Untargeted Metabolomics Analysis Reveals Differential Accumulation of Flavonoids Between Yellow-Seeded and Black-Seeded Rapeseed Varieties
Abstract
1. Introduction
2. Results
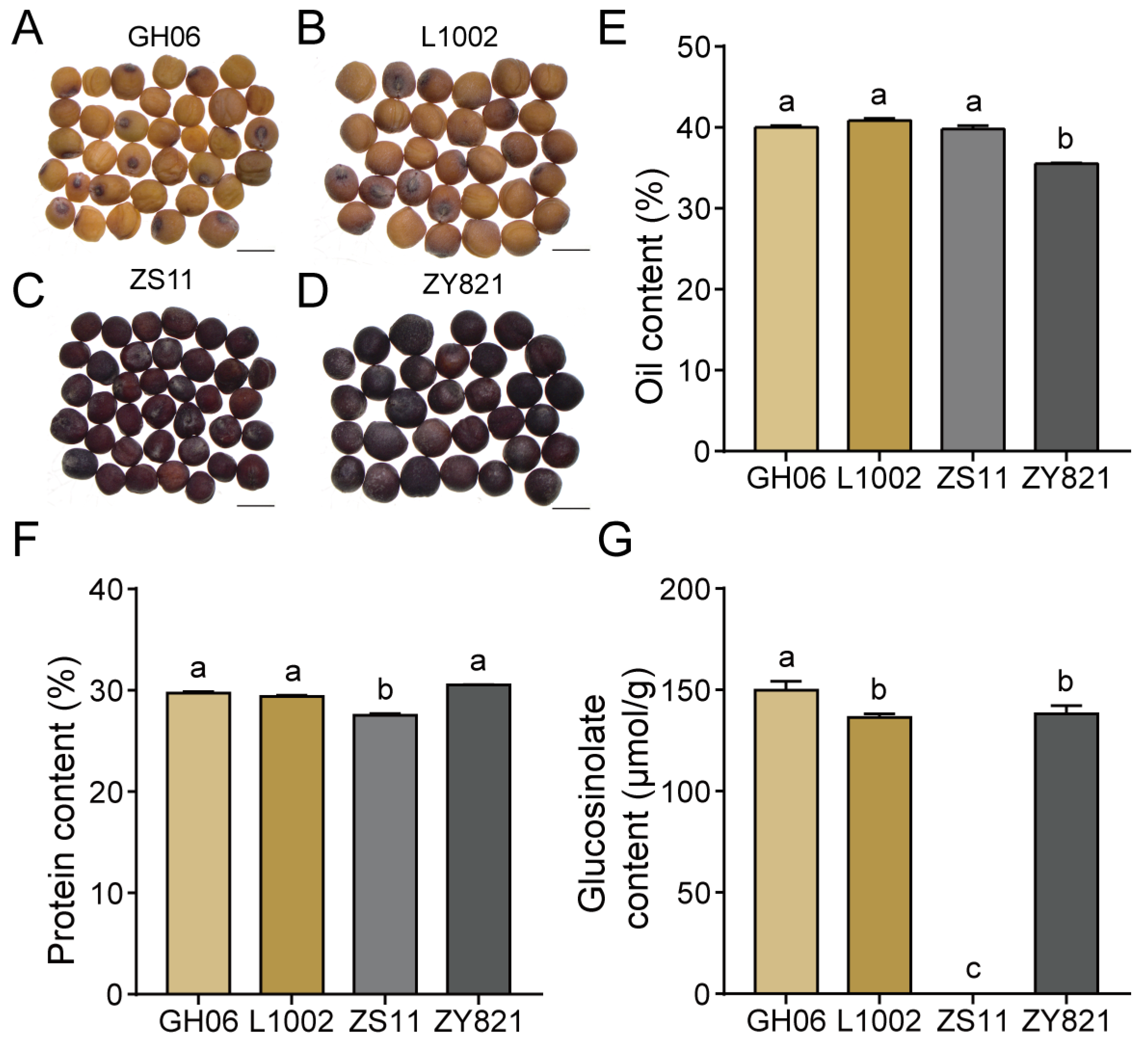
2.1. Differences in Quality Traits Among Mature B. napus Seeds
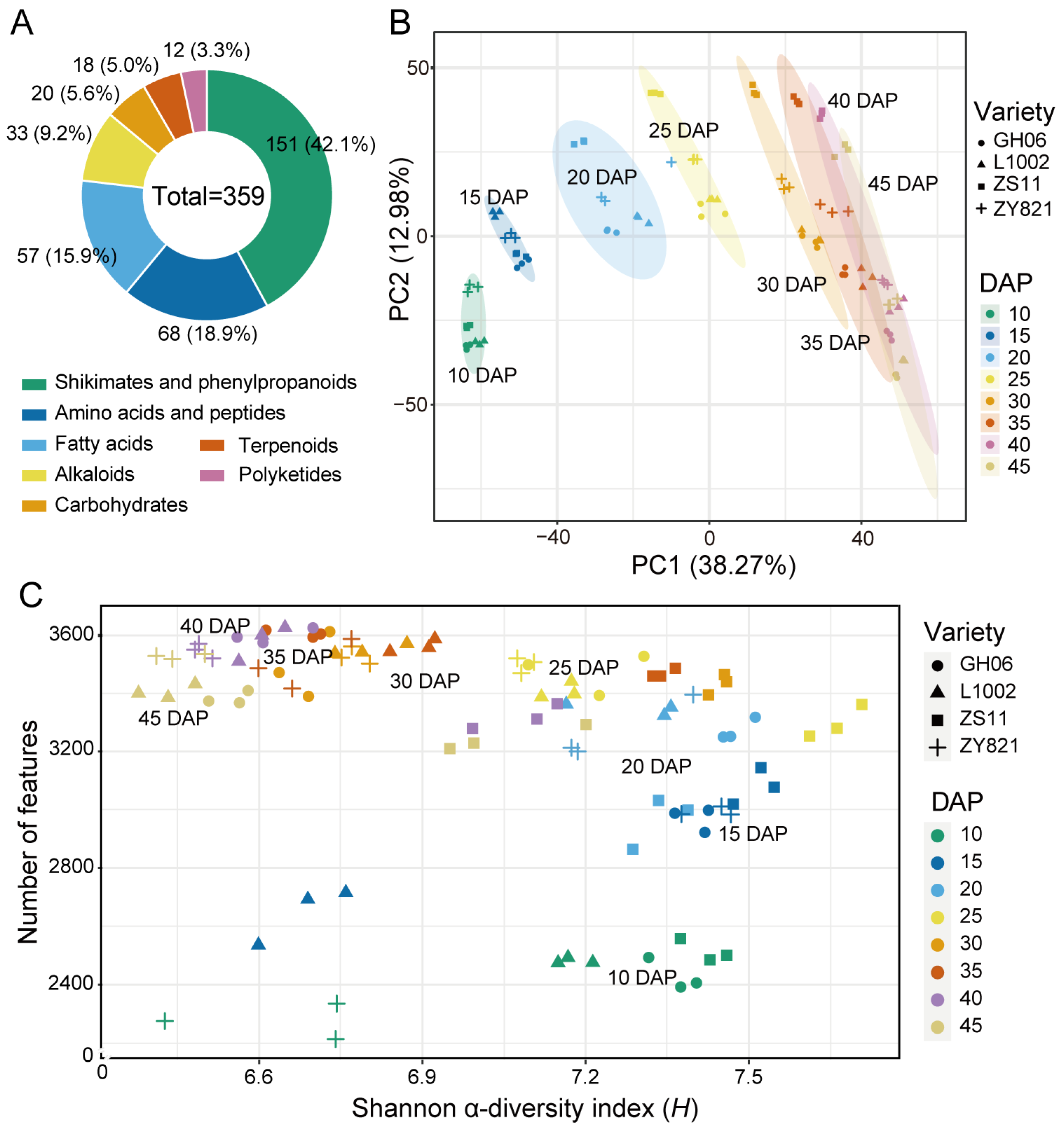
2.2. Metabolite Profiling of B. napus Seeds
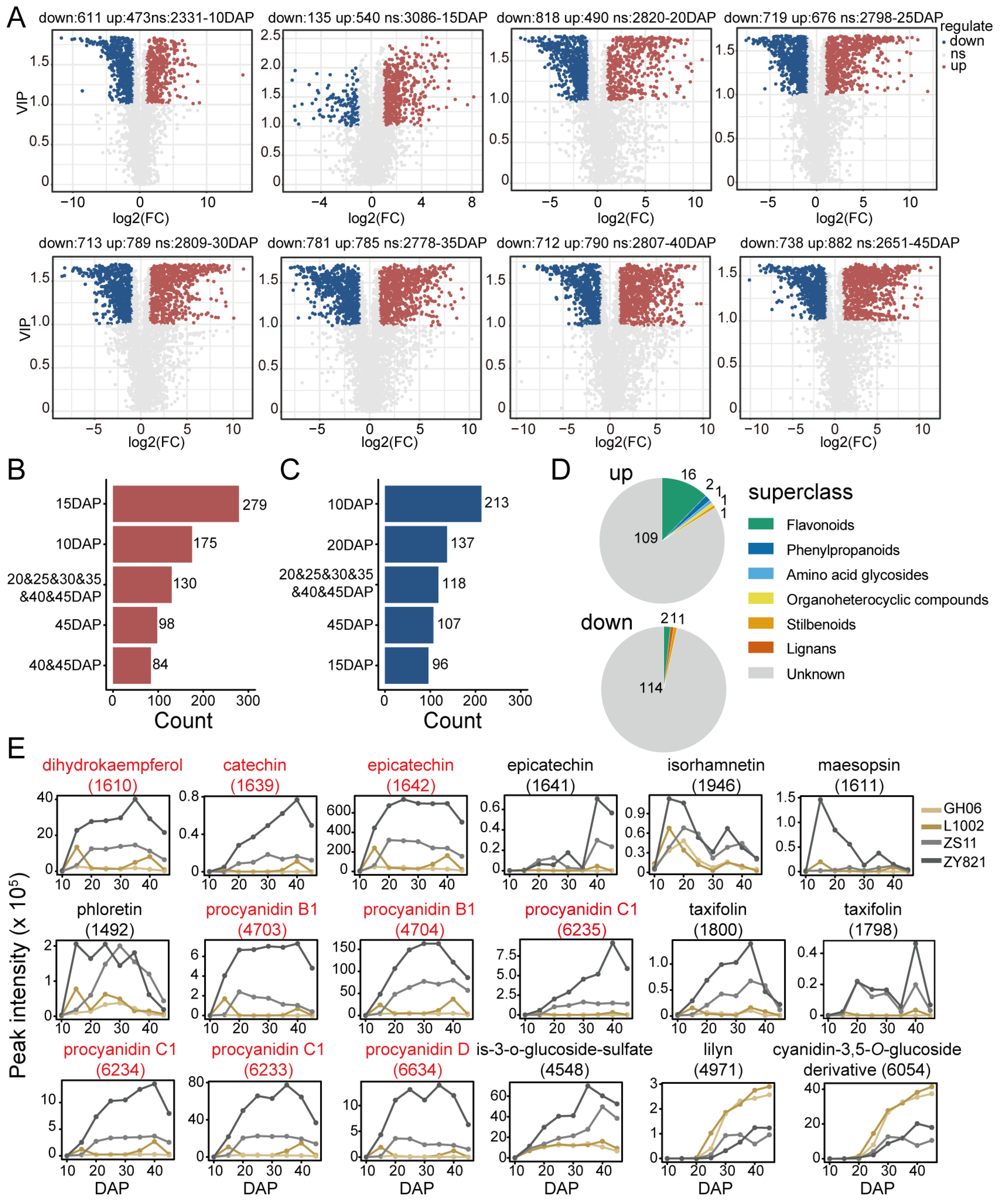
2.3. Differences in Detected Flavonoid Features Between Yellow-Seeded and Black-Seeded B. napus Varieties
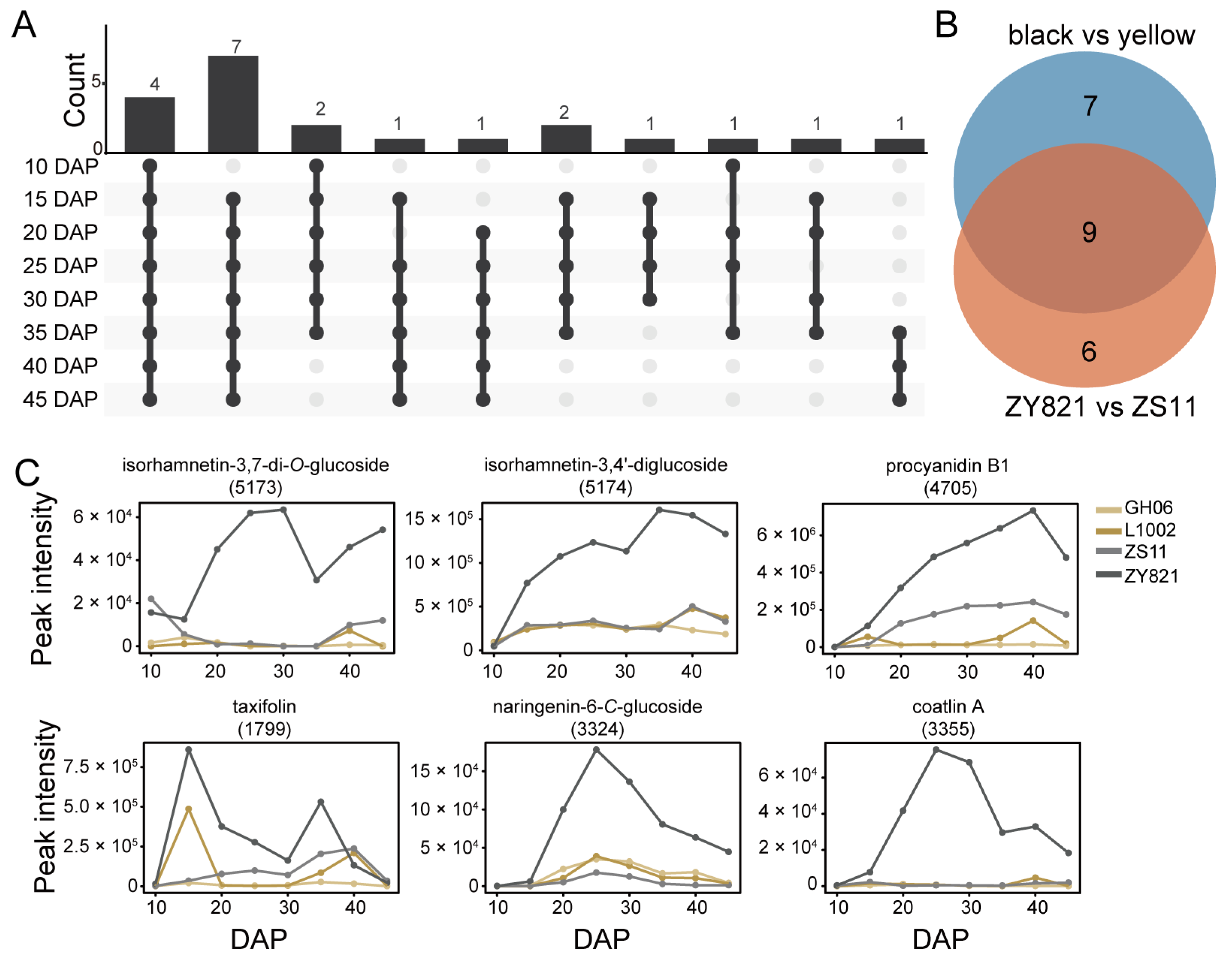
2.4. Differences in Flavonoid Features Between ZS11 and ZY821
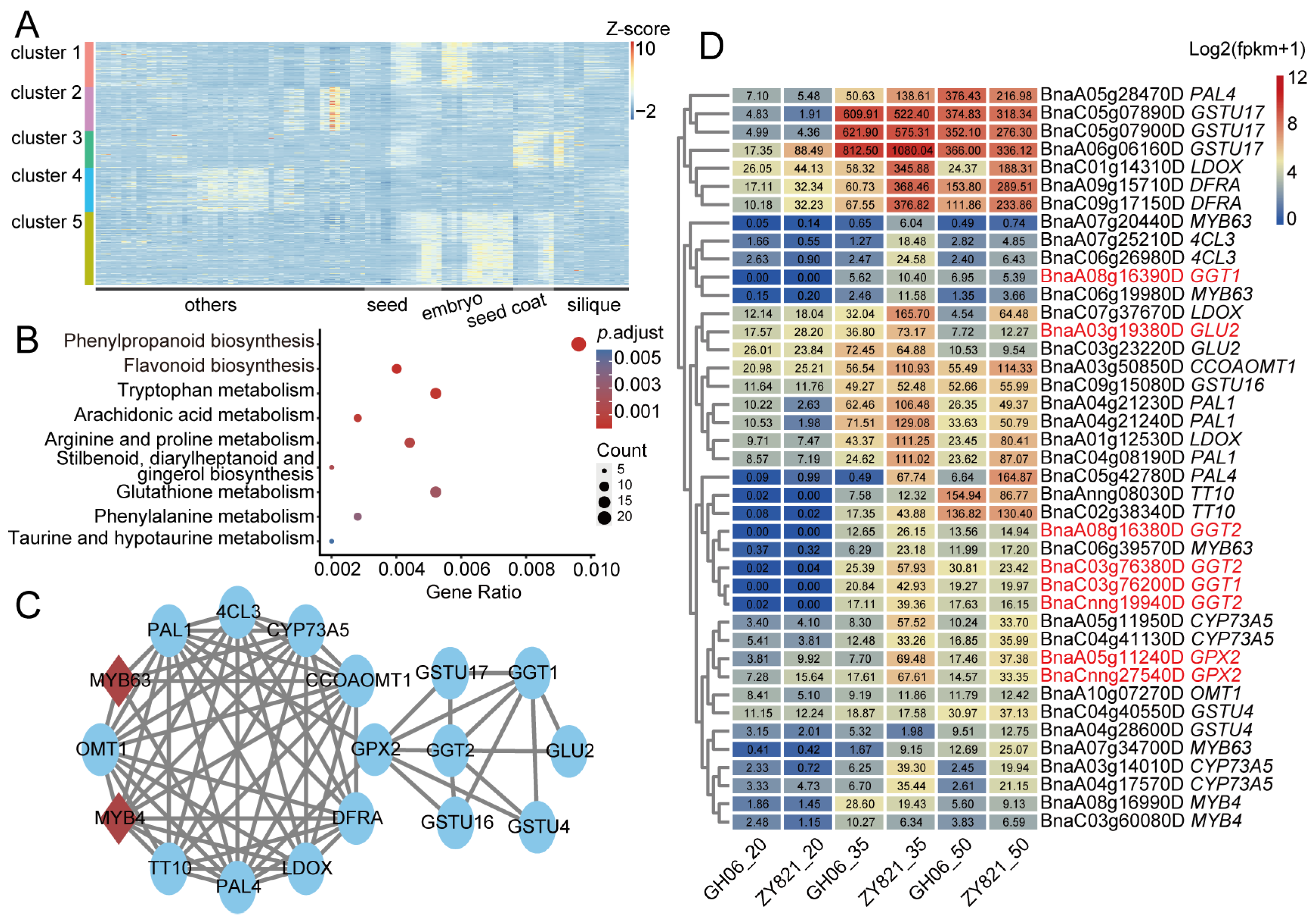
2.5. Combining Metabolome and Multiple Transcriptome Datasets to Identify Seed Pigmentation Regulatory Genes
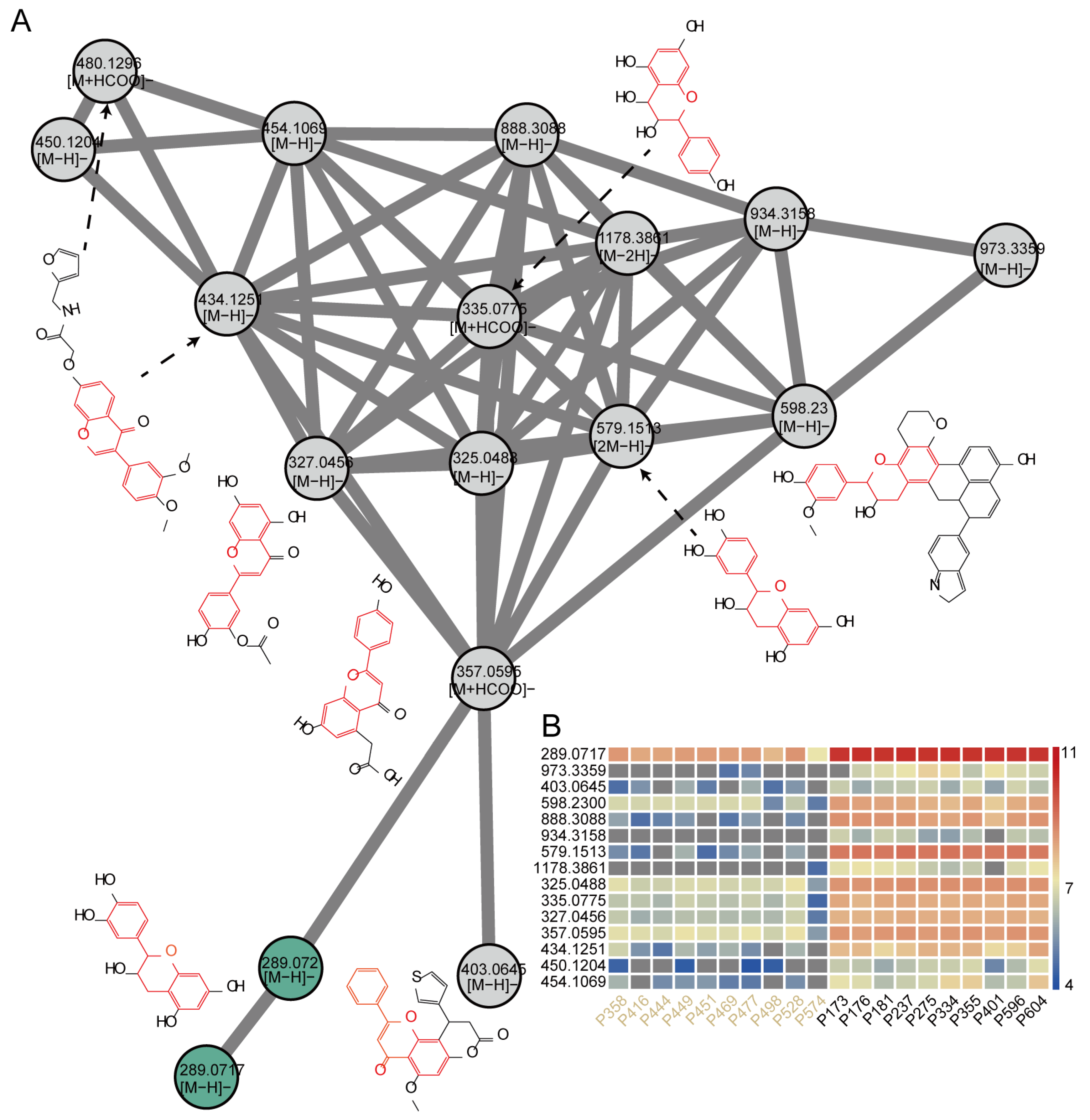
2.6. Construction of Molecular Networks to Explore Unknown Features
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Metabolite Extraction
4.3. UHPLC−HESI−MS/MS Analysis
4.4. Metabolomics Data Processing
4.5. Correlation Analysis of Metabolome and Transcriptome Data
4.6. Identification of Core Genes
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Maeda, H.A.; Fernie, A.R. Evolutionary History of Plant Metabolism. Annu. Rev. Plant Biol. 2021, 72, 185–216. [Google Scholar] [CrossRef] [PubMed]
- Erb, M.; Kliebenstein, D.J. Plant Secondary Metabolites as Defenses, Regulators, and Primary Metabolites: The Blurred Functional Trichotomy. Plant Physiol. 2020, 184, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.Y.; Fernie, A.R.; Luo, J. Exploring the Diversity of Plant Metabolism. Trends Plant Sci. 2019, 24, 83–98. [Google Scholar] [CrossRef]
- Wang, S.C.; Alseekh, S.; Fernie, A.R.; Luo, J. The Structure and Function of Major Plant Metabolite Modifications. Mol. Plant 2019, 12, 899–919. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, Y.; Ren, Y.; Xu, Y.; An, H.; Zhao, Q.; Wang, Y.; Li, H. Widely metabolomic combined with transcriptome analysis to build a bioactive compound regulatory network for the fruit growth cycle in Pseudocydonia sinensis. Food Chem. 2024, 456, 139933. [Google Scholar] [CrossRef]
- Zhou, L.; Wu, S.; Chen, Y.; Huang, R.; Cheng, B.; Mao, Q.; Liu, T.; Liu, Y.; Zhao, K.; Pan, H.; et al. Multi-omics analyzes of Rosa gigantea illuminate tea scent biosynthesis and release mechanisms. Nat. Commun. 2024, 15, 8469. [Google Scholar] [CrossRef]
- Cao, Y.; Han, Z.; Zhang, Z.; He, L.; Huang, C.; Chen, J.; Dai, F.; Xuan, L.; Yan, S.; Si, Z.; et al. UDP-glucosyltransferase 71C4 controls the flux of phenylpropanoid metabolism to shape cotton seed development. Plant Commun. 2024, 5, 100938. [Google Scholar] [CrossRef] [PubMed]
- Dan, Z.; Chen, Y.; Li, H.; Zeng, Y.; Xu, W.; Zhao, W.; He, R.; Huang, W. The metabolomic landscape of rice heterosis highlights pathway biomarkers for predicting complex phenotypes. Plant Physiol. 2021, 187, 1011–1025. [Google Scholar] [CrossRef]
- Chen, S.; Sun, B.; Shi, Z.Y.; Miao, X.X.; Li, H.C. Identification of the rice genes and metabolites involved in dual resistance against brown planthopper and rice blast fungus. Plant Cell Environ. 2022, 45, 1914–1929. [Google Scholar] [CrossRef]
- Peng, M.; Shahzad, R.; Gul, A.; Subthain, H.; Shen, S.; Lei, L.; Zheng, Z.; Zhou, J.; Lu, D.; Wang, S.; et al. Differentially evolved glucosyltransferases determine natural variation of rice flavone accumulation and UV-tolerance. Nat. Commun. 2017, 8, 1975. [Google Scholar] [CrossRef]
- Alseekh, S.; Scossa, F.; Wen, W.; Luo, J.; Yan, J.; Beleggia, R.; Klee, H.J.; Huang, S.; Papa, R.; Fernie, A.R. Domestication of Crop Metabolomes: Desired and Unintended Consequences. Trends Plant Sci. 2021, 26, 650–661. [Google Scholar] [CrossRef] [PubMed]
- Tohge, T.; de Souza, L.P.; Fernie, A.R. Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 2017, 68, 4013–4028. [Google Scholar] [CrossRef]
- Shen, N.; Wang, T.; Gan, Q.; Liu, S.; Wang, L.; Jin, B. Plant flavonoids: Classification, distribution, biosynthesis, and antioxidant activity. Food Chem. 2022, 383, 132531. [Google Scholar] [CrossRef]
- Liu, W.; Feng, Y.; Yu, S.; Fan, Z.; Li, X.; Li, J.; Yin, H. The Flavonoid Biosynthesis Network in Plants. Int. J. Mol. Sci. 2021, 22, 12824. [Google Scholar] [CrossRef]
- Zhao, J.; Dixon, R.A. The ’ins’ and ’outs’ of flavonoid transport. Trends Plant Sci. 2010, 15, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Lepiniec, L.; Debeaujon, I.; Routaboul, J.M.; Baudry, A.; Pourcel, L.; Nesi, N.; Caboche, M. Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 2006, 57, 405–430. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Lu, H.Q.; Jiang, K.X.; Wang, Y.R.; Wang, Y.P.; Jiang, J.J. The Flavonoid Biosynthesis and Regulation in Brassica napus: A Review. Int. J. Mol. Sci. 2022, 24, 357. [Google Scholar] [CrossRef] [PubMed]
- Petraru, A.; Amariei, S. Rapeseed—An Important Oleaginous Plant in the Oil Industry and the Resulting Meal a Valuable Source of Bioactive Compounds. Plants 2024, 13, 3085. [Google Scholar] [CrossRef]
- Li, H.; Yu, K.; Zhang, Z.; Yu, Y.; Wan, J.; He, H.; Fan, C. Targeted mutagenesis of flavonoid biosynthesis pathway genes reveals functional divergence in seed coat colour, oil content and fatty acid composition in Brassica napus L. Plant Biotechnol. J. 2024, 22, 445–459. [Google Scholar] [CrossRef]
- Zhao, H.; Shang, G.; Yin, N.; Chen, S.; Shen, S.; Jiang, H.; Tang, Y.; Sun, F.; Zhao, Y.; Niu, Y.; et al. Multi-omics analysis reveals the mechanism of seed coat color formation in Brassica rapa L. Theor. Appl. Genet. 2022, 135, 2083–2099. [Google Scholar] [CrossRef]
- Yu, C.Y. Molecular mechanism of manipulating seed coat coloration in oilseed Brassica species. J. Appl. Genet. 2013, 54, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Qu, C.; Yin, N.; Chen, S.; Wang, S.; Chen, X.; Zhao, H.; Shen, S.; Fu, F.; Zhou, B.; Xu, X.; et al. Comparative Analysis of the Metabolic Profiles of Yellow- versus Black-Seeded Rapeseed Using UPLC-HESI-MS/MS and Transcriptome Analysis. J. Agric. Food Chem. 2020, 68, 3033–3049. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Yu, K.; Cai, S.; Hu, L.; Amoo, O.; Xu, L.; Yang, Y.; Ma, B.; Jiao, Y.; Zhang, C.; et al. Targeted mutagenesis of BnTT8 homologs controls yellow seed coat development for effective oil production in Brassica napus L. Plant Biotechnol. J. 2020, 18, 1153–1168. [Google Scholar] [CrossRef]
- Xie, T.; Chen, X.; Guo, T.; Rong, H.; Chen, Z.; Sun, Q.; Batley, J.; Jiang, J.; Wang, Y. Targeted Knockout of BnTT2 Homologues for Yellow-Seeded Brassica napus with Reduced Flavonoids and Improved Fatty Acid Composition. J. Agric. Food Chem. 2020, 68, 5676–5690. [Google Scholar] [CrossRef]
- Lim, G.H.; Singhal, R.; Kachroo, A.; Kachroo, P. Fatty Acid- and Lipid-Mediated Signaling in Plant Defense. Annu. Rev. Phytopathol. 2017, 55, 505–536. [Google Scholar] [CrossRef] [PubMed]
- Marles, M.S.; Gruber, M.Y. Histochemical characterisation of unextractable seed coat pigments and quantification of extractable lignin in the Brassicaceae. J. Sci. Food Agric. 2004, 84, 251–262. [Google Scholar] [CrossRef]
- Yin, N.; Li, B.; Liu, X.; Liang, Y.; Lian, J.; Xue, Y.; Qu, C.; Lu, K.; Wei, L.; Wang, R.; et al. Two types of cinnamoyl-CoA reductase function divergently in accumulation of lignins, flavonoids and glucosinolates and enhance lodging resistance in Brassica napus. Crop J. 2022, 10, 647–660. [Google Scholar] [CrossRef]
- Li, B.; Jia, Y.; Xu, L.; Zhang, S.; Long, Z.; Wang, R.; Guo, Y.; Zhang, W.; Jiao, C.; Li, C.; et al. Transcriptional convergence after repeated duplication of an amino acid transporter gene leads to the independent emergence of the black husk/pericarp trait in barley and rice. Plant Biotechnol. J. 2024, 22, 1282–1298. [Google Scholar] [CrossRef]
- Yang, C.; Shen, S.; Zhou, S.; Li, Y.; Mao, Y.; Zhou, J.; Shi, Y.; An, L.; Zhou, Q.; Peng, W.; et al. Rice metabolic regulatory network spanning the entire life cycle. Mol. Plant 2022, 15, 258–275. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Y.; Zhou, L.; You, S.; Deng, H.; Chen, Y.; Alseekh, S.; Yuan, Y.; Fu, R.; Zhang, Z.; et al. MicroTom Metabolic Network: Rewiring Tomato Metabolic Regulatory Network throughout the Growth Cycle. Mol. Plant 2020, 13, 1203–1218. [Google Scholar] [CrossRef]
- Shu, P.; Zhang, Z.; Wu, Y.; Chen, Y.; Li, K.; Deng, H.; Zhang, J.; Zhang, X.; Wang, J.; Liu, Z.; et al. A comprehensive metabolic map reveals major quality regulations in red-flesh kiwifruit (Actinidia chinensis). New Phytol. 2023, 238, 2064–2079. [Google Scholar] [CrossRef]
- Zeng, Z.; Li, Y.; Zhu, M.; Wang, X.; Wang, Y.; Li, A.; Chen, X.; Han, Q.; Nieuwenhuizen, N.J.; Ampomah-Dwamena, C. Kiwifruit spatiotemporal multiomics networks uncover key tissue-specific regulatory processes throughout the life cycle. Plant Physiol. 2025, 197, kiae567. [Google Scholar] [CrossRef] [PubMed]
- Auger, B.; Marnet, N.; Gautier, V.; Maia-Grondard, A.; Leprince, F.; Renard, M.; Guyot, S.; Nesi, N.; Routaboul, J.-M. A Detailed Survey of Seed Coat Flavonoids in Developing Seeds of Brassica napus L. J. Agric. Food Chem. 2010, 58, 6246–6256. [Google Scholar] [CrossRef]
- Shao, Y.; Jiang, J.; Ran, L.; Lu, C.; Wei, C.; Wang, Y. Analysis of flavonoids and hydroxycinnamic acid derivatives in rapeseeds (Brassica napus L. var. napus) by HPLC-PDA--ESI(--)-MS(n)/HRMS. J. Agric. Food Chem. 2014, 62, 2935–2945. [Google Scholar] [CrossRef]
- Ren, Y.; Zhang, N.; Li, R.; Ma, X.; Zhang, L. Comparative transcriptome and flavonoids components analysis reveal the structural genes responsible for the yellow seed coat color of Brassica rapa L. PeerJ 2021, 9, e10770. [Google Scholar] [CrossRef]
- Shen, S.; Tang, Y.; Zhang, C.; Yin, N.; Mao, Y.; Sun, F.; Chen, S.; Hu, R.; Liu, X.; Shang, G.; et al. Metabolite Profiling and Transcriptome Analysis Provide Insight into Seed Coat Color in Brassica juncea. Int. J. Mol. Sci. 2021, 22, 7215. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Yang, L.; Liu, X.; Wang, T.; Kang, L.; Chen, H.; Lu, Y.; Zhang, Y.; Yang, S.; You, L.; et al. Natural variations in TT8 and its neighboring STK confer yellow seed with elevated oil content in Brassica juncea. Proc. Natl. Acad. Sci. USA 2025, 122, e2417264122. [Google Scholar] [CrossRef] [PubMed]
- Pourcel, L.; Routaboul, J.M.; Kerhoas, L.; Caboche, M.; Lepiniec, L.; Debeaujon, I. TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 2005, 17, 2966–2980. [Google Scholar] [CrossRef]
- Janusz, G.; Pawlik, A.; Świderska-Burek, U.; Polak, J.; Sulej, J.; Jarosz-Wilkołazka, A.; Paszczyński, A. Laccase Properties, Physiological Functions, and Evolution. Int. J. Mol. Sci. 2020, 21, 966. [Google Scholar] [CrossRef]
- Yuan, Z.; Li, G.; Zhang, H.; Peng, Z.; Ding, W.; Wen, H.; Zhou, H.; Zeng, J.; Chen, J.; Xu, J. Four novel Cit7GlcTs functional in flavonoid 7-O-glucoside biosynthesis are vital to flavonoid biosynthesis shunting in citrus. Hortic. Res. 2024, 11, uhae098. [Google Scholar] [CrossRef]
- Zhang, F.; Guo, H.; Huang, J.; Yang, C.; Li, Y.; Wang, X.; Qu, L.; Liu, X.; Luo, J. A UV-B-responsive glycosyltransferase, OsUGT706C2, modulates flavonoid metabolism in rice. Sci. China: Life Sci. 2020, 63, 1037–1052. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.; Yuan, L.; Luo, Y.; Jing, X.; Zhang, Z.; Wang, J.; Zhu, G. A freezing responsive UDP-glycosyltransferase improves potato freezing tolerance via modifying flavonoid metabolism. Hortic. Plant J. 2024. [Google Scholar] [CrossRef]
- Liu, X.; Wang, T.; Ruan, Y.; Xie, X.; Tan, C.; Guo, Y.; Li, B.; Qu, L.; Deng, L.; Li, M.; et al. Comparative Metabolome and Transcriptome Analysis of Rapeseed (Brassica napus L.) Cotyledons in Response to Cold Stress. Plants 2024, 13, 2212. [Google Scholar] [CrossRef]
- Wilson, A.E.; Tian, L. Phylogenomic analysis of UDP-dependent glycosyltransferases provides insights into the evolutionary landscape of glycosylation in plant metabolism. Plant J. 2019, 100, 1273–1288. [Google Scholar] [CrossRef]
- Li, L.; Tian, Z.; Chen, J.; Tan, Z.; Zhang, Y.; Zhao, H.; Wu, X.; Yao, X.; Wen, W.; Chen, W.; et al. Characterization of novel loci controlling seed oil content in Brassica napus by marker metabolite-based multi-omics analysis. Genome Biol. 2023, 24, 141. [Google Scholar] [CrossRef] [PubMed]
- Au, K.K.; Pérez-Gómez, J.; Neto, H.; Müller, C.; Meyer, A.J.; Fricker, M.D.; Moore, I. A perturbation in glutathione biosynthesis disrupts endoplasmic reticulum morphology and secretory membrane traffic in Arabidopsis thaliana. Plant J. 2012, 71, 881–894. [Google Scholar] [CrossRef]
- Shelton, D.; Stranne, M.; Mikkelsen, L.; Pakseresht, N.; Welham, T.; Hiraka, H.; Tabata, S.; Sato, S.; Paquette, S.; Wang, T.L.; et al. Transcription factors of Lotus: Regulation of isoflavonoid biosynthesis requires coordinated changes in transcription factor activity. Plant Physiol. 2012, 159, 531–547. [Google Scholar] [CrossRef] [PubMed]
- Francisco, R.M.; Regalado, A.; Ageorges, A.; Burla, B.J.; Bassin, B.; Eisenach, C.; Zarrouk, O.; Vialet, S.; Marlin, T.; Chaves, M.M.; et al. ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-Glucosides. Plant Cell 2013, 25, 1840–1854. [Google Scholar] [CrossRef]
- Tsugawa, H.; Ikeda, K.; Takahashi, M.; Satoh, A.; Mori, Y.; Uchino, H.; Okahashi, N.; Yamada, Y.; Tada, I.; Bonini, P.; et al. A lipidome atlas in MS-DIAL 4. Nat. Biotechnol. 2020, 38, 1159–1163. [Google Scholar] [CrossRef]
- Beck, L.C.; Masi, A.C.; Young, G.R.; Vatanen, T.; Lamb, C.A.; Smith, R.; Coxhead, J.; Butler, A.; Marsland, B.J.; Embleton, N.D. Strain-specific impacts of probiotics are a significant driver of gut microbiome development in very preterm infants. Nat. Microbiol. 2022, 7, 1525–1535. [Google Scholar] [CrossRef]
- Kim, H.W.; Wang, M.; Leber, C.A.; Nothias, L.F.; Reher, R.; Kang, K.B.; van der Hooft, J.J.J.; Dorrestein, P.C.; Gerwick, W.H.; Cottrell, G.W. NPClassifier: A Deep Neural Network-Based Structural Classification Tool for Natural Products. J. Nat. Prod. 2021, 84, 2795–2807. [Google Scholar] [CrossRef] [PubMed]
- Petrén, H.; Köllner, T.G.; Junker, R.R. Quantifying chemodiversity considering biochemical and structural properties of compounds with the R package chemodiv. New Phytol. 2023, 237, 2478–2492. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Lai, Z.; Tsugawa, H.; Wohlgemuth, G.; Mehta, S.; Mueller, M.; Zheng, Y.; Ogiwara, A.; Meissen, J.; Showalter, M.; Takeuchi, K.; et al. Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nat. Methods 2018, 15, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Chao, H.; Li, T.; Luo, C.; Huang, H.; Ruan, Y.; Li, X.; Niu, Y.; Fan, Y.; Sun, W.; Zhang, K.; et al. BrassicaEDB: A Gene Expression Database for Brassica Crops. Int. J. Mol. Sci. 2020, 21, 5831. [Google Scholar] [CrossRef]
- Giorgino, T. Computing and Visualizing Dynamic Time Warping Alignments in R: The dtw Package. J. Stat. Softw. 2009, 31, 1–24. [Google Scholar] [CrossRef]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, S.; Wei, L.; Huang, Y.; Liu, D.; Jia, Y.; Luo, C.; Lin, Y.; Liang, C.; Hu, Y.; et al. BnIR: A multi-omics database with various tools for Brassica napus research and breeding. Mol. Plant 2023, 16, 775–789. [Google Scholar] [CrossRef]
- Bader, G.D.; Hogue, C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef]
- Pang, Z.; Zhou, G.; Ewald, J.; Chang, L.; Hacariz, O.; Basu, N.; Xia, J. Using MetaboAnalyst 5.0 for LC-HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 2022, 17, 1735–1761. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, S.; Tang, Y.; Liu, D.; Chen, L.; Zhang, Y.; Ye, K.; Sun, F.; Wei, X.; Du, H.; Zhao, H.; et al. Untargeted Metabolomics Analysis Reveals Differential Accumulation of Flavonoids Between Yellow-Seeded and Black-Seeded Rapeseed Varieties. Plants 2025, 14, 753. https://doi.org/10.3390/plants14050753
Shen S, Tang Y, Liu D, Chen L, Zhang Y, Ye K, Sun F, Wei X, Du H, Zhao H, et al. Untargeted Metabolomics Analysis Reveals Differential Accumulation of Flavonoids Between Yellow-Seeded and Black-Seeded Rapeseed Varieties. Plants. 2025; 14(5):753. https://doi.org/10.3390/plants14050753
Chicago/Turabian StyleShen, Shulin, Yunshan Tang, Daiqin Liu, Lulu Chen, Yi Zhang, Kaijie Ye, Fujun Sun, Xingzhi Wei, Hai Du, Huiyan Zhao, and et al. 2025. "Untargeted Metabolomics Analysis Reveals Differential Accumulation of Flavonoids Between Yellow-Seeded and Black-Seeded Rapeseed Varieties" Plants 14, no. 5: 753. https://doi.org/10.3390/plants14050753
APA StyleShen, S., Tang, Y., Liu, D., Chen, L., Zhang, Y., Ye, K., Sun, F., Wei, X., Du, H., Zhao, H., Li, J., Qu, C., & Yin, N. (2025). Untargeted Metabolomics Analysis Reveals Differential Accumulation of Flavonoids Between Yellow-Seeded and Black-Seeded Rapeseed Varieties. Plants, 14(5), 753. https://doi.org/10.3390/plants14050753







