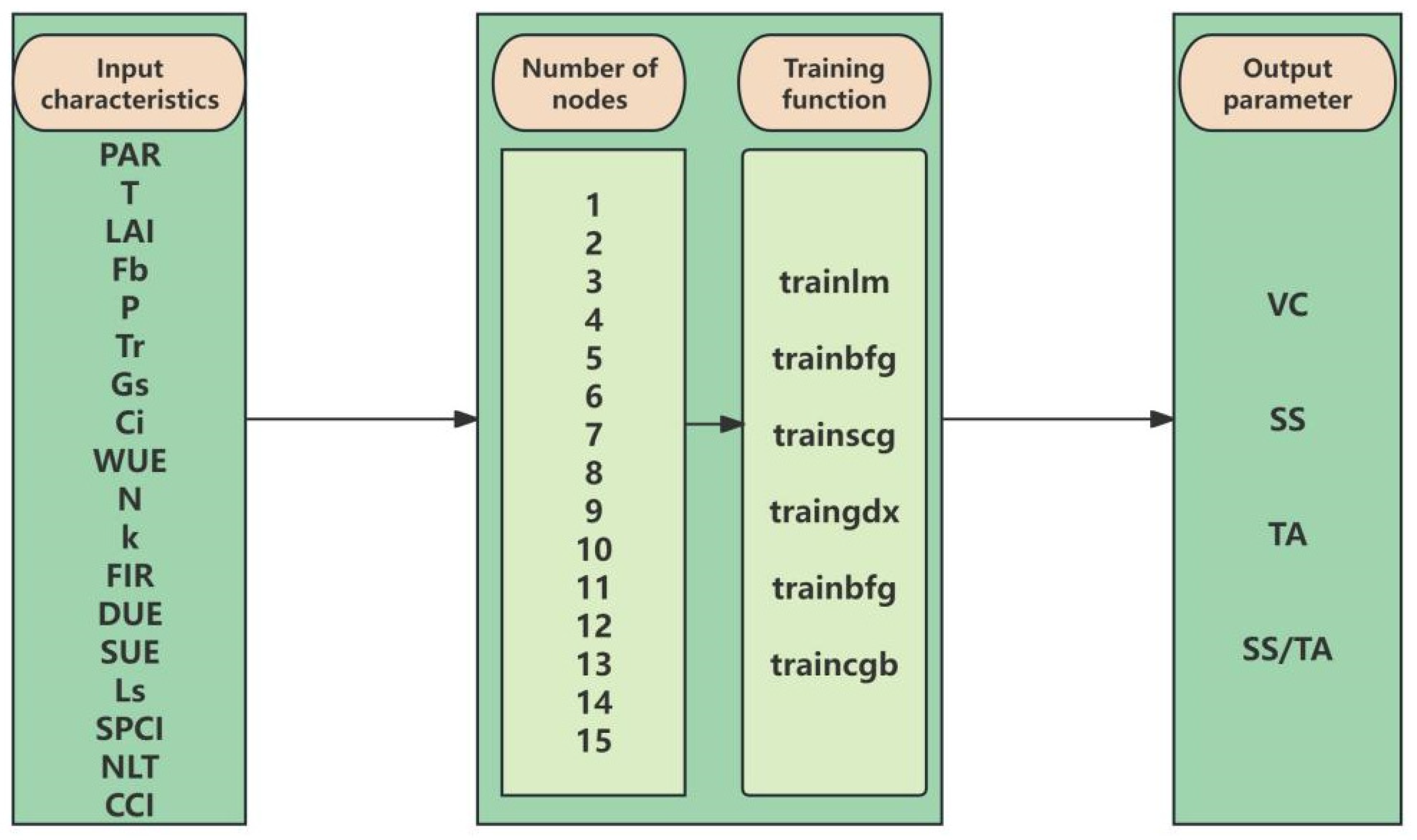
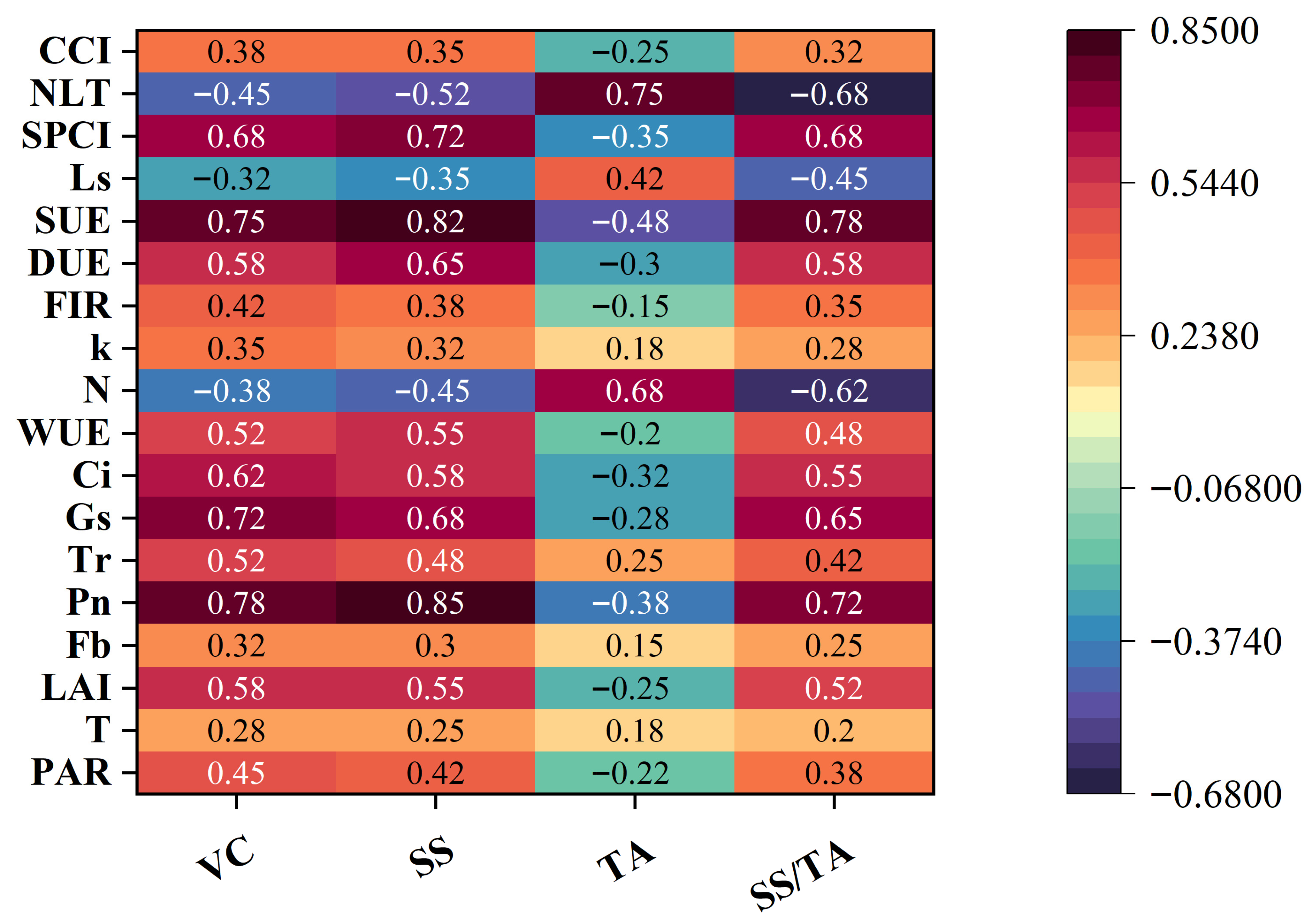
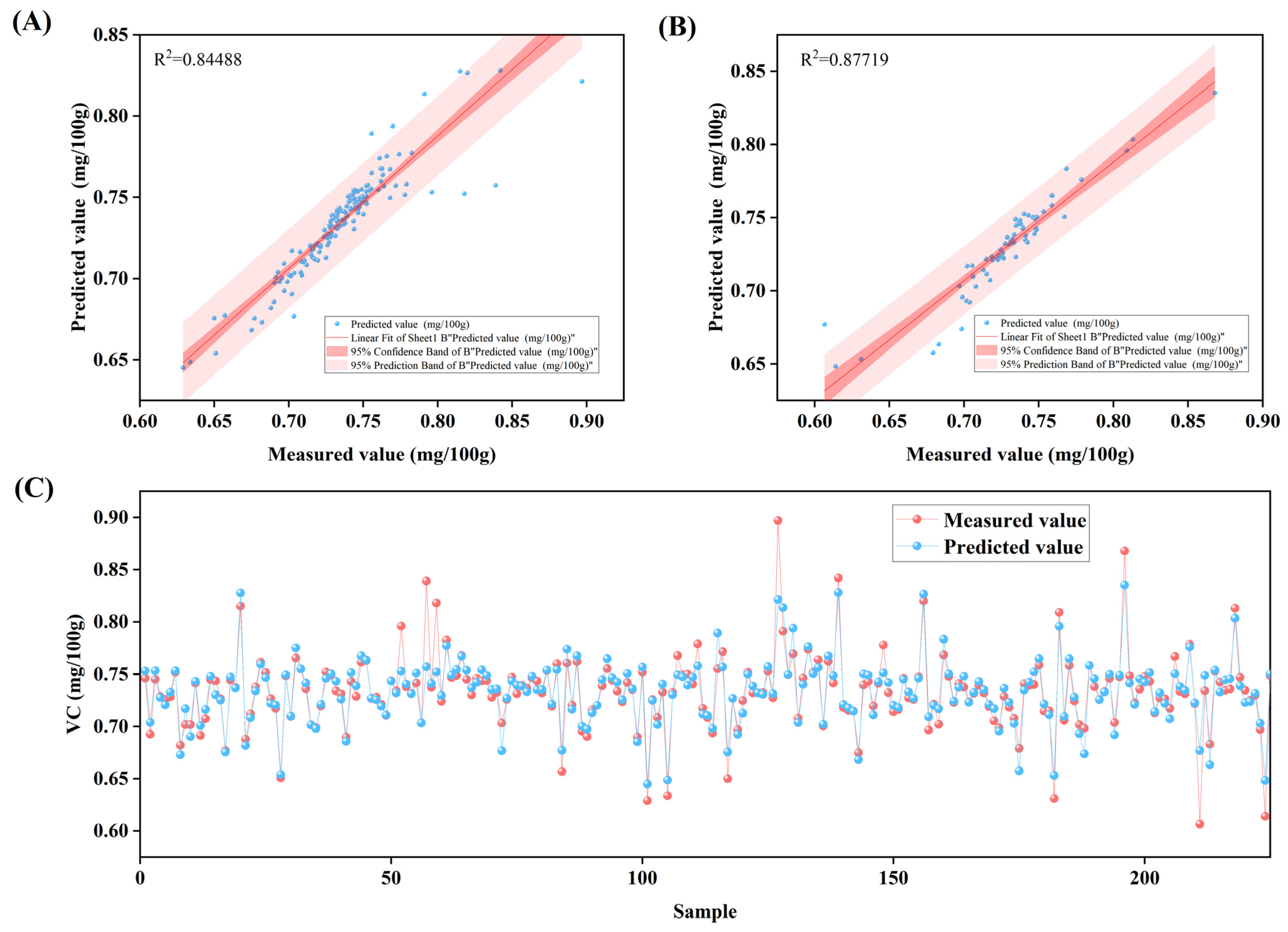
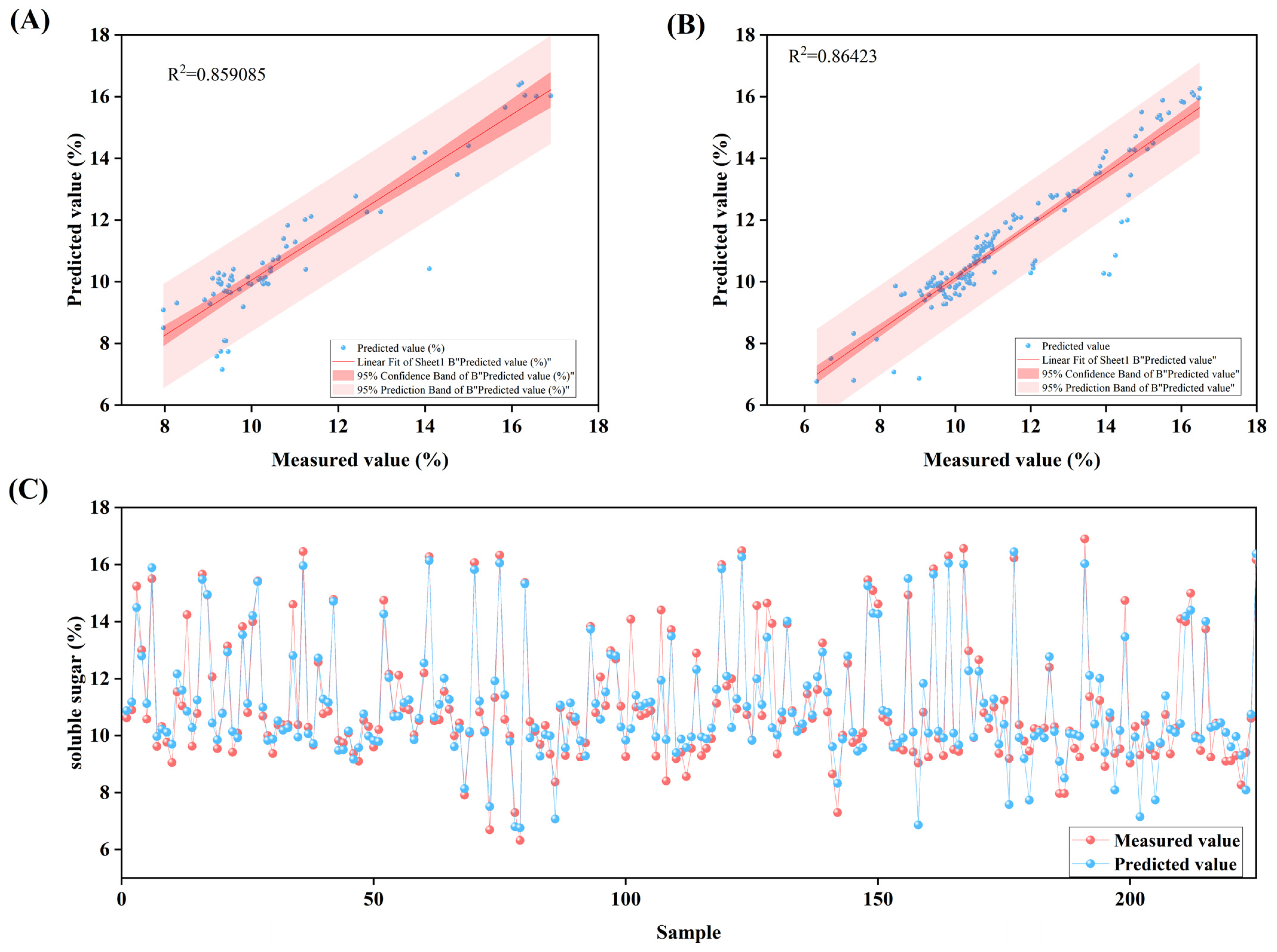
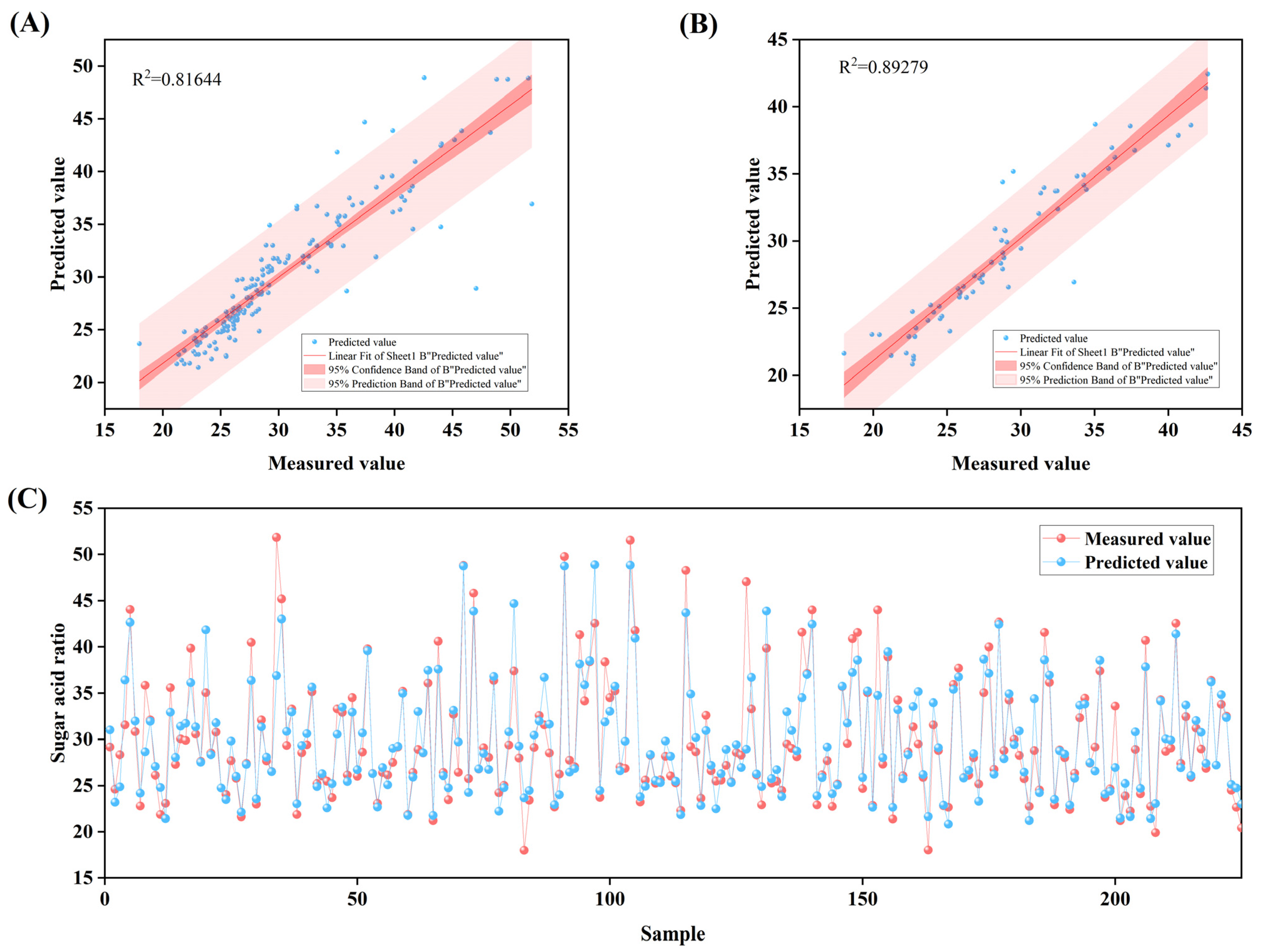
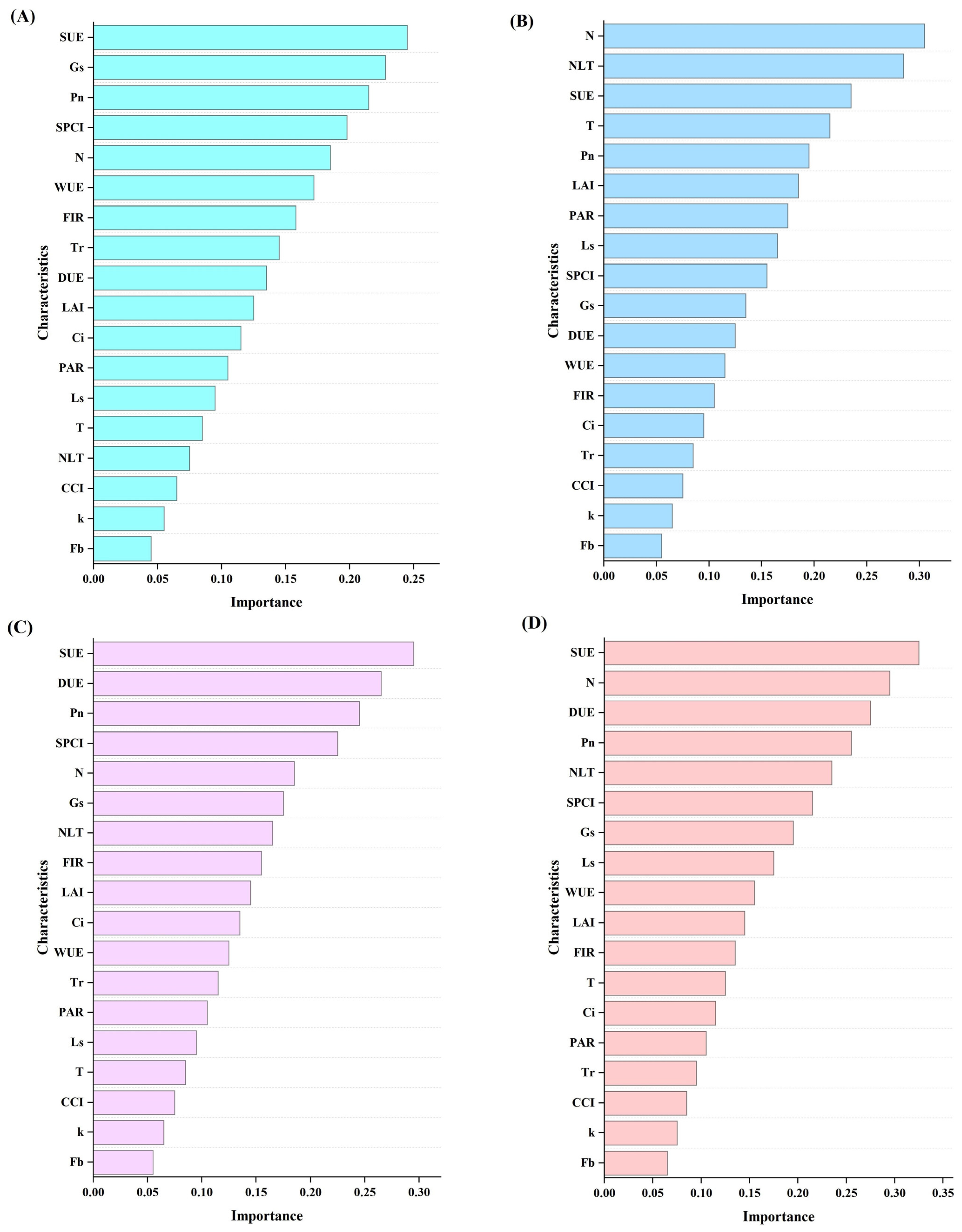
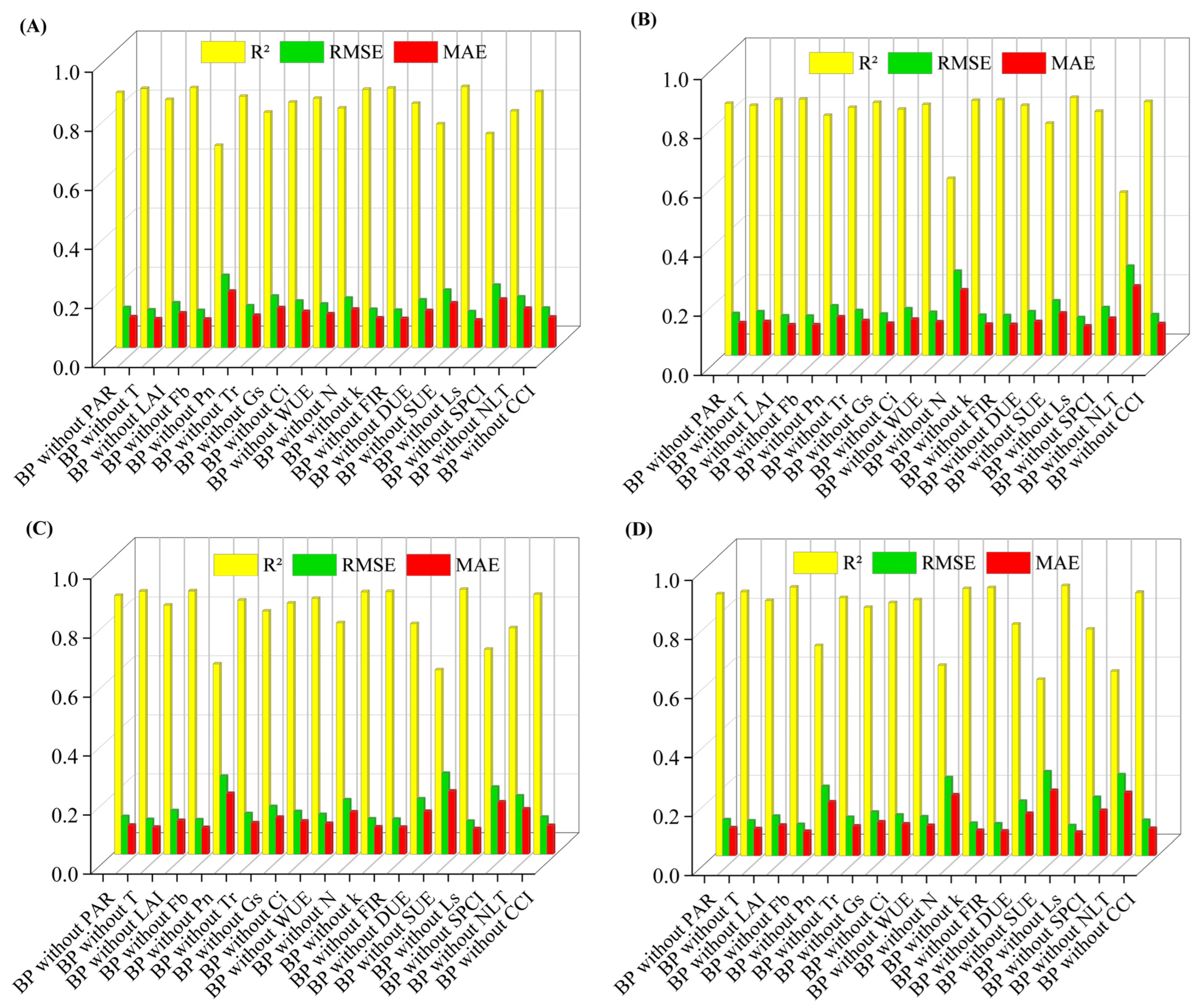
2.3.1. Determination of Fruit Quality Parameters
The vitamin C content was determined using the molybdenum blue colorimetric method [
17]. First, a 1 mg/mL ascorbic acid standard stock solution was prepared: 100 mg of ascorbic acid standard was accurately weighed and diluted to 100 mL with a 50 g/L oxalic acid solution. Aliquots of 0, 0.2, 0.4, 0.6, 0.8, and 1.0 mL of the stock solution were separately transferred into 10 mL centrifuge tubes, and each was adjusted to a final volume of 1.0 mL by adding 50 g/L oxalic acid solution. Subsequently, 1 mL of 5% sulfuric acid-ethanol solution and 1 mL of 5% ammonium molybdate solution were added sequentially to each tube. After thorough mixing, the reaction was carried out in a water bath at 30 °C for 30 min. The absorbance was measured at a wavelength of 705 nm, and a standard curve was plotted.
For sample analysis, 2.00 g of fruit homogenate was accurately weighed and mixed with 10 mL of pre-chilled 50 g/L oxalic acid solution. The mixture was homogenized in an ice bath and then centrifuged at 4 °C and 8000 r/min for 15 min. The resulting supernatant was collected as the test solution. A 1.0 mL aliquot of the test solution was taken, and sulfuric acid-ethanol solution and ammonium molybdate solution were added following the same procedure described above. The mixture was reacted in a water bath at 30 °C for 30 min before measurement.
The absorbance of the reaction solution was measured at a wavelength of 705 nm using a UV-Vis spectrophotometer. The vitamin C content in the sample was calculated based on the standard curve, as shown in Formula (1).
where C is the ascorbic acid concentration (μg/mL) obtained from the standard curve; V is the total volume of the extraction solution (mL); n is the dilution factor; and m is the sample mass (g).
The soluble sugar content was determined by the anthrone colorimetric method [
18]. First, a 1 mg/mL glucose standard solution was prepared by accurately weighing 100 mg of anhydrous glucose, dissolving it in distilled water, and diluting to a final volume of 100 mL. Aliquots of 0, 0.1, 0.2, 0.4, 0.6, and 0.8 mL of this solution were separately transferred into stoppered test tubes, and distilled water was added to bring the total volume in each tube to 1.0 mL. Subsequently, 4.0 mL of a freshly prepared anthrone-ethyl acetate solution (0.2 g anthrone dissolved in 100 mL ethyl acetate) and 5.0 mL of concentrated sulfuric acid were added sequentially. After thorough shaking, the mixtures were reacted in a boiling water bath for 10 min, immediately cooled in an ice bath, and the absorbance was measured at a wavelength of 630 nm to construct the standard curve.
For sample processing, 1.00 g of fruit homogenate was accurately weighed, mixed with 10 mL of an 80% ethanol solution, and extracted in an 80 °C water bath for 30 min with intermittent shaking. After cooling, the mixture was centrifuged at 4000 r/min for 10 min, and the supernatant was collected. The residue was extracted once more, and the combined supernatants were decolorized with activated carbon and then diluted to 25 mL with 80% ethanol. A 0.5 mL aliquot of the extract was taken and subjected to the color development procedure described above. The absorbance at 630 nm was measured, and the soluble sugar content was calculated according to Formula (2).
The titratable acidity (TA) content was determined by neutralization titration with NaOH [
19]. Precisely 5.00 g of fruit homogenate sample was weighed into a 150 mL conical flask. Then, 50 mL of freshly boiled and cooled distilled water was added, and the mixture was extracted in an 80 °C water bath for 30 min with occasional shaking. After cooling, the mixture was filtered, and the residue was washed with distilled water 3–4 times. The filtrate was collected and diluted to volume in a 100 mL volumetric flask.
Exactly 20.0 mL of the sample extract was pipetted into a 150 mL conical flask, followed by the addition of 2 drops of 1% phenolphthalein indicator. The solution was titrated with a 0.01 mol/L NaOH standard solution until a faint pink color appeared and persisted for 30 s as the endpoint. The volume of NaOH solution consumed was recorded. A blank test was performed simultaneously using distilled water.
The titratable acidity, expressed as malic acid, was calculated according to the following formula (Formula (3)).
V is the volume of NaOH consumed in sample titration (mL); V0 is the volume of NaOH consumed in blank titration (mL); C is the concentration of the NaOH standard solution (mol/L); K is the conversion factor for malic acid (0.067); V1 is the total volume of the sample extract (mL); V2 is the volume of sample solution used in titration (mL); m is the sample mass (g).
Sugar-acid ratio (SS/TA): Calculated as (SS/TA) [
20].
2.3.2. Determination of Photosynthetic and Canopy Structural Indices in Fruit Trees
Photosynthetically active radiation (PAR) was measured using an LAI-2200C Plant Canopy Analyzer (LI-COR Biosciences, Lincoln, NE, USA) or an equivalent model [
21]. The instrument was equipped with a 180° fisheye lens and calculated PAR by comparing the radiation difference above and below the canopy. Measurements were conducted under clear, cloudless weather conditions, within the time window from 2 h after sunrise to 2 h before sunset.
Prior to formal measurement, the instrument was preheated for 20 min and uniformly set to the UTC + 8 time zone to automatically record timestamps. Initial calibration was performed at an open, unobstructed area to establish a reference point, and reference PAR data above the canopy were collected. Subsequently, five measurement points were arranged beneath the canopy of each sample tree, located at the four quadrant points and the center point of the crown projection. The detector was placed horizontally at a height of 1.5 m above the ground, with the fisheye lens oriented toward the zenith. At each measurement point, three consecutive readings were recorded, and their average was taken as the PAR value for that point.
During the measurement process, radiation data were simultaneously recorded both above the canopy (in an open area) and below the canopy. The instrument automatically calculated the canopy transmittance and output the PAR value (unit: μmol·m
−2·s
−1). Finally, the distribution of PAR within the canopy was analyzed based on the instrument’s built-in model, with the specific calculation formula provided in Formula (4).
is the measurement value at the reference point above the canopy; is the total transmittance of the canopy (calculated by the instrument using the gap fraction model).
Leaf area index (LAI) and direct transmittance coefficient (T) were measured using an LAI-2200C plant canopy analyzer (LI-COR Biosciences, USA) [
22]. The instrument is equipped with a 180° fisheye lens and a light sensor, enabling simultaneous acquisition of canopy digital images and radiation data. Measurements were conducted under clear, cloudless weather conditions between sunrise and sunset, specifically during periods when the solar elevation angle exceeded 30° and light intensity was stable (10:00–14:00), with no strong wind interference (wind speed < 3 m/s).
Prior to measurement, the instrument was preheated for 15 min, and the measurement parameters were set to 5 zenith angle rings and 4 azimuth angles. A reference point measurement was performed in an open area above the canopy: the sensor was placed horizontally at a height of 1.8 m above the ground, and three sets of PAR data were recorded consecutively. The average of these readings was taken as the reference value above the canopy (PAR
0). For each sample tree, measurement points were arranged using the five-point method, located at the center and the east, west, south, and north directions, respectively. Each point was positioned at a distance of two-thirds of the crown radius from the main trunk. The sensor was kept horizontal with the lens facing the zenith, and three sets of valid data were acquired at each measurement point.
In Equation (5), G is the leaf projection function (default value is 0.85); Ω is the aggregation index (default value is 0.95).
In Equation (6), T is the direct transmittance coefficient; PARbelow is the average photosynthetically active radiation below the canopy; PAR0 is the reference value above the canopy.
Net photosynthetic rate (Pn), transpiration rate (Tr), stomatal conductance (Gs), and intercellular CO
2 concentration (Ci) were measured using an LI-6800 portable photosynthesis system (LI-COR Biosciences, USA) [
23,
24,
25]. The instrument was equipped with a standard 2 × 3 cm transparent leaf chamber and a red-blue light source. All measurements were conducted between 9:00 and 11:00 AM under clear weather conditions.
Prior to measurement, the instrument was preheated for 30 min. The CO2 absorbent and desiccant were replaced, and the CO2/H2O infrared gas analyzer was calibrated. The LED light source was also checked and preheated. The measurement parameters were set as follows: photosynthetic photon flux density (PPFD) at 1000 μmol·m−2·s−1, reference chamber CO2 concentration maintained at 400 μmol·mol−1, leaf chamber temperature controlled at 25 ± 1 °C, airflow rate at 500 μmol·s−1, and relative humidity kept at 60 ± 5%. During measurement, current-year branches facing east within the canopy were selected. From these, fully expanded, well-illuminated, and disease-free functional leaves were chosen, and the measurement points were marked (avoiding the midrib). The leaf was flattened and clamped into the leaf chamber, ensuring complete coverage of the measurement window. After allowing parameters to stabilize (typically requiring 2–3 min), recording commenced when the rates of change for both the CO2 concentration difference (ΔCO2) and the H2O concentration difference (ΔH2O) fell below 0.5%/min. Three sets of stable data were collected consecutively at 30 S intervals, with the measured values for Pn, Tr, Gs, and Ci automatically output by the instrument.
Water use efficiency (WUE) was calculated using Equation (7) [
26].
The total nitrogen content in leaves was determined using the Kjeldahl method [
27]. After drying and grinding the leaf samples, 0.2000 g was accurately weighed, mixed with a combined catalyst and concentrated sulfuric acid, and digested at 420 °C for 2 h. The digest was then distilled in a Kjeldahl distillation apparatus, absorbed with boric acid, and titrated with a 0.01 mol/L HCl standard solution. The total nitrogen content was calculated according to Formula (8).
where V is the volume of hydrochloric acid consumed by the sample (mL); V0 is the volume of hydrochloric acid consumed by the blank (mL); C is the concentration of the hydrochloric acid standard solution (mol/L); D is the dilution factor (10 in this method); m is the sample mass (g); and 0.014 is the millimolar mass of nitrogen (g/mmol).
The stomatal limitation value (Ls) was calculated using Equation (9) [
28].
is the measured intercellular CO2 concentration (μmol·mol−1); is the atmospheric CO2 concentration, using the average measured value of 412 μmol·mol−1 above the canopy in this study.
Extinction coefficient (k): This parameter was calculated using the Beer-Lambert law in conjunction with LAI and the light intensity beneath the canopy, as shown in Equation (10) [
29].
where I is the average light intensity below the canopy (μmol·m
−2·s
−1); I
0 is the natural light intensity above the canopy (μmol·m
−2·s
−1); and LAI is the leaf area index.
Canopy fractional interception of radiation (FIR) [
30], direct light use efficiency (DUE) [
31], and scattered light use efficiency (SUE) [
32] were calculated using Formulas (11), (12), and (13), respectively, based on measurements of photosynthetically active radiation (PAR) inside and outside the canopy.
In the formula, PARbelow is the average photosynthetically active radiation below the canopy (μ mol·m−2·s−1); PARabove is the natural photosynthetically active radiation above the canopy (μ mol·m−2·s−1); ηdirect is the direct light conversion efficiency coefficient, with a value of 0.85; ηdiffuse is the diffuse light conversion efficiency coefficient, with a value of 0.92.
Canopy openness (Fb) was measured using a plant canopy analyzer [
33]. Multiple measurement points were evenly distributed beneath the canopy of the fruit trees. An LAI-2200C canopy analyzer (LI-COR Biosciences, Lincoln, NE, USA) was used to simultaneously record light data from a reference point above the canopy and from each measurement point below it. The instrument’s dedicated analysis software automatically calculated the canopy openness value based on the canopy porosity in the 0° zenith angle direction.