Establishment of a Genetic Transformation and Gene Editing Method by Floral Dipping in Descurainia sophia
Abstract
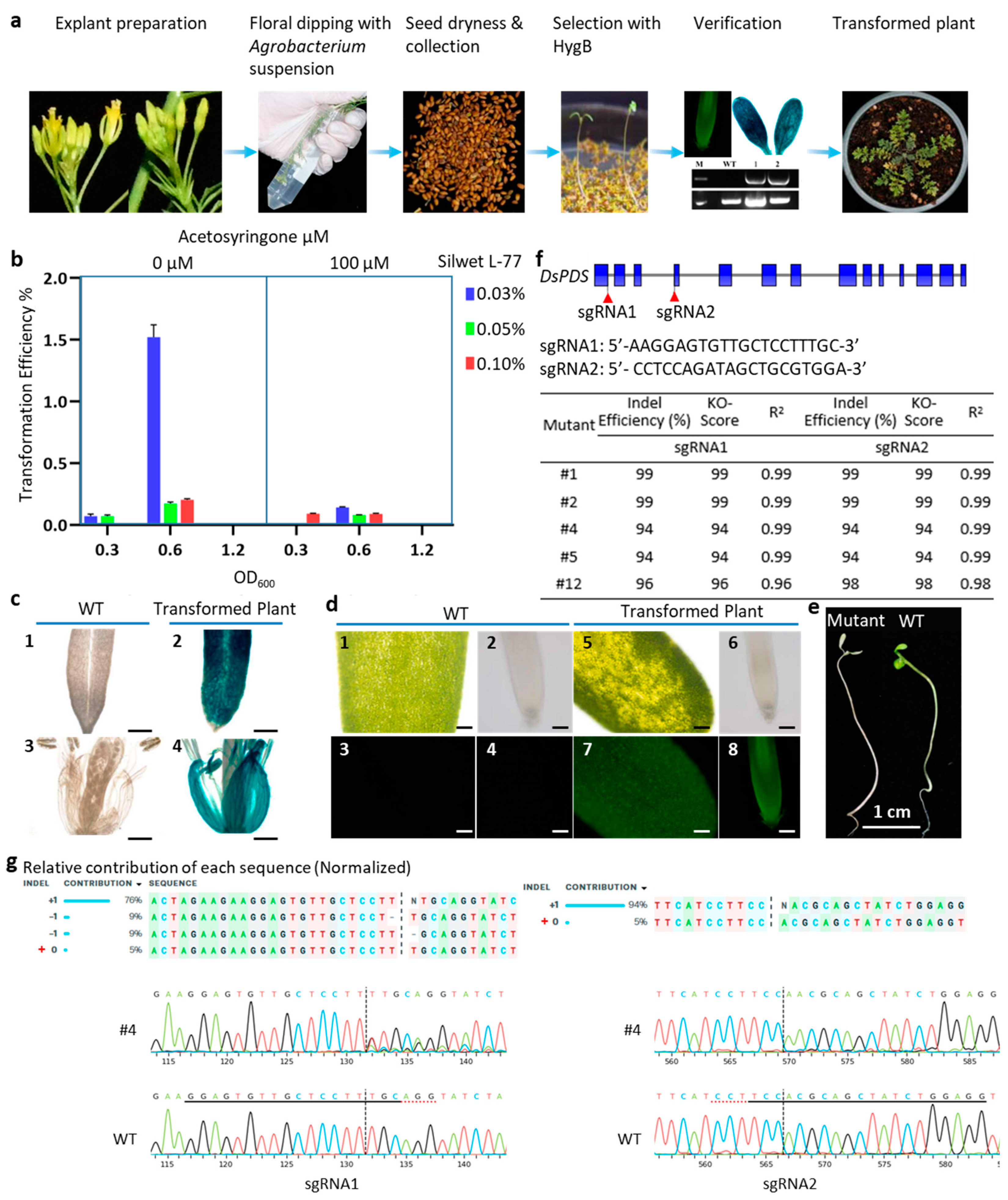
1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. Plant Materials and Growth Conditions
3.2. Vector Construction
3.3. A. tumefaciens-Mediated Delivery System of D. sophia
3.4. GFP Detection and GUS Staining
3.5. Evaluation of the Editing Efficiency in D. sophia
3.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Khan, M.; Wang, N. Descurainia sophia (L.): A weed with multiple medicinal uses. Punjab Univ. J. Zool. 2012, 27, 45–51. [Google Scholar]
- Nimrouzi, M.; Zarshenas, M.M. Phytochemical and pharmacological aspects of Descurainia sophia Webb ex Prantl: Modern and traditional applications. Avicenna J. Phytomed. 2016, 6, 266–272. [Google Scholar] [PubMed]
- Hsieh, P.C.; Kuo, C.Y.; Lee, Y.H.; Wu, Y.K.; Yang, M.C.; Tzeng, I.S.; Lan, C.C. Therapeutic effects and mechanisms of actions of Descurainia sophia. Int. J. Med. Sci. 2020, 17, 2163–2170. [Google Scholar] [CrossRef]
- HadiNezhad, M.; Rowland, O.; Hosseinian, F. The Fatty Acid Profile and Phenolic Composition of Descurainia sophia Seeds Extracted by Supercritical CO2. J. Am. Oil Chem. Soc. 2015, 92, 1379–1390. [Google Scholar] [CrossRef]
- Tavakoli, R.; Salarian, R. Steroids and Phenolic Compounds from Descurainia sophia. Chem. Nat. Compd. 2017, 53, 192–193. [Google Scholar] [CrossRef]
- Mohamed, N.H.; Mahrous, A.E. Chemical Constituents of Descurainia sophia L. and its Biological Activity. Rec. Nat. Prod. 2009, 3, 58. [Google Scholar]
- Bekker, N.P.; Ul’chenko, N.T.; Glushenkova, A.I. Lipids from Descurainia Sophia Seeds. Chem. Nat. Compd. 2005, 41, 346–347. [Google Scholar] [CrossRef]
- Gong, J.H.; Zhang, Y.L.; He, J.L.; Zheng, X.K.; Feng, W.S.; Wang, X.L.; Kuang, H.X.; Li, C.G.; Cao, Y.G. Extractions of oil from Descurainia sophia seed using supercritical CO2, chemical compositions by GC-MS and evaluation of the anti-tussive, expectorant and anti-asthmatic activities. Molecules 2015, 20, 13296–13312. [Google Scholar] [CrossRef]
- Park, J.S.; Lim, C.J.; Bang, O.S.; Kim, N.S. Ethanolic extract of Descurainia sophia seeds sensitizes A549 human lung cancer cells to TRAIL cytotoxicity by upregulating death receptors. BMC Complement. Altern. Med. 2016, 16, 115. [Google Scholar] [CrossRef]
- Dastres, E.; Jahangiri, E.; Edalat, M.; Zamani, A.; Amiri, M.; Pourghasemi, H.R. Habitat suitability modeling of Descurainia sophia medicinal plant using three bivariate models. Environ. Monit. Assess. 2023, 195, 392. [Google Scholar] [CrossRef]
- Feng, W.S.; Li, C.G.; Zheng, X.K.; Li, L.L.; Chen, W.J.; Zhang, Y.L.; Cao, Y.G.; Gong, J.H.; Kuang, H.X. Three new sulphur glycosides from the seeds of Descurainia sophia. Nat. Prod. Res. 2016, 30, 1675–1681. [Google Scholar] [CrossRef]
- Zeng, M.; Zhang, L.; Li, M.; Zhang, B.; Feng, W.; Kuang, H.-X.; Zheng, X. Mechanism of the diuretic activity of Descurainia sophia seed. Bangladesh J. Pharmacol. 2018, 13, 157–2018. [Google Scholar] [CrossRef]
- Ke, Y.; Wang, M.; Li, Y.; Shan, Z.; Mi, W.; Yuan, P.; Feng, W.; Zheng, X. Oligosaccharides composition of Descurainiae sophia exerts anti-heart failure by improving heart function and water-liquid metabolism in rats with heart failure. Biomed. Pharmacother. 2020, 129, 110487. [Google Scholar] [CrossRef]
- Sparrow, P.A.C.; Goldsack, C.M.P.; Østergaard, L. Transformation Technology in the Brassicaceae. In Genetics and Genomics of the Brassicaceae; Schmidt, R., Bancroft, I., Eds.; Springer: New York, NY, USA, 2011; pp. 505–525. [Google Scholar]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Hu, D.; Bent, A.F.; Hou, X.; Li, Y. Agrobacterium-mediated vacuum infiltration and floral dip transformation of rapid-cycling Brassica rapa. BMC Plant Biol. 2019, 19, 246. [Google Scholar] [CrossRef]
- Verma, S.S.; Chinnusamy, V.; Bansa, K.C. A Simplified Floral Dip Method for Transformation of Brassica napus and B. carinata. J. Plant Biochem. Biotechnol. 2008, 17, 197–200. [Google Scholar] [CrossRef]
- Mishra, R.; Agarwal, P.; Mohanty, A. Applications of Genome Editing Techniques for the Improvement of Medicinal Plants. In Phytochemical Genomics: Plant Metabolomics and Medicinal Plant Genomics; Swamy, M.K., Kumar, A., Eds.; Springer Nature: Singapore, 2022; pp. 545–569. [Google Scholar]
- Arshid Shabir, P. Chapter 9—CRISPR/Cas9-mediated genome editing in medicinal and aromatic plants: Developments and applications. In Medicinal and Aromatic Plants; Aftab, T., Hakeem, K.R., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 209–221. [Google Scholar]
- Feng, S.; Song, W.; Fu, R.; Zhang, H.; Xu, A.; Li, J. Application of the CRISPR/Cas9 system in Dioscorea zingiberensis. Plant Cell Tiss. Org. Cult. 2018, 135, 133–141. [Google Scholar] [CrossRef]
- Zhou, Z.; Tan, H.; Li, Q.; Chen, J.; Gao, S.; Wang, Y.; Chen, W.; Zhang, L. CRISPR/Cas9-mediated efficient targeted mutagenesis of RAS in Salvia miltiorrhiza. Phytochemistry 2018, 148, 63–70. [Google Scholar] [CrossRef]
- Zakaria, M.M.; Schemmerling, B.; Ober, D. CRISPR/Cas9-Mediated Genome Editing in Comfrey (Symphytum officinale) Hairy Roots Results in the Complete Eradication of Pyrrolizidine Alkaloids. Molecules 2021, 26, 1498. [Google Scholar] [CrossRef]
- Ziemienowicz, A. Agrobacterium-mediated plant transformation: Factors, applications and recent advances. Biocatal. Agric. Biotech. 2014, 3, 95–102. [Google Scholar] [CrossRef]
- Liu, S.; Ma, J.; Liu, H.; Guo, Y.; Li, W.; Niu, S. An efficient system for Agrobacterium-mediated transient transformation in Pinus tabuliformis. Plant Methods 2020, 16, 52. [Google Scholar] [CrossRef]
- Lv, Q.; Chen, C.; Xu, Y.; Hu, S.; Wang, L.; Sun, K.; Chen, X.; Li, X. Optimization of Agrobacterium tumefaciens-Mediated Transformation Systems in Tea Plant (Camellia sinensis). Hortic. Plant J. 2017, 3, 105–109. [Google Scholar] [CrossRef]
- Kumar, R.; Mamrutha, H.M.; Kaur, A.; Venkatesh, K.; Sharma, D.; Singh, G.P. Optimization of Agrobacterium-mediated transformation in spring bread wheat using mature and immature embryos. Mol. Biol. Rep. 2019, 46, 1845–1853. [Google Scholar] [CrossRef]
- Sheikholeslam, S.N.; Weeks, D.P. Acetosyringone promotes high efficiency transformation of Arabidopsis thaliana explants by Agrobacterium tumefaciens. Plant Mol. Biol. 1987, 8, 291–298. [Google Scholar] [CrossRef]
- Godwin, I.; Todd, G.; Ford-Lloyd, B.; Newbury, H.J. The effects of acetosyringone and pH on Agrobacterium-mediated transformation vary according to plant species. Plant Cell Rep. 1991, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Jia, Y.; Cao, Y.; Wang, Y. Improving T-DNA Transfer to Tamarix hispida by Adding Chemical Compounds During Agrobacterium tumefaciens Culture. Front. Plant Sci. 2020, 11, 501358. [Google Scholar] [CrossRef]
- Desfeux, C.; Clough, S.J.; Bent, A.F. Female reproductive tissues are the primary target of Agrobacterium-mediated transformation by the Arabidopsis floral-dip method. Plant Physiol. 2000, 123, 895–904. [Google Scholar] [CrossRef]
- Bastaki, N.K.; Cullis, C.A. Floral-dip transformation of flax (Linum usitatissimum) to generate transgenic progenies with a high transformation rate. J. Vis. Exp. 2014, 94, e52189. [Google Scholar]
- Purwantoro, A.; Irsyadi, M.B.; Sawitri, W.D.; Fatumi, N.C.; Fajrina, S.N. Efficient floral dip transformation method using Agrobacterium tumefaciens on Cosmos sulphureus Cav. Saudi J. Biol. Sci. 2023, 30, 103702. [Google Scholar] [CrossRef]
- Jain, N.; Khurana, P.; Khurana, J.P. A rapid and efficient protocol for genotype-independent, Agrobacterium-mediated transformation of indica and japonica rice using mature seed-derived embryogenic calli. Plant Cell Tiss. Org. Cult. 2022, 151, 59–73. [Google Scholar] [CrossRef]
- Bélanger, J.G.; Copley, T.R.; Hoyos-Villegas, V.; Charron, J.-B.; O’Donoughue, L. A comprehensive review of in planta stable transformation strategies. Plant Methods 2024, 20, 79. [Google Scholar] [CrossRef]
- Ziemienowicz, A. Plant selectable markers and reporter genes. Acta Physiol. Plant. 2001, 23, 363–374. [Google Scholar] [CrossRef]
- Kain, S.R.; Adams, M.; Kondepudi, A.; Yang, T.T.; Ward, W.W.; Kitts, P. Green fluorescent protein as a reporter of gene expression and protein localization. BioTechniques 1995, 19, 650–655. [Google Scholar] [PubMed]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Gu, H.; Ma, L.; Peng, Y.; Deng, X.W.; Chen, Z.; Qu, L.J. Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res. 2007, 17, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Lou, H.; Liu, Q.; Pei, S.; Liao, Q.; Li, Z. Efficient and transformation-free genome editing in pepper enabled by RNA virus-mediated delivery of CRISPR/Cas9. J. Integr. Plant Biol. 2024. [Google Scholar] [CrossRef]
- Li, F.; Kawato, N.; Sato, H.; Kawaharada, Y.; Henmi, M.; Shinoda, A.; Hasunuma, T.; Nishitani, C.; Osakabe, Y.; Osakabe, K.; et al. Release of chimeras and efficient selection of editing mutants by CRISPR/Cas9-mediated gene editing in apple. Sci. Hortic. 2023, 316, 112011. [Google Scholar] [CrossRef]
- Ding, L.; Chen, Y.; Ma, Y.; Wang, H.; Wei, J. Effective reduction in chimeric mutants of poplar trees produced by CRISPR/Cas9 through a second round of shoot regeneration. Plant Biotechnol. Rep. 2020, 14, 549–558. [Google Scholar] [CrossRef]
- Xing, H.L.; Dong, L.; Wang, Z.P.; Zhang, H.Y.; Han, C.Y.; Liu, B.; Wang, X.C.; Chen, Q.J. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014, 14, 327. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, T.; Yang, H.; Zhou, D.; Zhao, S.; Wang, J.; Zhang, T.; Huang, M.; Kong, D.; Liu, Y. Establishment of a Genetic Transformation and Gene Editing Method by Floral Dipping in Descurainia sophia. Plants 2024, 13, 2833. https://doi.org/10.3390/plants13202833
Jia T, Yang H, Zhou D, Zhao S, Wang J, Zhang T, Huang M, Kong D, Liu Y. Establishment of a Genetic Transformation and Gene Editing Method by Floral Dipping in Descurainia sophia. Plants. 2024; 13(20):2833. https://doi.org/10.3390/plants13202833
Chicago/Turabian StyleJia, Tianjiao, Hua Yang, Dingding Zhou, Sanzeng Zhao, Jianyong Wang, Tao Zhang, Mingkun Huang, Danyu Kong, and Yi Liu. 2024. "Establishment of a Genetic Transformation and Gene Editing Method by Floral Dipping in Descurainia sophia" Plants 13, no. 20: 2833. https://doi.org/10.3390/plants13202833
APA StyleJia, T., Yang, H., Zhou, D., Zhao, S., Wang, J., Zhang, T., Huang, M., Kong, D., & Liu, Y. (2024). Establishment of a Genetic Transformation and Gene Editing Method by Floral Dipping in Descurainia sophia. Plants, 13(20), 2833. https://doi.org/10.3390/plants13202833





