Transcriptomic Analysis Reveals That the Photosynthesis and Carotenoid Metabolism Pathway Is Involved in the Salinity Stress Response in Brassica rapa L. ssp. Pekinensis
Abstract
1. Introduction
2. Results
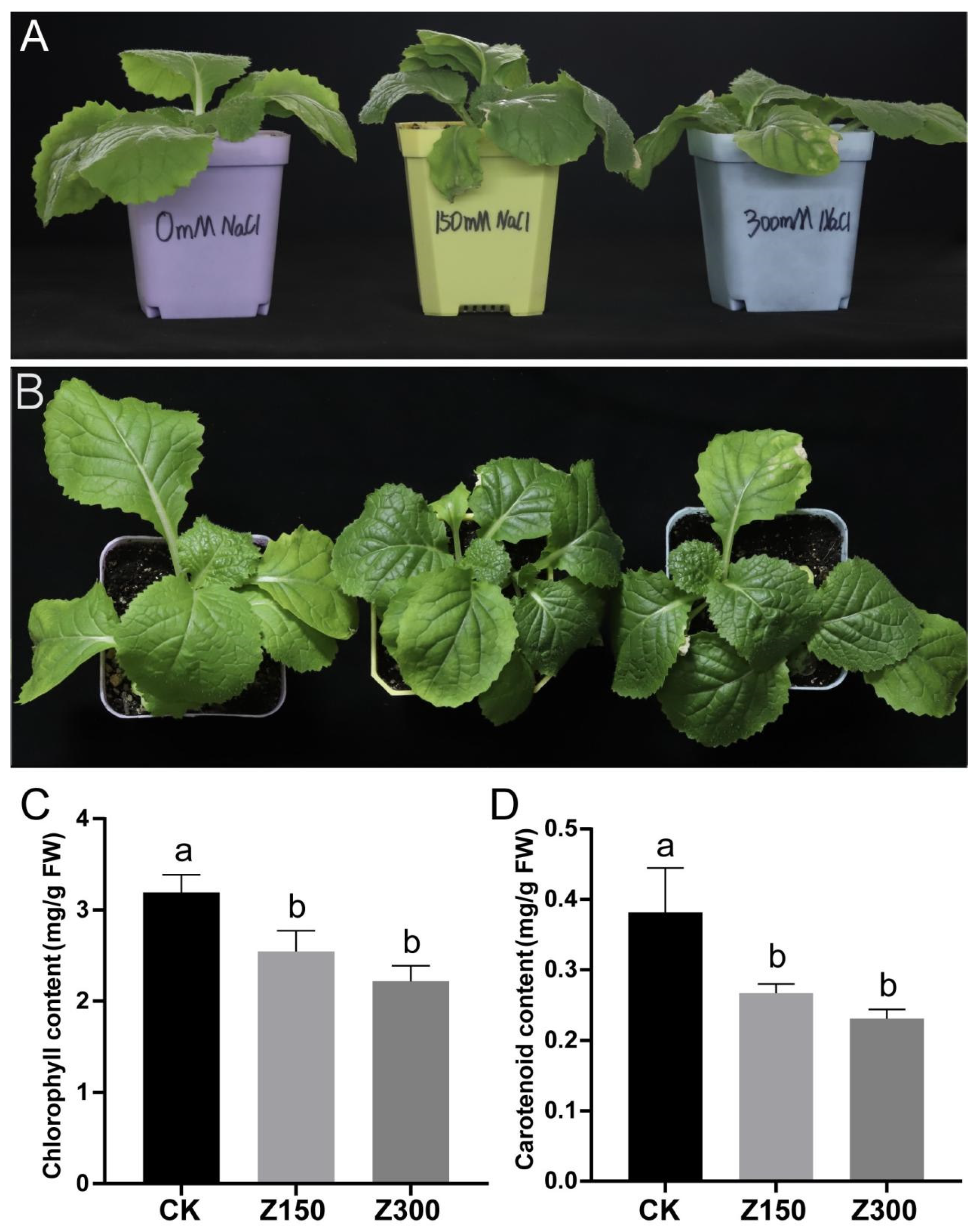
2.1. Salinity Stress Causes Chlorosis and Decreased Contents of Chlorophyll and Carotenoids in the Leaves of B. rapa L. ssp. Pekinensis
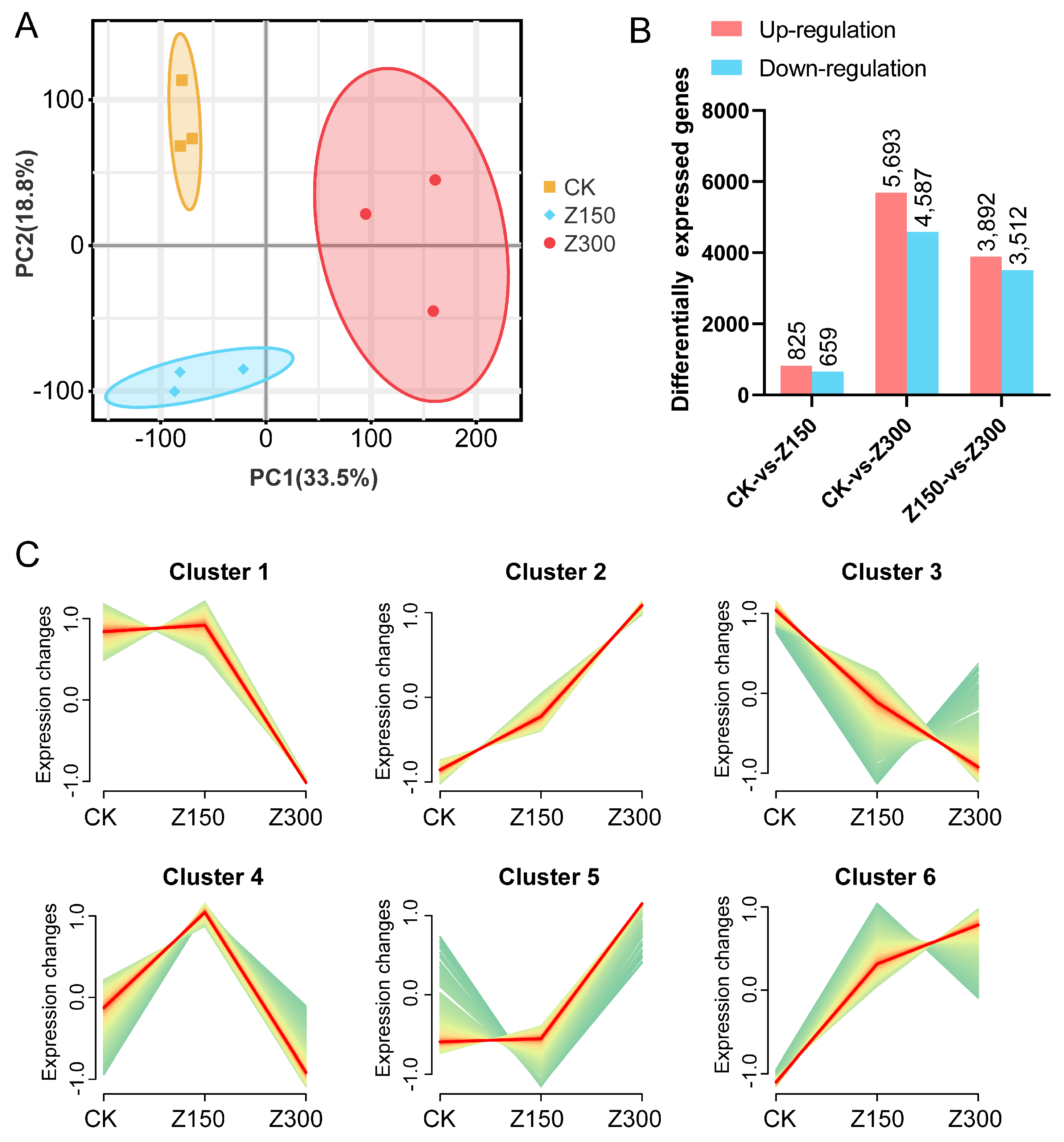
2.2. Salinity Stress Triggers Reprogramming of Gene Expression
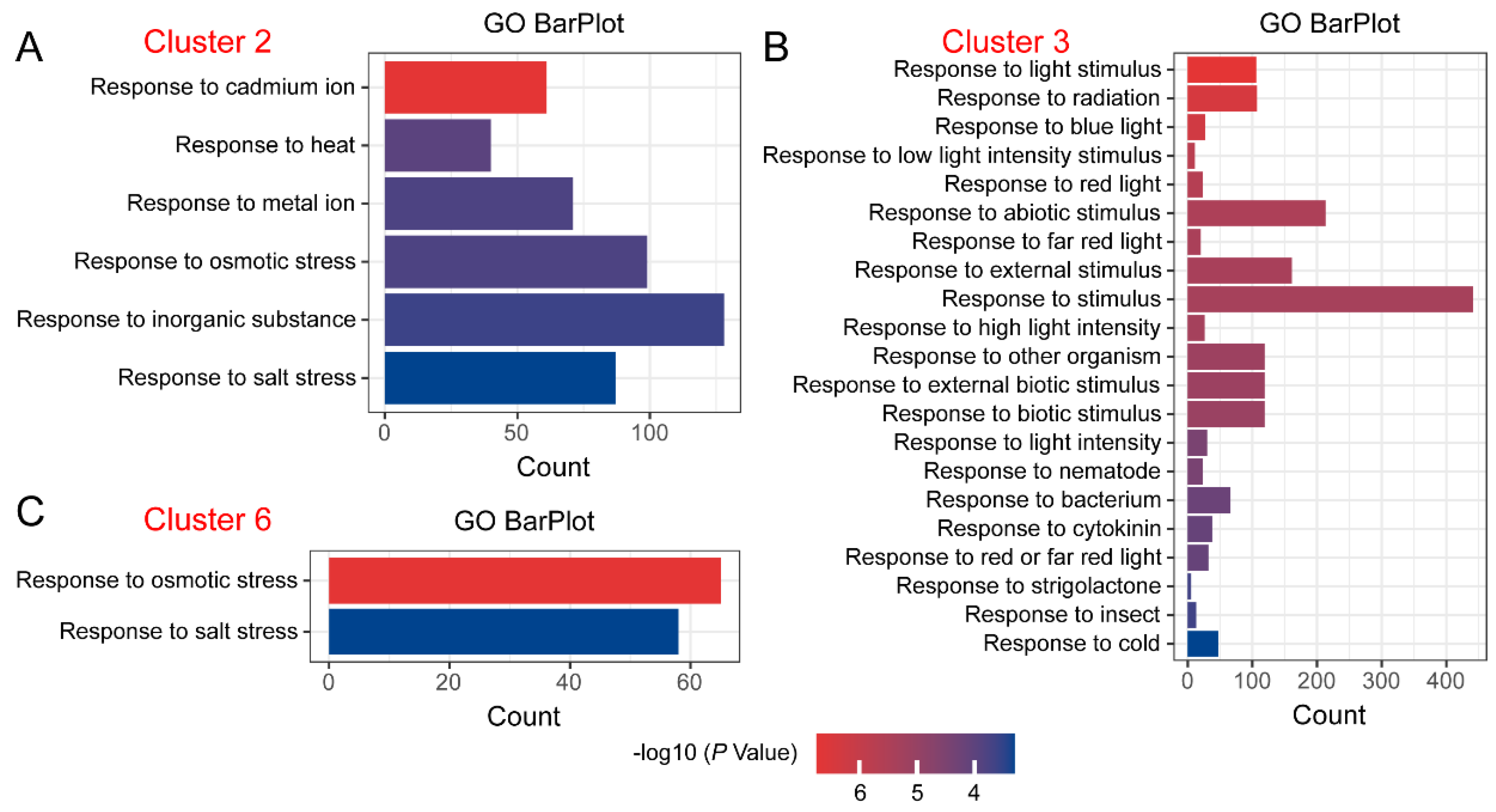
2.3. GO Enrichment Analysis of DEGs with Specific Expression Trends
2.4. DEGs’ Response to Salt Stress
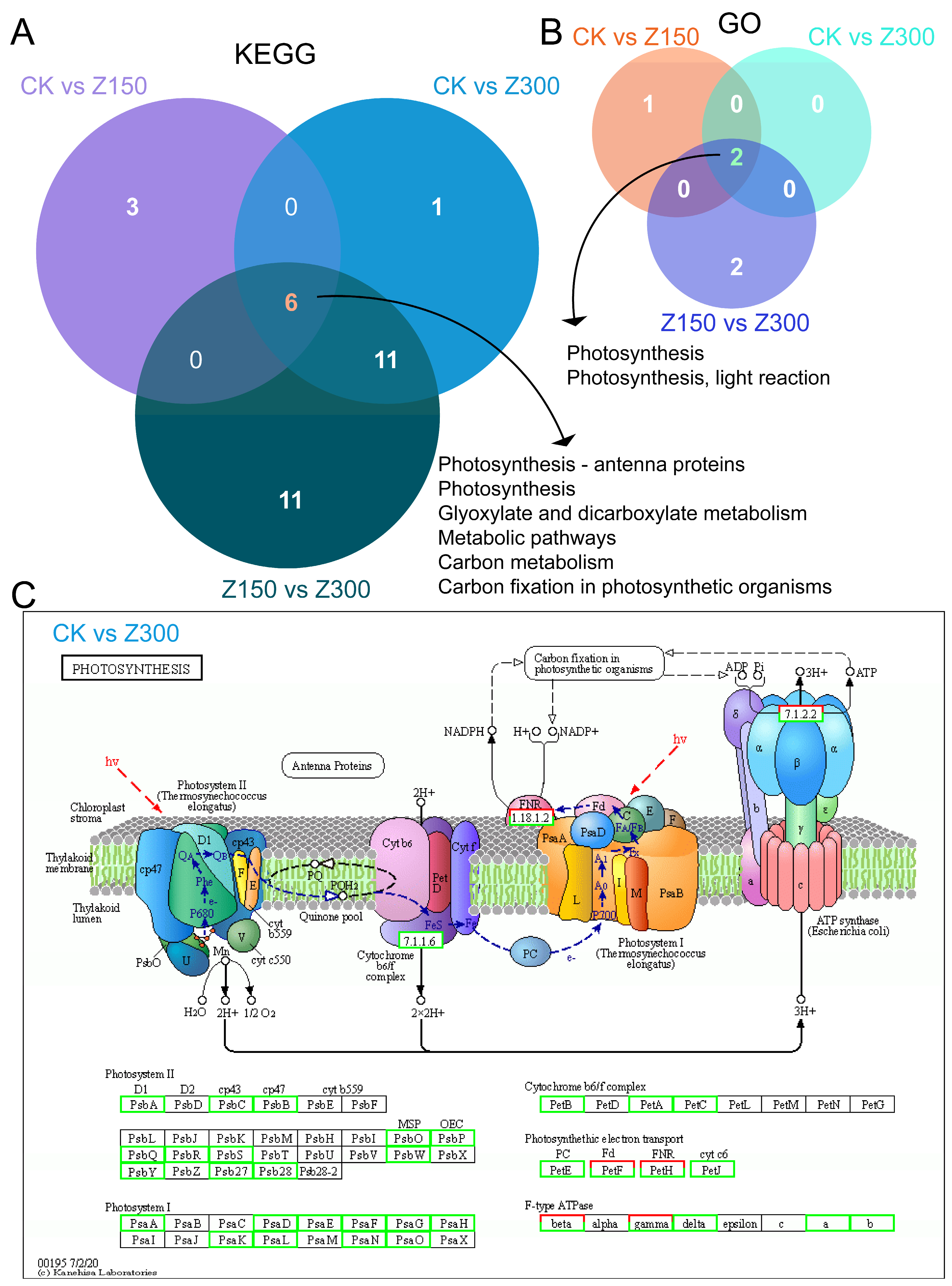
2.5. Salinity Stress Inhibits Photosynthesis and Carbon Metabolism
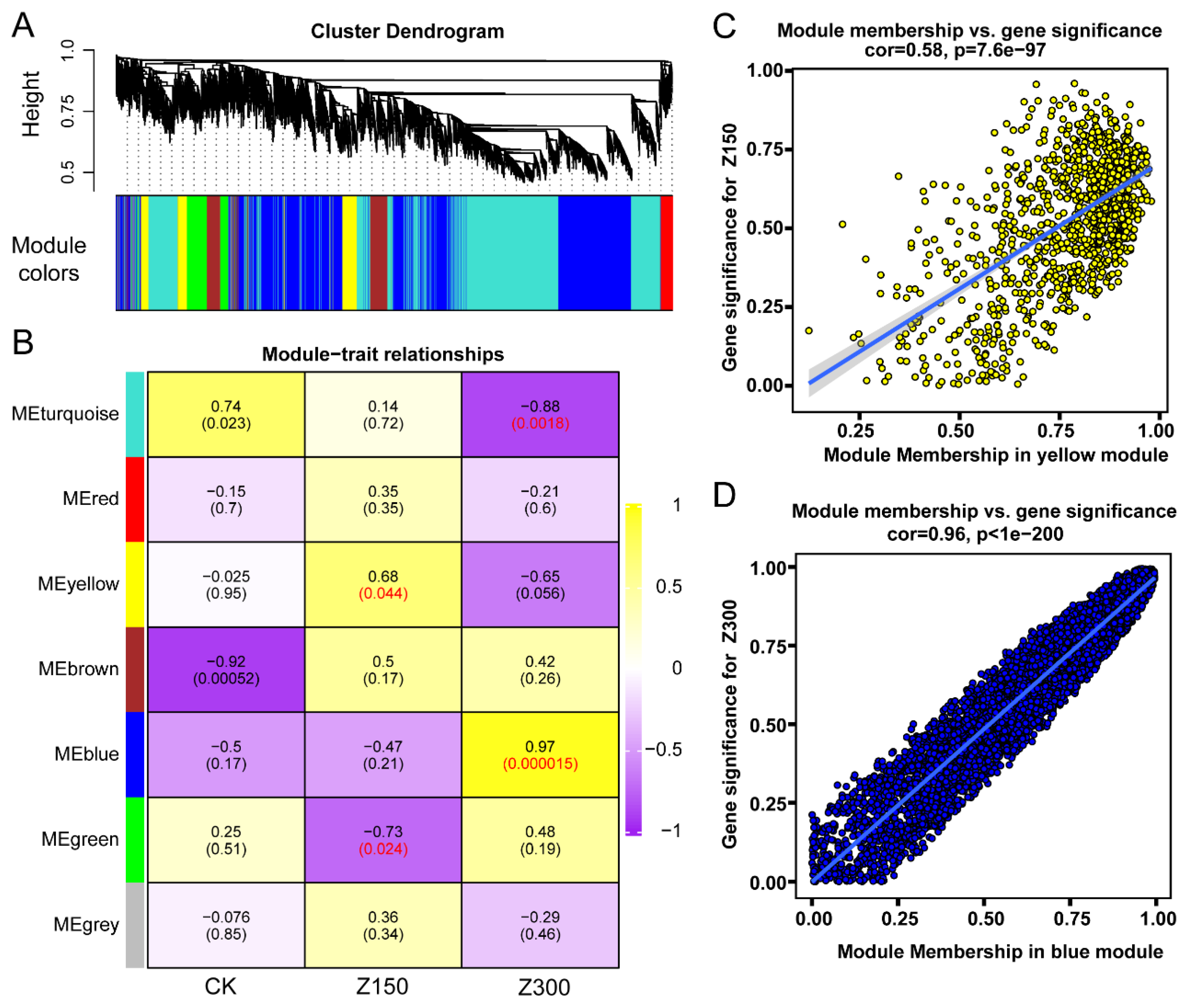
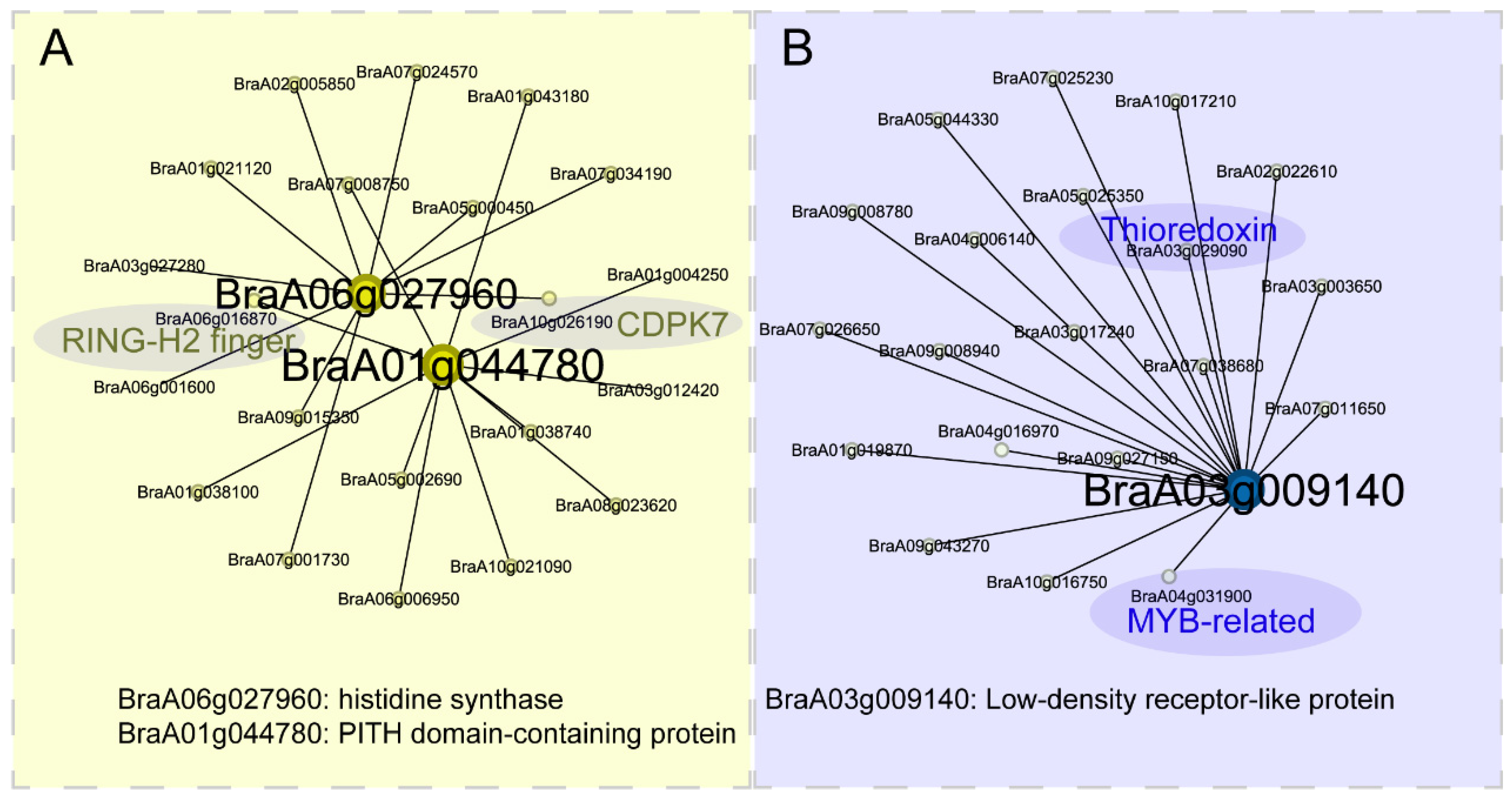
2.6. WGCNA Identifies Hub Genes Involved in Salinity Stress Response
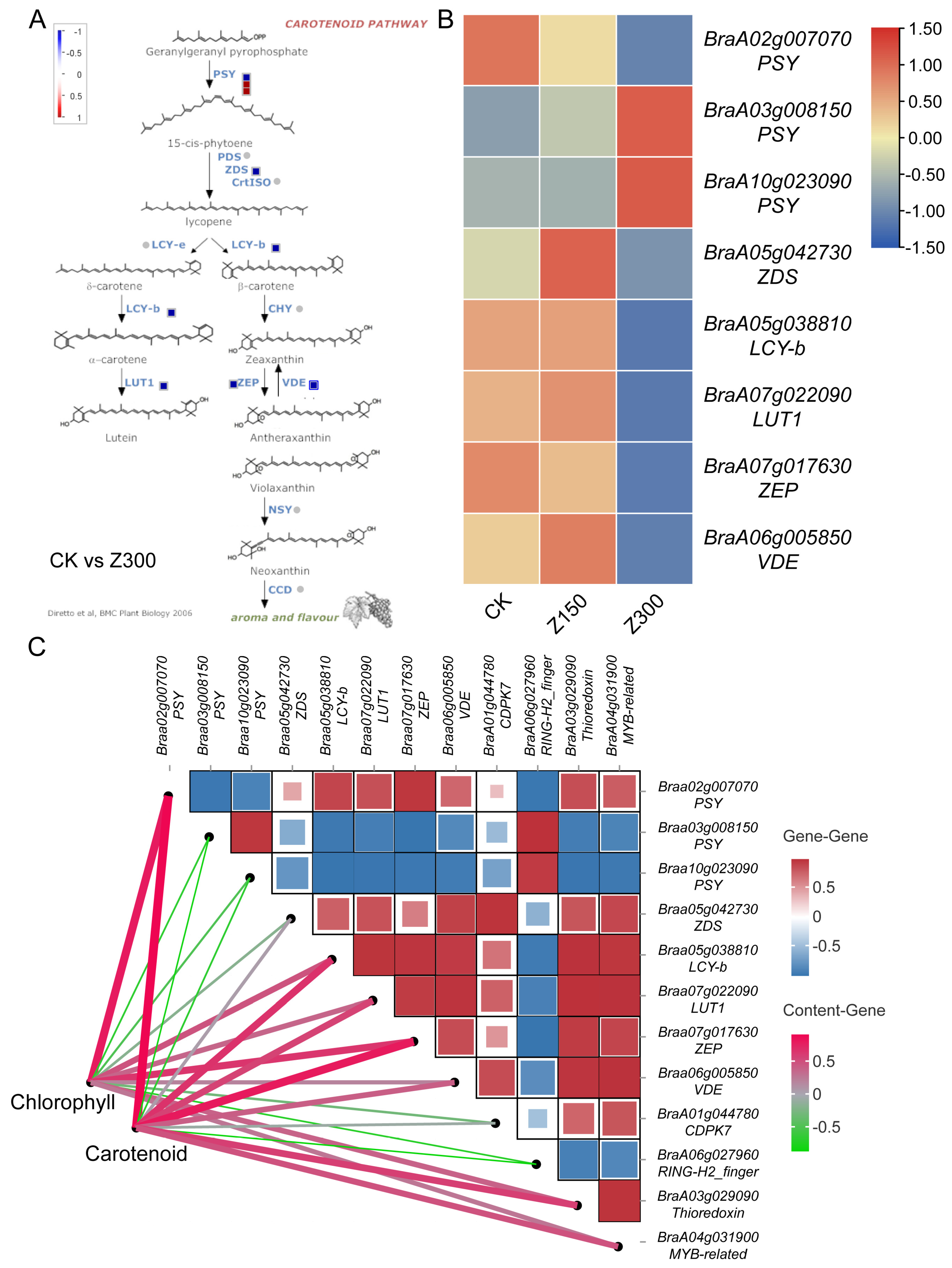
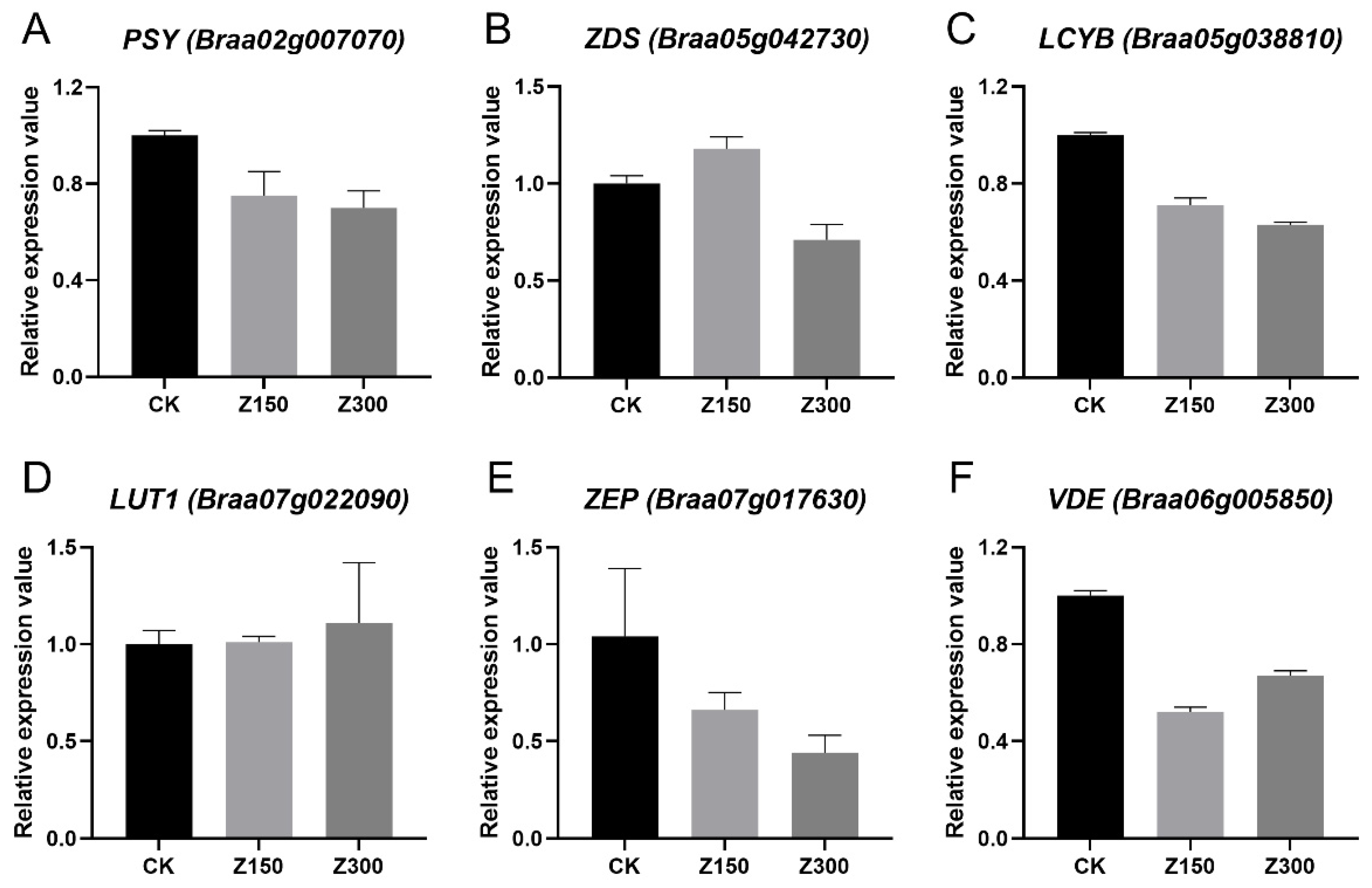
2.7. Salinity Stress Inhibits Carotenoid Metabolism
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
- The following abbreviations are used in this manuscript:
| DEGs | Differentially expressed genes |
| GO | Gene Ontology |
| WGCNA | Weighted Gene Co-expression Network Analysis |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PCA | Principal-component analysis |
| HSPs | Heat shock proteins |
| PODs | Peroxidases |
| CIPKs | CBL-interacting protein kinases |
| GSTs | Glutathione S-transferases |
| MAPKs | Mitogen-activated protein kinases |
References
- Favela-González, K.M.; Hernández-Almanza, A.Y.; De la Fuente-Salcido, N.M. The value of bioactive compounds of cruciferous vegetables (Brassica) as antimicrobials and antioxidants: A review. J. Food Biochem. 2020, 44, e13414. [Google Scholar] [CrossRef]
- Sharma, S.; Priyanka; Shree, B.; Ramachandran, P.; Kumar, V.; Thakur, R.; Kumar, S. Cabbage and Red Cabbage Sprouts: Powerhouse of Nutrients. In Advances in Plant Sprouts: Phytochemistry and Biofunctionalities; Majid, I., Kehinde, B.A., Dar, B., Nanda, V., Eds.; Springer International Publishing: Cham, Switzerland, 2023; pp. 363–382. [Google Scholar] [CrossRef]
- Ding, Y.; Liu, Y.; Zhao, L.; Zhou, M.; Zhang, L.; Wang, G.; Jia, J. Effects of salt stress on nutritional quality of orange-heading Chinese cabbage seedlings. Pak. J. Bot. 2023, 55, 837–841. [Google Scholar] [CrossRef]
- Li, Z.; Zhu, L.; Zhao, F.; Li, J.; Zhang, X.; Kong, X.; Wu, H.; Zhang, Z. Plant Salinity Stress Response and Nano-Enabled Plant Salt Tolerance. Front. Plant Sci. 2022, 13, 843994. [Google Scholar] [CrossRef]
- Tahjib-Ul-Arif, M.; Hasan, M.T.; Rahman, M.A.; Nuruzzaman, M.; Rahman, A.M.S.; Hasanuzzaman, M.; Haque, M.R.; Hossain, M.A.; Abdel Latef, A.A.H.; Murata, Y.; et al. Plant response to combined salinity and waterlogging stress: Current research progress and future prospects. Plant Stress 2023, 7, 100137. [Google Scholar] [CrossRef]
- Jha, U.C.; Bohra, A.; Jha, R.; Parida, S.K. Salinity stress response and ‘omics’ approaches for improving salinity stress tolerance in major grain legumes. Plant Cell Rep. 2019, 38, 255–277. [Google Scholar] [CrossRef] [PubMed]
- Ali, E.; Hussain, S.; Jalal, F.; Khan, M.A.; Imtiaz, M.; Said, F.; Ismail, M.; Khan, S.; Ali, H.M.; Hatamleh, A.A.; et al. Salicylic acid-mitigates abiotic stress tolerance via altering defense mechanisms in Brassica napus (L.). Front. Plant Sci. 2023, 14, 1187260. [Google Scholar] [CrossRef] [PubMed]
- Hualpa-Ramirez, E.; Carrasco-Lozano, E.C.; Madrid-Espinoza, J.; Tejos, R.; Ruiz-Lara, S.; Stange, C.; Norambuena, L. Stress salinity in plants: New strategies to cope with in the foreseeable scenario. Plant Physiol. Biochem. 2024, 208, 108507. [Google Scholar] [CrossRef] [PubMed]
- Balasubramaniam, T.; Shen, G.; Esmaeili, N.; Zhang, H. Plants’ Response Mechanisms to Salinity Stress. Plants 2023, 12, 2253. [Google Scholar] [CrossRef] [PubMed]
- Ludwiczak, A.; Osiak, M.; Cárdenas-Pérez, S.; Lubińska-Mielińska, S.; Piernik, A. Osmotic Stress or Ionic Composition: Which Affects the Early Growth of Crop Species More? Agronomy 2021, 11, 435. [Google Scholar] [CrossRef]
- Katerji, N.; van Hoorn, J.W.; Hamdy, A.; Mastrorilli, M. Comparison of corn yield response to plant water stress caused by salinity and by drought. Agric. Water Manag. 2004, 65, 95–101. [Google Scholar] [CrossRef]
- Das, P.; Agarwala, N.; Gill, S.S.; Varshney, R.K. Emerging role of plant long non coding RNAs (lncRNAs) in salinity stress response. Plant Stress 2023, 10, 100265. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Bhat, B.A.; Mir, R.A.; Mir, W.R.; Hamdani, S.S.; Mir, M.A. Transcription factors-golden keys to modulate the plant metabolism to develop salinity tolerance. Plant Stress 2024, 11, 100409. [Google Scholar] [CrossRef]
- Wei, S.; Gao, L.; Zhang, Y.; Zhang, F.; Yang, X.; Huang, D. Genome-wide investigation of the NAC transcription factor family in melon (Cucumis melo L.) and their expression analysis under salt stress. Plant Cell Rep. 2016, 35, 1827–1839. [Google Scholar] [CrossRef]
- Xu, T.; Zhang, L.; Yang, Z.; Wei, Y.; Dong, T. Identification and Functional Characterization of Plant MiRNA Under Salt Stress Shed Light on Salinity Resistance Improvement Through MiRNA Manipulation in Crops. Front. Plant Sci. 2021, 12, 665439. [Google Scholar] [CrossRef]
- Gao, Z.; Ma, C.; Zheng, C.; Yao, Y.; Du, Y. Advances in the regulation of plant salt-stress tolerance by miRNA. Mol. Biol. Rep. 2022, 49, 5041–5055. [Google Scholar] [CrossRef]
- Park, C.-M. Auxin Homeostasis in Plant Stress Adaptation Response. Plant Signal. Behav. 2007, 2, 306–307. [Google Scholar] [CrossRef]
- Verma, S.; Negi, N.P.; Pareek, S.; Mudgal, G.; Kumar, D. Auxin response factors in plant adaptation to drought and salinity stress. Physiol. Plant. 2022, 174, e13714. [Google Scholar] [CrossRef] [PubMed]
- Angon, P.B.; Mondal, S.; Akter, S.; Sakil, M.A.; Jalil, M.A. Roles of CRISPR to mitigate drought and salinity stresses on plants. Plant Stress 2023, 8, 100169. [Google Scholar] [CrossRef]
- Alam, M.S.; Kong, J.; Tao, R.; Ahmed, T.; Alamin, M.; Alotaibi, S.S.; Abdelsalam, N.R.; Xu, J.-H. CRISPR/Cas9 Mediated Knockout of the OsbHLH024 Transcription Factor Improves Salt Stress Resistance in Rice (Oryza sativa L.). Plants 2022, 11, 1184. [Google Scholar] [CrossRef]
- Wani, S.H.; Kumar, V.; Khare, T.; Guddimalli, R.; Parveda, M.; Solymosi, K.; Suprasanna, P.; Kavi Kishor, P.B. Engineering salinity tolerance in plants: Progress and prospects. Planta 2020, 251, 76. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.J.; Negrão, S.; Tester, M. Salt resistant crop plants. Curr. Opin. Biotechnol. 2014, 26, 115–124. [Google Scholar] [CrossRef]
- DiCara, C.; Gedan, K. Distinguishing the Effects of Stress Intensity and Stress Duration in Plant Responses to Salinity. Plants 2023, 12, 2522. [Google Scholar] [CrossRef] [PubMed]
- Thabet, S.G.; Elkelish, A.; Alwutayd, K.M.; Alqudah, A.M. Genetic Insights into Stress Resilience via Enhancing Salinity Adaptation in Barley. Plant Mol. Biol. Rep. 2024, 42, 424–434. [Google Scholar] [CrossRef]
- Sharma, R.; Mishra, M.; Gupta, B.; Parsania, C.; Singla-Pareek, S.L.; Pareek, A. De Novo Assembly and Characterization of Stress Transcriptome in a Salinity-Tolerant Variety CS52 of Brassica juncea. PLoS ONE 2015, 10, e0126783. [Google Scholar] [CrossRef] [PubMed]
- Yong, H.-Y.; Zou, Z.; Kok, E.-P.; Kwan, B.-H.; Chow, K.; Nasu, S.; Nanzyo, M.; Kitashiba, H.; Nishio, T. Comparative Transcriptome Analysis of Leaves and Roots in Response to Sudden Increase in Salinity in Brassica napus by RNA-seq. BioMed Res. Int. 2014, 2014, 467395. [Google Scholar] [CrossRef]
- Mohamed, I.A.A.; Shalby, N.; El-Badri, A.M.; Batool, M.; Wang, C.; Wang, Z.; Salah, A.; Rady, M.M.; Jie, K.; Wang, B.; et al. RNA-seq analysis revealed key genes associated with salt tolerance in rapeseed germination through carbohydrate metabolism, hormone, and MAPK signaling pathways. Ind. Crops Prod. 2022, 176, 114262. [Google Scholar] [CrossRef]
- Chen, B.; Bian, X.; Tu, M.; Yu, T.; Jiang, L.; Lu, Y.; Chen, X. Moderate Salinity Stress Increases the Seedling Biomass in Oilseed Rape (Brassica napus L.). Plants 2023, 12, 1650. [Google Scholar] [CrossRef]
- Nazari, M.; Ghasemi-Soloklui, A.A.; Kordrostami, M.; Abdel Latef, A.A.H. Deciphering the response of medicinal plants to abiotic stressors: A focus on drought and salinity. Plant Stress 2023, 10, 100255. [Google Scholar] [CrossRef]
- Diretto, G.; Tavazza, R.; Welsch, R.; Pizzichini, D.; Mourgues, F.; Papacchioli, V.; Beyer, P.; Giuliano, G. Metabolic engineering of potato tuber carotenoids through tuber-specific silencing of lycopene epsilon cyclase. BMC Plant Biology 2006, 6, 13. [Google Scholar] [CrossRef]
- Shahid, M.A.; Sarkhosh, A.; Khan, N.; Balal, R.M.; Ali, S.; Rossi, L.; Gómez, C.; Mattson, N.; Nasim, W.; Garcia-Sanchez, F. Insights into the Physiological and Biochemical Impacts of Salt Stress on Plant Growth and Development. Agronomy 2020, 10, 938. [Google Scholar] [CrossRef]
- Adil, M.; Bashir, S.; Bashir, S.; Aslam, Z.; Ahmad, N.; Younas, T.; Asghar, R.M.A.; Alkahtani, J.; Dwiningsih, Y.; Elshikh, M.S. Zinc oxide nanoparticles improved chlorophyll contents, physical parameters, and wheat yield under salt stress. Front. Plant Sci. 2022, 13, 932861. [Google Scholar] [CrossRef]
- Hand, M.J.; Taffouo, V.D.; Nouck, A.E.; Nyemene, K.P.J.; Tonfack, B.; Meguekam, T.L.; Youmbi, E. Effects of Salt Stress on Plant Growth, Nutrient Partitioning, Chlorophyll Content, Leaf Relative Water Content, Accumulation of Osmolytes and Antioxidant Compounds in Pepper (Capsicum annuum L.) Cultivars. Not. Bot. Horti Agrobot. Cluj-Napoca 2017, 45, 481–490. [Google Scholar] [CrossRef]
- Santos, C.V. Regulation of chlorophyll biosynthesis and degradation by salt stress in sunflower leaves. Sci. Hortic. 2004, 103, 93–99. [Google Scholar] [CrossRef]
- Zahra, N.; Al Hinai, M.S.; Hafeez, M.B.; Rehman, A.; Wahid, A.; Siddique, K.H.M.; Farooq, M. Regulation of photosynthesis under salt stress and associated tolerance mechanisms. Plant Physiol. Biochem. 2022, 178, 55–69. [Google Scholar] [CrossRef]
- Borghesi, E.; González-Miret, M.L.; Escudero-Gilete, M.L.; Malorgio, F.; Heredia, F.J.; Meléndez-Martínez, A.J. Effects of Salinity Stress on Carotenoids, Anthocyanins, and Color of Diverse Tomato Genotypes. J. Agric. Food Chem. 2011, 59, 11676–11682. [Google Scholar] [CrossRef] [PubMed]
- Ben Abdallah, S.; Aung, B.; Amyot, L.; Lalin, I.; Lachâal, M.; Karray-Bouraoui, N.; Hannoufa, A. Salt stress (NaCl) affects plant growth and branch pathways of carotenoid and flavonoid biosyntheses in Solanum nigrum. Acta Physiol. Plant. 2016, 38, 72. [Google Scholar] [CrossRef]
- Lim, J.-H.; Park, K.-J.; Kim, B.-K.; Jeong, J.-W.; Kim, H.-J. Effect of salinity stress on phenolic compounds and carotenoids in buckwheat (Fagopyrum esculentum M.) sprout. Food Chem. 2012, 135, 1065–1070. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Ahn, Y.O.; Ahn, M.-J.; Lee, H.-S.; Kwak, S.-S. Down-regulation of β-carotene hydroxylase increases β-carotene and total carotenoids enhancing salt stress tolerance in transgenic cultured cells of sweetpotato. Phytochemistry 2012, 74, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Saleem, S.; Ul Mushtaq, N.; Shah, W.H.; Rasool, A.; Hakeem, K.R.; Ul Rehman, R. Beneficial Role of Phytochemicals in Oxidative Stress Mitigation in Plants. In Antioxidant Defense in Plants: Molecular Basis of Regulation; Aftab, T., Hakeem, K.R., Eds.; Springer: Singapore, 2022; pp. 435–451. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef]
- Chen, H.-C.; Huang, S.-C.; Chen, Y.-F.; Kuo, C.-W.; Chen, Y.-H.; Chang, M.-C. Overexpression of OsERF106MZ promotes parental root growth in rice seedlings by relieving the ABA-mediated inhibition of root growth under salinity stress conditions. BMC Plant Biol. 2023, 23, 144. [Google Scholar] [CrossRef] [PubMed]
- Rong, W.; Qi, L.; Wang, A.; Ye, X.; Du, L.; Liang, H.; Xin, Z.; Zhang, Z. The ERF transcription factor TaERF3 promotes tolerance to salt and drought stresses in wheat. Plant Biotechnol. J. 2014, 12, 468–479. [Google Scholar] [CrossRef]
- Pan, Y.; Seymour, G.B.; Lu, C.; Hu, Z.; Chen, X.; Chen, G. An ethylene response factor (ERF5) promoting adaptation to drought and salt tolerance in tomato. Plant Cell Rep. 2012, 31, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Chen, M.; Li, L.; Xu, Z.; Chen, X.; Guo, J.; Ma, Y. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 2009, 60, 3781–3796. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Bao, X.; Zhi, Y.; Wu, Q.; Guo, Y.; Yin, X.; Zeng, L.; Li, J.; Zhang, J.; He, W.; et al. Overexpression of a MYB Family Gene, OsMYB6, Increases Drought and Salinity Stress Tolerance in Transgenic Rice. Front. Plant Sci. 2019, 10, 168. [Google Scholar] [CrossRef]
- Zhu, N.; Cheng, S.; Liu, X.; Du, H.; Dai, M.; Zhou, D.-X.; Yang, W.; Zhao, Y. The R2R3-type MYB gene OsMYB91 has a function in coordinating plant growth and salt stress tolerance in rice. Plant Sci. 2015, 236, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Khoso, M.A.; Hussain, A.; Ritonga, F.N.; Ali, Q.; Channa, M.M.; Alshegaihi, R.M.; Meng, Q.; Ali, M.; Zaman, W.; Brohi, R.D.; et al. WRKY transcription factors (TFs): Molecular switches to regulate drought, temperature, and salinity stresses in plants. Front. Plant Sci. 2022, 13, 1039329. [Google Scholar] [CrossRef]
- Uarrota, V.G.; Stefen, D.L.V.; Leolato, L.S.; Gindri, D.M.; Nerling, D. Revisiting Carotenoids and Their Role in Plant Stress Responses: From Biosynthesis to Plant Signaling Mechanisms During Stress. In Antioxidants and Antioxidant Enzymes in Higher Plants; Gupta, D.K., Palma, J.M., Corpas, F.J., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 207–232. [Google Scholar] [CrossRef]
- Ismaiel, M.M.S.; El-Ayouty, Y.M.; Said, A.A.; Fathey, H.A. Transformation of Dunaliella parva with PSY gene: Carotenoids show enhanced antioxidant activity under polyethylene glycol and calcium treatments. Biocatal. Agric. Biotechnol. 2018, 16, 378–384. [Google Scholar] [CrossRef]
- Li, X.; Wang, Y.; Chen, S.; Tian, H.; Fu, D.; Zhu, B.; Luo, Y.; Zhu, H. Lycopene Is Enriched in Tomato Fruit by CRISPR/Cas9-Mediated Multiplex Genome Editing. Front. Plant Sci. 2018, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ren, P.; Ji, L.; Zhu, B.; Xie, G. OsVDE, a xanthophyll cycle key enzyme, mediates abscisic acid biosynthesis and negatively regulates salinity tolerance in rice. Planta 2021, 255, 6. [Google Scholar] [CrossRef]
- Song, J.; Sun, B.; Chen, C.; Ning, Z.; Zhang, S.; Cai, Y.; Zheng, X.; Cao, B.; Chen, G.; Jin, D.; et al. An R-R-type MYB transcription factor promotes non-climacteric pepper fruit carotenoid pigment biosynthesis. Plant J. 2023, 115, 724–741. [Google Scholar] [CrossRef] [PubMed]
- Meier, S.; Tzfadia, O.; Vallabhaneni, R.; Gehring, C.; Wurtzel, E.T. A transcriptional analysis of carotenoid, chlorophyll and plastidial isoprenoid biosynthesis genes during development and osmotic stress responses in Arabidopsis thaliana. BMC Syst. Biol. 2011, 5, 77. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Li, S.; Asim, M.; Mao, J.; Xu, D.; Ullah, Z.; Liu, G.; Wang, Q.; Liu, H. The Arabidopsis Calcium-Dependent Protein Kinases (CDPKs) and Their Roles in Plant Growth Regulation and Abiotic Stress Responses. Int. J. Mol. Sci. 2018, 19, 1900. [Google Scholar] [CrossRef]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How Plant Hormones Mediate Salt Stress Responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef] [PubMed]
- Arif, Y.; Singh, P.; Siddiqui, H.; Bajguz, A.; Hayat, S. Salinity induced physiological and biochemical changes in plants: An omic approach towards salt stress tolerance. Plant Physiol. Biochem. 2020, 156, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H.K. Chlorophyll Fluorescence Signatures of Leaves during the Autumnal Chlorophyll Breakdown. J. Plant Physiol. 1987, 131, 101–110. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Chen, H.; Wang, T.; He, X.; Cai, X.; Lin, R.; Liang, J.; Wu, J.; King, G.; Wang, X. BRAD V3.0: An upgraded Brassicaceae database. Nucleic Acids Res. 2022, 50, D1432–D1441. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, R.; Harris, M.A.; Huntley, R.; Van Auken, K.; Cherry, J.M. A guide to best practices for Gene Ontology (GO) manual annotation. Database 2013, 2013, bat054. [Google Scholar] [CrossRef] [PubMed]
- Mu, H.; Chen, J.; Huang, W.; Huang, G.; Deng, M.; Hong, S.; Ai, P.; Gao, C.; Zhou, H. OmicShare tools: A zero-code interactive online platform for biological data analysis and visualization. iMeta 2024, 3, e228. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Yu, S.; Zhang, F.; Shen, X.; Zhao, X.; Yu, Y.; Zhang, D. Reference Gene Selection for Real-Time Quantitative Polymerase Chain Reaction of mRNA Transcript Levels in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Plant Mol. Biol. Rep. 2010, 28, 597–604. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, J.; Wang, F.; Yuan, M.; Wang, Z.; Qin, Z.; Zhang, X.; Shao, Y.; Pei, H. Transcriptomic Analysis Reveals That the Photosynthesis and Carotenoid Metabolism Pathway Is Involved in the Salinity Stress Response in Brassica rapa L. ssp. Pekinensis. Plants 2025, 14, 566. https://doi.org/10.3390/plants14040566
Jia J, Wang F, Yuan M, Wang Z, Qin Z, Zhang X, Shao Y, Pei H. Transcriptomic Analysis Reveals That the Photosynthesis and Carotenoid Metabolism Pathway Is Involved in the Salinity Stress Response in Brassica rapa L. ssp. Pekinensis. Plants. 2025; 14(4):566. https://doi.org/10.3390/plants14040566
Chicago/Turabian StyleJia, Jin, Fengshuo Wang, Mengmeng Yuan, Zhiying Wang, Zhe Qin, Xiaoli Zhang, Yutao Shao, and Haixia Pei. 2025. "Transcriptomic Analysis Reveals That the Photosynthesis and Carotenoid Metabolism Pathway Is Involved in the Salinity Stress Response in Brassica rapa L. ssp. Pekinensis" Plants 14, no. 4: 566. https://doi.org/10.3390/plants14040566
APA StyleJia, J., Wang, F., Yuan, M., Wang, Z., Qin, Z., Zhang, X., Shao, Y., & Pei, H. (2025). Transcriptomic Analysis Reveals That the Photosynthesis and Carotenoid Metabolism Pathway Is Involved in the Salinity Stress Response in Brassica rapa L. ssp. Pekinensis. Plants, 14(4), 566. https://doi.org/10.3390/plants14040566





