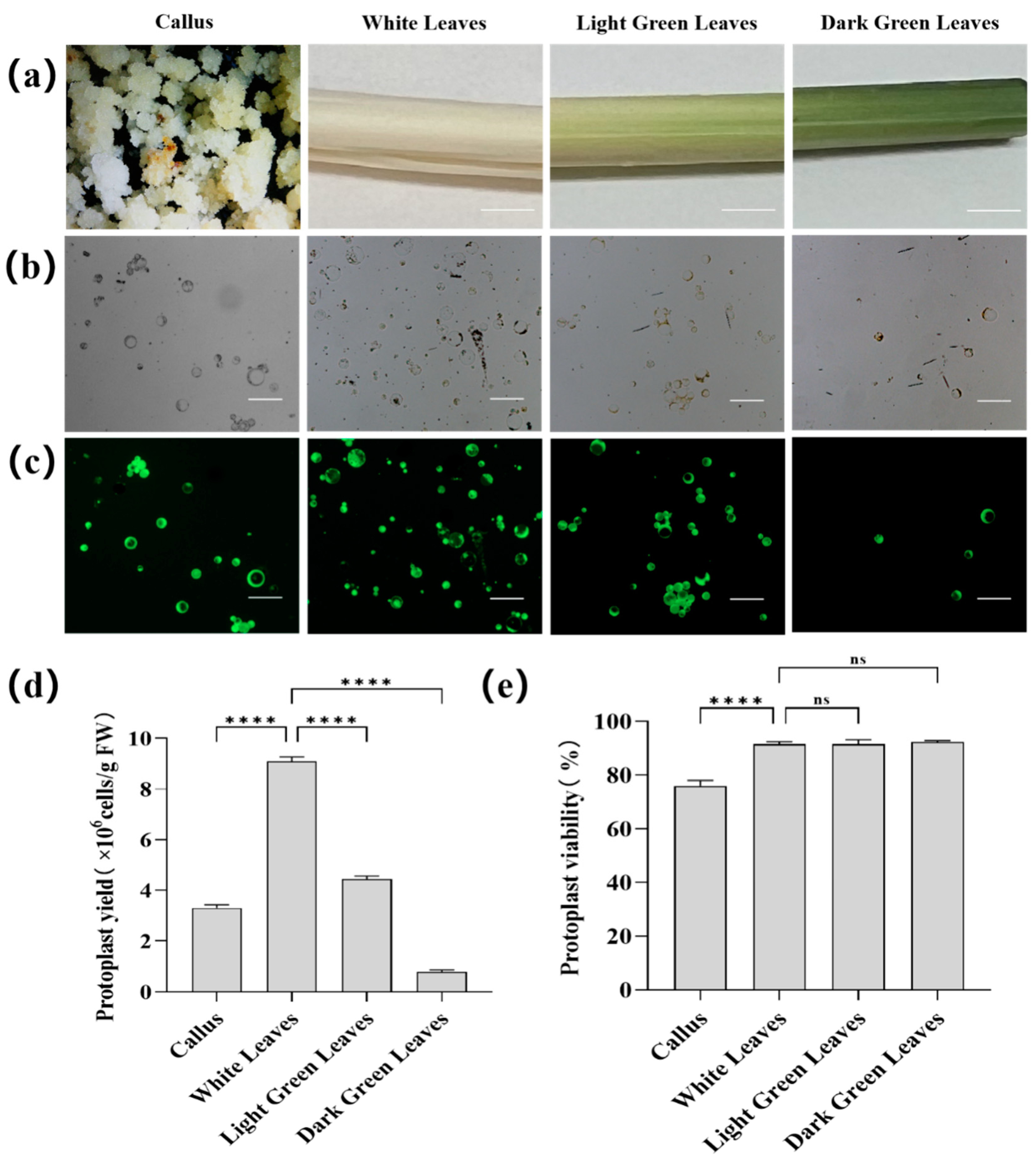
3.1. Protoplast Isolation from Areca Palm Tissues
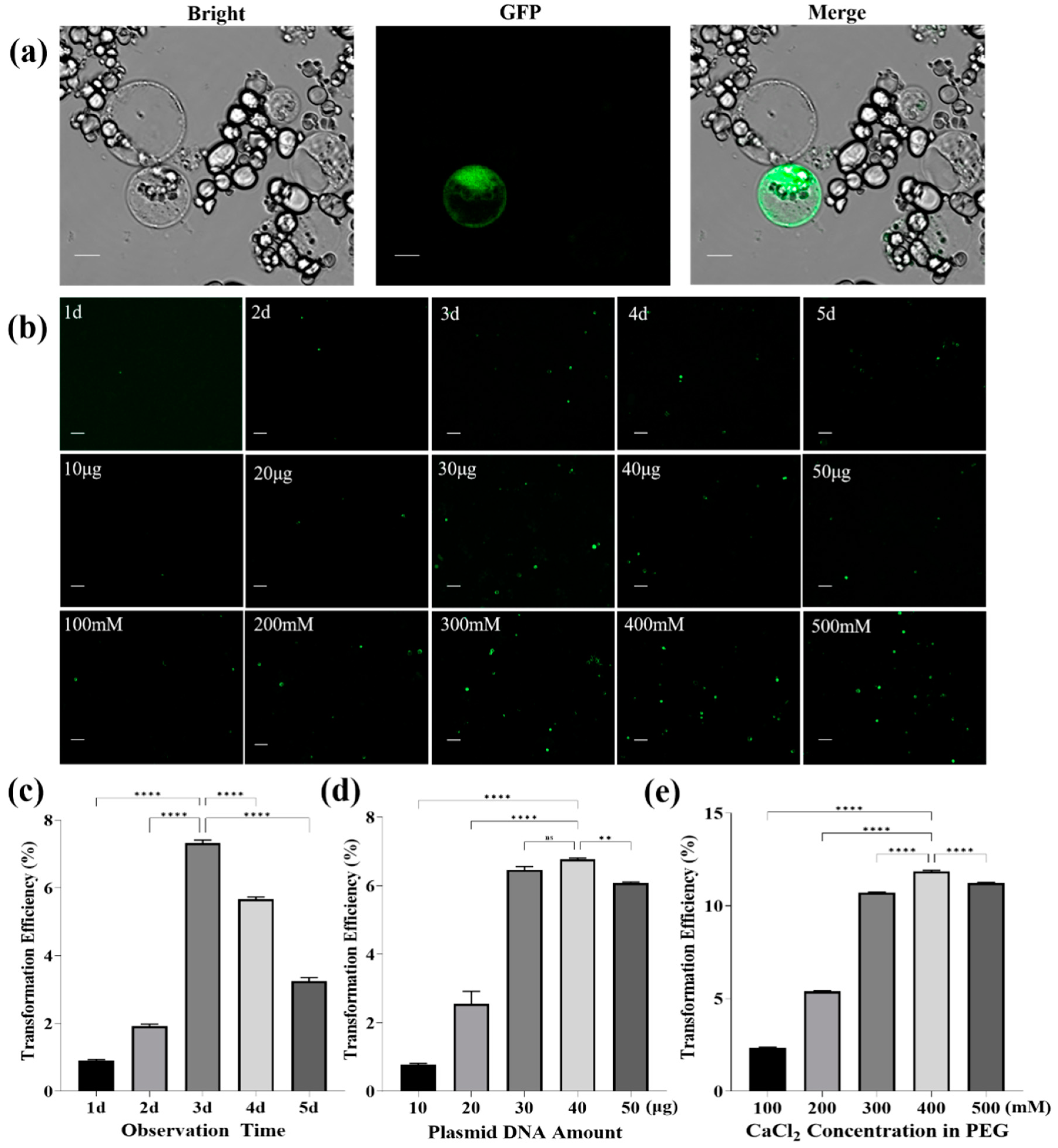
3.2. Optimization of PEG-Mediated Protoplast Transformation
3.3. Subcellular Localization Analysis
3.4. CRISPR/Cas9-Mediated Gene Editing
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Ding, H.; Zhou, G.; Zhao, L.; Li, X.; Wang, Y.; Xia, C.; Xia, Z.; Wan, Y. Genome-Wide Association Analysis of Fruit Shape-Related Traits in Areca catechu. Int. J. Mol. Sci. 2023, 24, 4686. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Huang, L.; Xu, C.; Qi, L.; Wu, Z.; Li, J.; Chen, H.; Wu, Y.; Fu, T.; Zhu, H.; et al. Chromosome-Scale Genome Assembly of Areca Palm (Areca catechu). Mol. Ecol. Resour. 2021, 21, 2504–2519. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; Liu, Y.-J.; Wu, N.; Sun, T.; He, X.-Y.; Gao, Y.-X.; Wu, C.-J. Areca catechu L. (Arecaceae): A Review of Its Traditional Uses, Botany, Phytochemistry, Pharmacology and Toxicology. J. Ethnopharmacol. 2015, 164, 340–356. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cao, X.; Jia, X.; Liu, L.; Cao, H.; Qin, W.; Li, M. Iron Deficiency Leads to Chlorosis Through Impacting Chlorophyll Synthesis and Nitrogen Metabolism in Areca catechu L. Front. Plant Sci. 2021, 12, 710093. [Google Scholar] [CrossRef]
- Zhou, G.; Yin, H.; Chen, F.; Wang, Y.; Gao, Q.; Yang, F.; He, C.; Zhang, L.; Wan, Y. The Genome of Areca catechu Provides Insights into Sex Determination of Monoecious Plants. New Phytol. 2022, 236, 2327–2343. [Google Scholar] [CrossRef]
- Davey, M.R.; Anthony, P.; Power, J.B.; Lowe, K.C. Plant Protoplasts: Status and Biotechnological Perspectives. Biotechnol. Adv. 2005, 23, 131–171. [Google Scholar] [CrossRef]
- Chen, K.; Chen, J.; Pi, X.; Huang, L.-J.; Li, N. Isolation, Purification, and Application of Protoplasts and Transient Expression Systems in Plants. Int. J. Mol. Sci. 2023, 24, 16892. [Google Scholar] [CrossRef]
- Naing, A.H.; Adedeji, O.S.; Kim, C.K. Protoplast Technology in Ornamental Plants: Current Progress and Potential Applications on Genetic Improvement. Sci. Hortic. 2021, 283, 110043. [Google Scholar] [CrossRef]
- Li, S.; Zhao, R.; Ye, T.; Guan, R.; Xu, L.; Ma, X.; Zhang, J.; Xiao, S.; Yuan, D. Isolation, Purification and PEG-Mediated Transient Expression of Mesophyll Protoplasts in Camellia Oleifera. Plant Methods 2022, 18, 141. [Google Scholar] [CrossRef]
- Ozyigit, I.I. Gene Transfer to Plants by Electroporation: Methods and Applications. Mol. Biol. Rep. 2020, 47, 3195–3210. [Google Scholar] [CrossRef]
- Reich, T.J.; Iyer, V.N.; Miki, B.L. Efficient Transformation of Alfalfa Protoplasts by the Intranuclear Microinjection of Ti Plasmids. Bio/Technology 1986, 4, 1001–1004. [Google Scholar] [CrossRef]
- Liu, Z.; Friesen, T.L. Polyethylene Glycol (PEG)-Mediated Transformation in Filamentous Fungal Pathogens. In Plant Fungal Pathogens: Methods and Protocols; Bolton, M.D., Thomma, B.P.H.J., Eds.; Humana Press: Totowa, NJ, USA, 2012; pp. 365–375. ISBN 978-1-61779-501-5. [Google Scholar]
- Herzog, R.; Solovyeva, I.; Bölker, M.; Lugones, L.G.; Hennicke, F. Exploring Molecular Tools for Transformation and Gene Expression in the Cultivated Edible Mushroom Agrocybe aegerita. Mol. Genet. Genom. MGG 2019, 294, 663–677. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, L.M.; Kaur, P.; Stanton, D.; Grosser, J.W.; Dutt, M. A Cationic Lipid Mediated CRISPR/Cas9 Technique for the Production of Stable Genome Edited Citrus Plants. Plant Methods 2022, 18, 33. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, L.; He, C.; Luo, H. An Efficient Transient Mesophyll Protoplast System for Investigation of the Innate Immunity Responses in the Rubber Tree (Hevea brasiliensis). Plant Cell Tissue Organ Cult. PCTOC 2016, 126, 281–290. [Google Scholar] [CrossRef]
- Masani, M.Y.A.; Noll, G.A.; Parveez, G.K.A.; Sambanthamurthi, R.; Prüfer, D. Efficient Transformation of Oil Palm Protoplasts by PEG-Mediated Transfection and DNA Microinjection. PLoS ONE 2014, 9, e96831. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, L.; Liu, H.; Gou, B.; Hu, W.; Qin, L.; Shen, W.; Wang, A.; Cui, H.; Dai, Z. Direct Leaf-Peeling Method for Areca Protoplasts: A Simple and Efficient System for Protoplast Isolation and Transformation in Areca Palm (Areca catechu). BMC Plant Biol. 2023, 23, 56. [Google Scholar] [CrossRef]
- Yu, X.; Yu, J.; Lu, Y.; Li, W.; Huo, G.; Zhang, J.; Li, Y.; Zhao, J.; Li, J. An Efficient and Universal Protoplast-Based Transient Gene Expression System for Genome Editing in Brassica Crops. Hortic. Plant J. 2024, 10, 983–994. [Google Scholar] [CrossRef]
- Gaillochet, C.; Peña Fernández, A.; Goossens, V.; D’Halluin, K.; Drozdzecki, A.; Shafie, M.; Van Duyse, J.; Van Isterdael, G.; Gonzalez, C.; Vermeersch, M.; et al. Systematic Optimization of Cas12a Base Editors in Wheat and Maize Using the ITER Platform. Genome Biol. 2023, 24, 6. [Google Scholar] [CrossRef]
- Hiranniramol, K.; Chen, Y.; Liu, W.; Wang, X. Generalizable sgRNA Design for Improved CRISPR/Cas9 Editing Efficiency. Bioinformatics 2020, 36, 2684–2689. [Google Scholar] [CrossRef]
- Karvelis, T.; Gasiunas, G.; Young, J.; Bigelyte, G.; Silanskas, A.; Cigan, M.; Siksnys, V. Rapid Characterization of CRISPR-Cas9 Protospacer Adjacent Motif Sequence Elements. Genome Biol. 2015, 16, 253. [Google Scholar] [CrossRef]
- Zhang, Y.; Massel, K.; Godwin, I.D.; Gao, C. Applications and Potential of Genome Editing in Crop Improvement. Genome Biol. 2018, 19, 210. [Google Scholar] [CrossRef] [PubMed]
- Xiang, X.; Corsi, G.I.; Anthon, C.; Qu, K.; Pan, X.; Liang, X.; Han, P.; Dong, Z.; Liu, L.; Zhong, J.; et al. Enhancing CRISPR-Cas9 gRNA Efficiency Prediction by Data Integration and Deep Learning. Nat. Commun. 2021, 12, 3238. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, G.W.; Kogenaru, M.; Keegan, S.; Haase, M.A.B.; Kagermazova, L.; Arias, M.A.; Onyebeke, K.; Adams, S.; Beyer, D.K.; Fenyö, D.; et al. Engineered Transcription-Associated Cas9 Targeting in Eukaryotic Cells. Nat. Commun. 2024, 15, 10287. [Google Scholar] [CrossRef] [PubMed]
- Sagawa, C.H.D.; Thomson, G.; Mermaz, B.; Vernon, C.; Liu, S.; Jacob, Y.; Irish, V.F. An Efficient Multiplex Approach to CRISPR/Cas9 Gene Editing in Citrus. Plant Methods 2024, 20, 148. [Google Scholar] [CrossRef]
- Bahariah, B.; Masani, M.Y.A.; Fizree, M.P.M.A.A.; Rasid, O.A.; Parveez, G.K.A. Multiplex CRISPR/Cas9 Gene-Editing Platform in Oil Palm Targeting Mutations in EgFAD2 and EgPAT Genes. J. Genet. Eng. Biotechnol. 2023, 21, 3. [Google Scholar] [CrossRef]
- Fan, Y.; Xin, S.; Dai, X.; Yang, X.; Huang, H.; Hua, Y. Efficient Genome Editing of Rubber Tree (Hevea Brasiliensis) Protoplasts Using CRISPR/Cas9 Ribonucleoproteins. Ind. Crops Prod. 2020, 146, 112146. [Google Scholar] [CrossRef]
- Zhang, D.; Li, Z.; Htwe, Y.M.; Shi, P.; Wei, X.; Nie, H.; Nin, J.; Wu, L.; Khan, F.S.; Yu, Q.; et al. Insights into the Developmental Trajectories of Zygotic Embryo, Embryogenic Callus and Somatic Embryo in Coconut by Single-Cell Transcriptomic Analysis. Ind. Crops Prod. 2024, 212, 118338. [Google Scholar] [CrossRef]
- Costa, N.; Carvalho, C.; Clarindo, W. Improved Procedures to Assess Plant Protoplast Viability: Evidencing Cytological and Genomic Damage. Cytologia 2018, 83, 397–405. [Google Scholar] [CrossRef]
- Yoo, S.-D.; Cho, Y.-H.; Sheen, J. Arabidopsis Mesophyll Protoplasts: A Versatile Cell System for Transient Gene Expression Analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Sun, T.; Liu, Q.; Chen, X.; Hu, F.; Wang, K. Hi-TOM 2.0: An Improved Platform for High-Throughput Mutation Detection. Sci. China Life Sci. 2024, 67, 1532–1534. [Google Scholar] [CrossRef]
- Chen, S.; Tao, L.; Zeng, L.; Vega-Sanchez, M.E.; Umemura, K.; Wang, G.-L. A Highly Efficient Transient Protoplast System for Analyzing Defence Gene Expression and Protein-Protein Interactions in Rice. Mol. Plant Pathol. 2006, 7, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Rong, H.; Feng, Y.; Xin, Y.; Luan, X.; Zhou, Q.; Xu, M.; Xu, L. Protoplast Isolation and Transient Transformation System for Ginkgo biloba L. Front. Plant Sci. 2023, 14, 1145754. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-C.; Li, W.; Chen, H.; Li, Q.; Sun, Y.-H.; Shi, R.; Lin, C.-Y.; Wang, J.P.; Chen, H.-C.; Chuang, L.; et al. A Simple Improved-Throughput Xylem Protoplast System for Studying Wood Formation. Nat. Protoc. 2014, 9, 2194–2205. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhu, H.; Ren, Y.; Feng, C.; Ye, Z.; Cai, H.; Wan, X.; Peng, C. Efficient Isolation and Purification of Tissue-Specific Protoplasts from Tea Plants (Camellia sinensis (L.) O. Kuntze). Plant Methods 2021, 17, 84. [Google Scholar] [CrossRef]
- Guo, Q.; Wang, Y.; Zou, J.; Jing, H.; Li, D. Efficient Isolation and Transformation of Protoplasts in Coconut Endosperm and Leaves for Gene Function Studies. Trop. Plants 2023, 2, 16. [Google Scholar] [CrossRef]
- Adjei, M.O.; Zhao, H.; Tao, X.; Yang, L.; Deng, S.; Li, X.; Mao, X.; Li, S.; Huang, J.; Luo, R.; et al. Using A Protoplast Transformation System to Enable Functional Studies in Mangifera indica L. Int. J. Mol. Sci. 2023, 24, 11984. [Google Scholar] [CrossRef]
- Yang, W.; Ren, J.; Liu, W.; Liu, D.; Xie, K.; Zhang, F.; Wang, P.; Guo, W.; Wu, X. An Efficient Transient Gene Expression System for Protein Subcellular Localization Assay and Genome Editing in Citrus Protoplasts. Hortic. Plant J. 2023, 9, 425–436. [Google Scholar] [CrossRef]
- Shao, Y.; Mu, D.; Pan, L.; Wilson, I.W.; Zheng, Y.; Zhu, L.; Lu, Z.; Wan, L.; Fu, J.; Wei, S.; et al. Optimization of Isolation and Transformation of Protoplasts from Uncaria rhynchophylla and Its Application to Transient Gene Expression Analysis. Int. J. Mol. Sci. 2023, 24, 3633. [Google Scholar] [CrossRef]
- Yang, P.; Sun, Y.; Sun, X.; Li, Y.; Wang, L. Optimization of Preparation and Transformation of Protoplasts from Populus Simonii × P. Nigra Leaves and Subcellular Localization of the Major Latex Protein 328 (MLP328). Plant Methods 2024, 20, 3. [Google Scholar] [CrossRef]
- Zhang, Y.; Su, J.; Duan, S.; Ao, Y.; Dai, J.; Liu, J.; Wang, P.; Li, Y.; Liu, B.; Feng, D.; et al. A Highly Efficient Rice Green Tissue Protoplast System for Transient Gene Expression and Studying Light/Chloroplast-Related Processes. Plant Methods 2011, 7, 30. [Google Scholar] [CrossRef]
- Liu, X.; Yang, J.; Song, Y.; Zhang, X.; Wang, X.; Wang, Z. Effects of sgRNA Length and Number on Gene Editing Efficiency and Predicted Mutations Generated in Rice. Crop J. 2022, 10, 577–581. [Google Scholar] [CrossRef]
- Masani, M.Y.A.; Noll, G.; Parveez, G.K.A.; Sambanthamurthi, R.; Prüfer, D. Regeneration of Viable Oil Palm Plants from Protoplasts by Optimizing Media Components, Growth Regulators and Cultivation Procedures. Plant Sci. 2013, 210, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Chabane, D.; Assani, A.; Bouguedoura, N.; Haïcour, R.; Ducreux, G. Induction of Callus Formation from Difficile Date Palm Protoplasts by Means of Nurse Culture. Comptes Rendus Biol. 2007, 330, 392–401. [Google Scholar] [CrossRef] [PubMed]
| Plasmid | Primer | Sequences (5′-3′) |
|---|---|---|
| pNLS-mNeon | p6-UBI F | gagctcggtacccggggatctgcagcgtgacccggtcgtg |
| UBI-NLS R | ccatggtggcctgcagaagtaacaccaaacaacagg | |
| UBI-NLS F | acttctgcaggccaccatggactataaggaccac | |
| NLS-mNeon R | tgctcaccatggctgctgggactccgtggata | |
| NLS-mNeon F | cccagcagccatggtgagcaagggcgaggagg | |
| mNeon-NLS R | ccggccttttcttgtacagctcgtccatgcc | |
| mNeon-NLS F | gctgtacaagaaaaggccggcggccacgaa | |
| NLS-p6 R | acgacggccagtgccaagctactccccatgggaattcgta | |
| pNLS-mCherry | p6-UBI F | acgacggccagtgccaagcttgcagcgtgacccggtcgtg |
| UBI-NLS R | ccatggtggcctgcagaagtaacaccaaac | |
| UBI-NLS F | acttctgcaggccaccatggactataagga | |
| NLS-mCherry R | tgctcaccatggctgctgggactccgtgga | |
| NLS-mCherry F | cccagcagccatggtgagcaagggcgagga | |
| mCherry-NLS R | ccggccttttcttatacagctcgtccatgc | |
| mCherry-NLS F | gctgtataagaaaaggccggcggccacgaa | |
| NLS-p6 R | gagctcggtacccggggatccgatcgggaggatcttactt |
| Plasmid | sgRNA | Sequences (5′-3′) |
|---|---|---|
| pAtU6-ZmCas9 | AcPDS-sgRNA | caaagtggagatggatccaa |
| pAtU6-2gR-ZmCas9 | AcPDS-sgRNA1 | cactttgagtttaagtggaa |
| AcPDS-sgRNA2 | atgaaacttgagatttcaag |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nie, H.; Batool, S.; Htwe, Y.M.; Fang, X.; Zhang, D.; Shi, P.; Li, Z.; Ma, M.; Su, H.; Yu, Q.; et al. Development of Protoplast-Based Gene Editing System for Areca Palm. Plants 2025, 14, 832. https://doi.org/10.3390/plants14060832
Nie H, Batool S, Htwe YM, Fang X, Zhang D, Shi P, Li Z, Ma M, Su H, Yu Q, et al. Development of Protoplast-Based Gene Editing System for Areca Palm. Plants. 2025; 14(6):832. https://doi.org/10.3390/plants14060832
Chicago/Turabian StyleNie, Hao, Saira Batool, Yin Min Htwe, Xiaomeng Fang, Dapeng Zhang, Peng Shi, Zhiying Li, Mingjun Ma, Hanlu Su, Qun Yu, and et al. 2025. "Development of Protoplast-Based Gene Editing System for Areca Palm" Plants 14, no. 6: 832. https://doi.org/10.3390/plants14060832
APA StyleNie, H., Batool, S., Htwe, Y. M., Fang, X., Zhang, D., Shi, P., Li, Z., Ma, M., Su, H., Yu, Q., He, X., & Wang, Y. (2025). Development of Protoplast-Based Gene Editing System for Areca Palm. Plants, 14(6), 832. https://doi.org/10.3390/plants14060832






