Evaluating Species Delimitation Methods in Chloroidium (Trebouxiophyceae, Chlorophyta): Efficacy of DNA Barcodes and Description of Chloroidium pseudoellipsoideum sp. nov. from Arctic Soils
Abstract
1. Introduction
2. Results
2.1. Light Microscopy
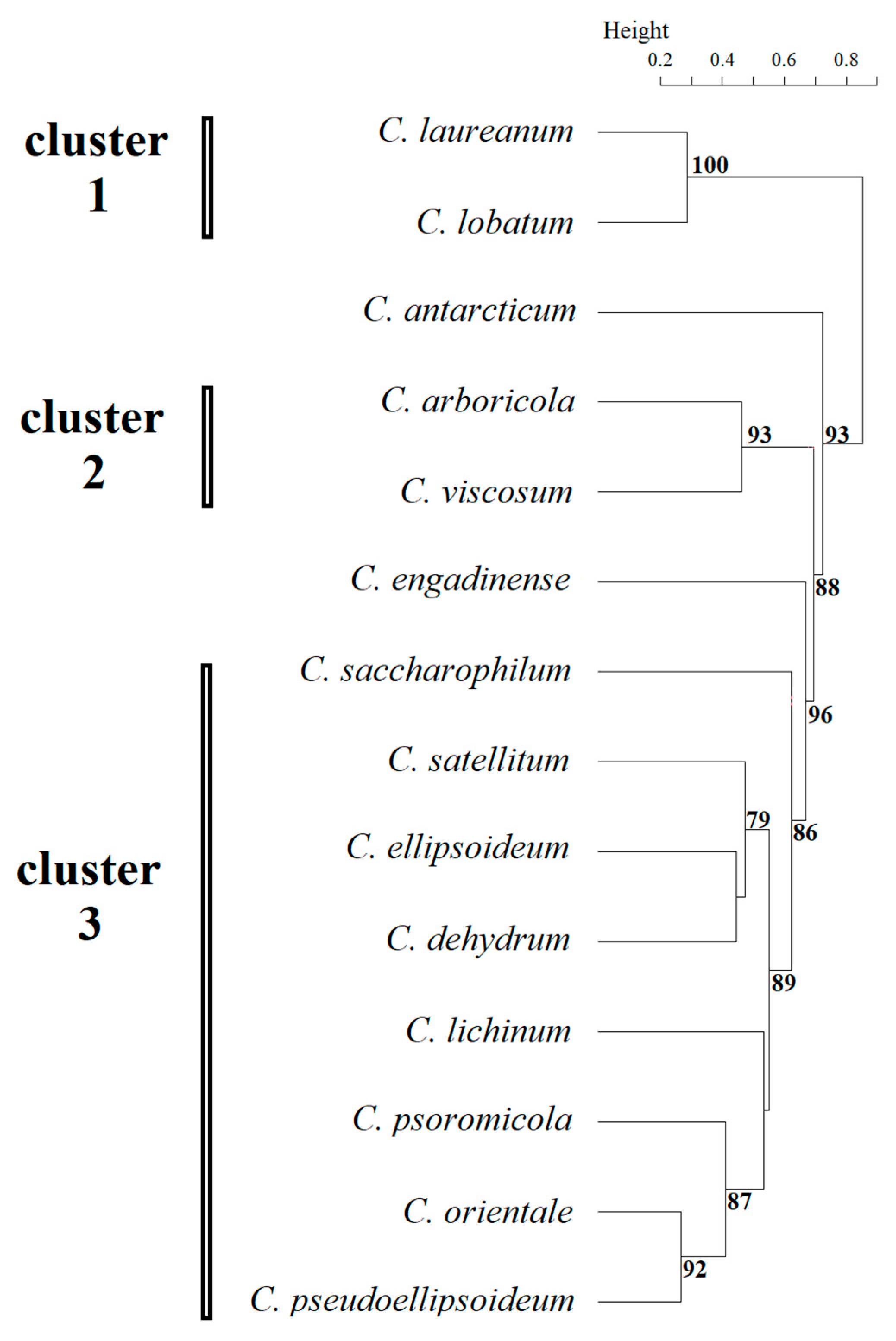
2.2. Morphological Clustering and Phylogenetic Incongruence
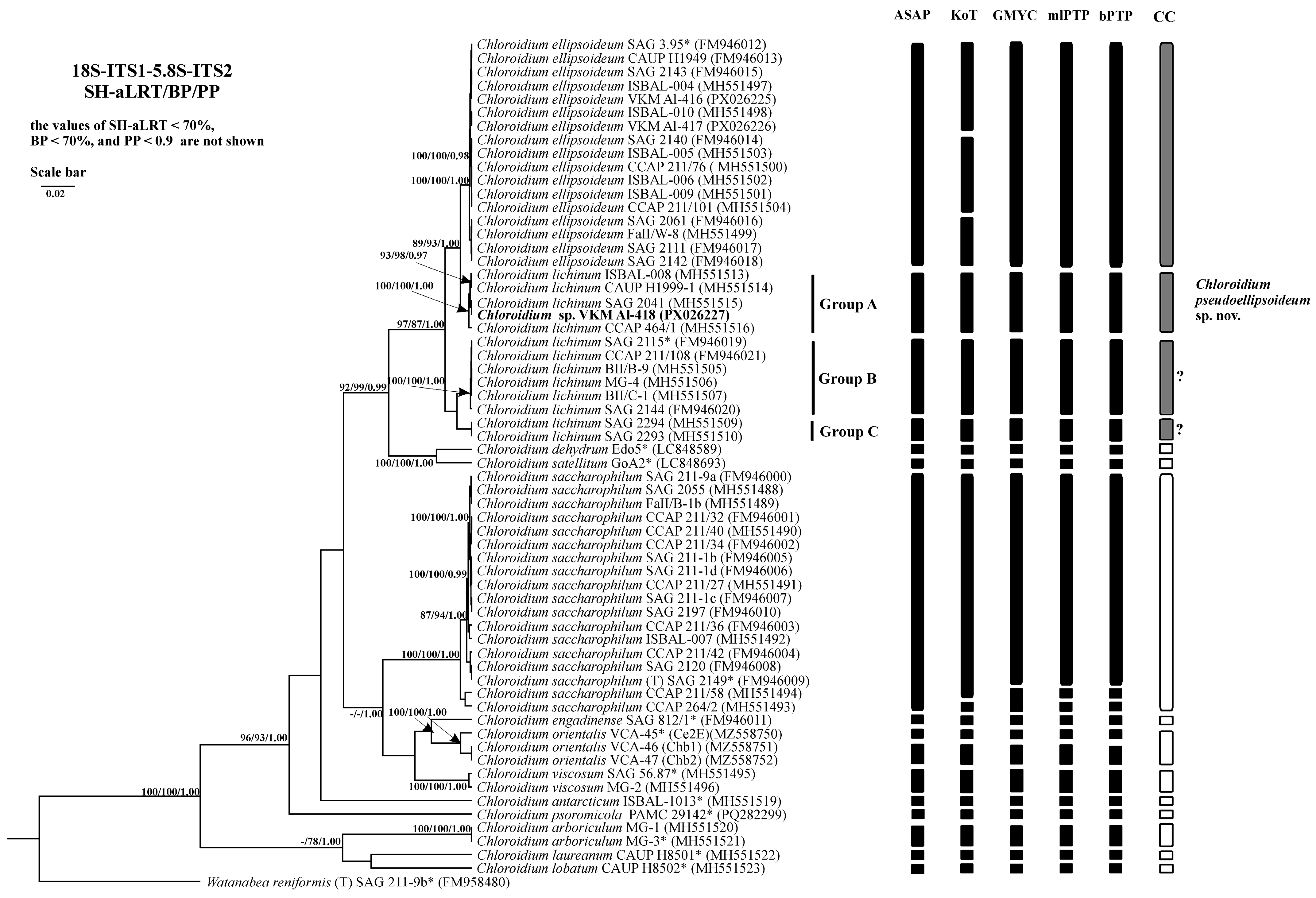
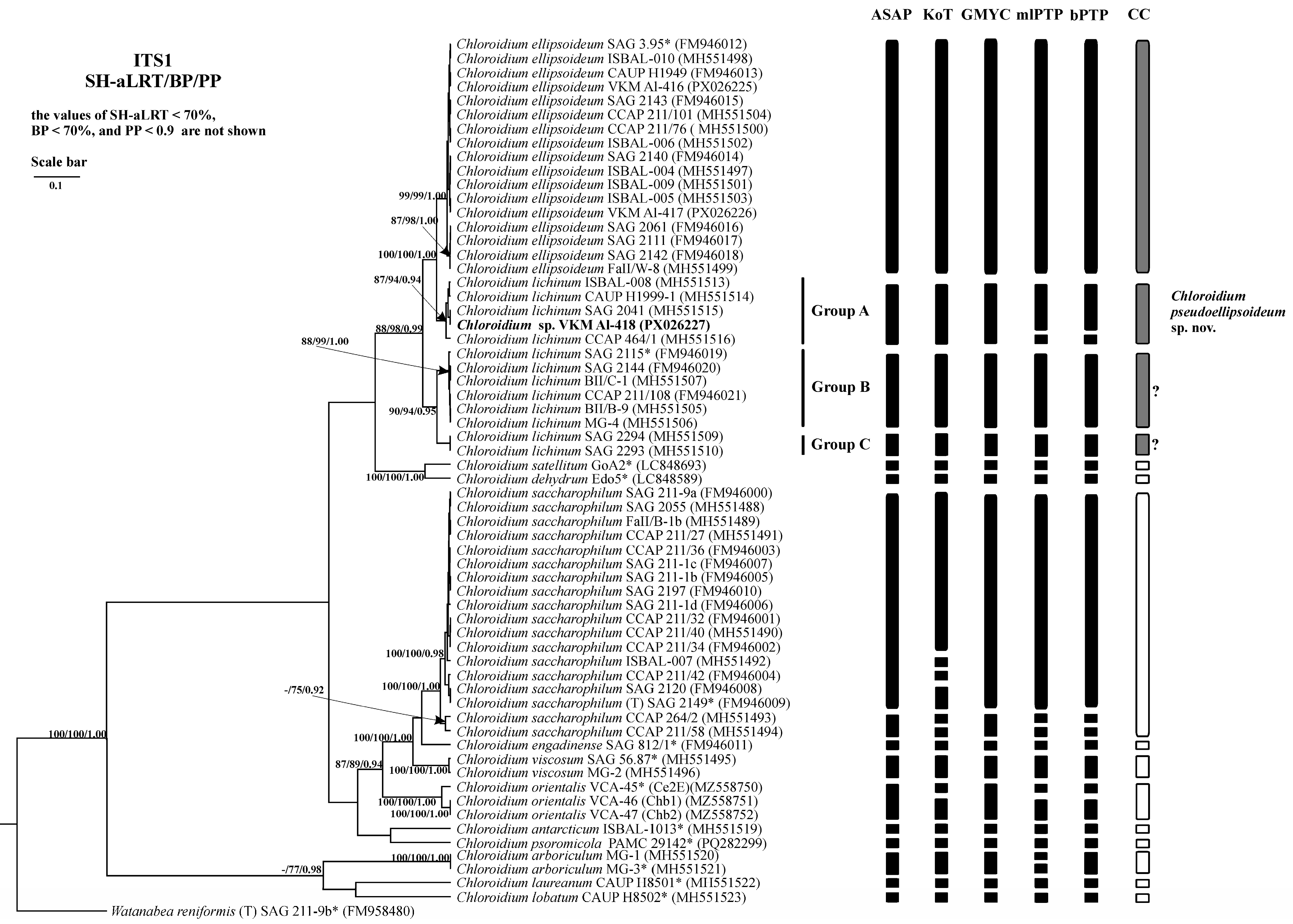
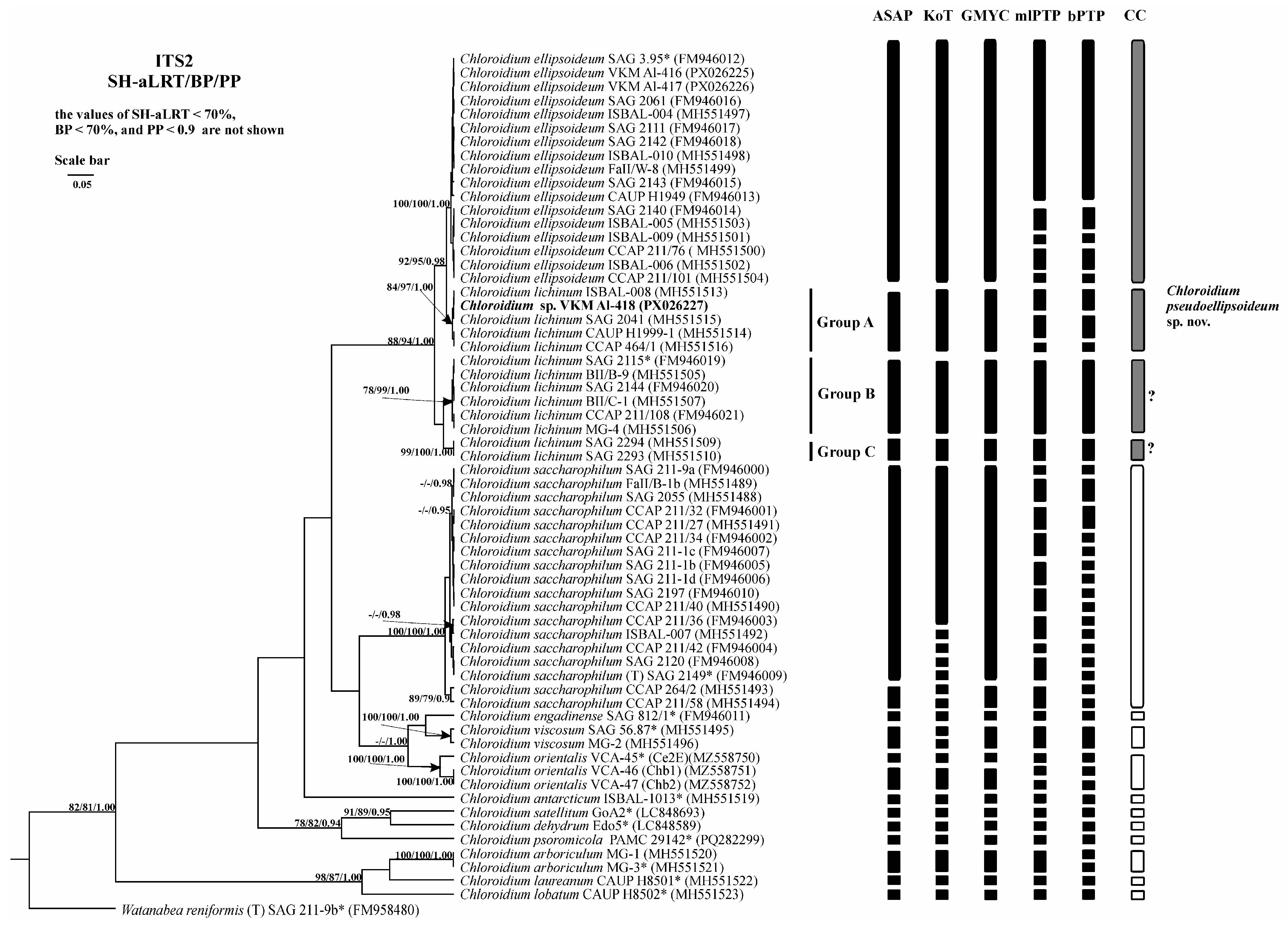
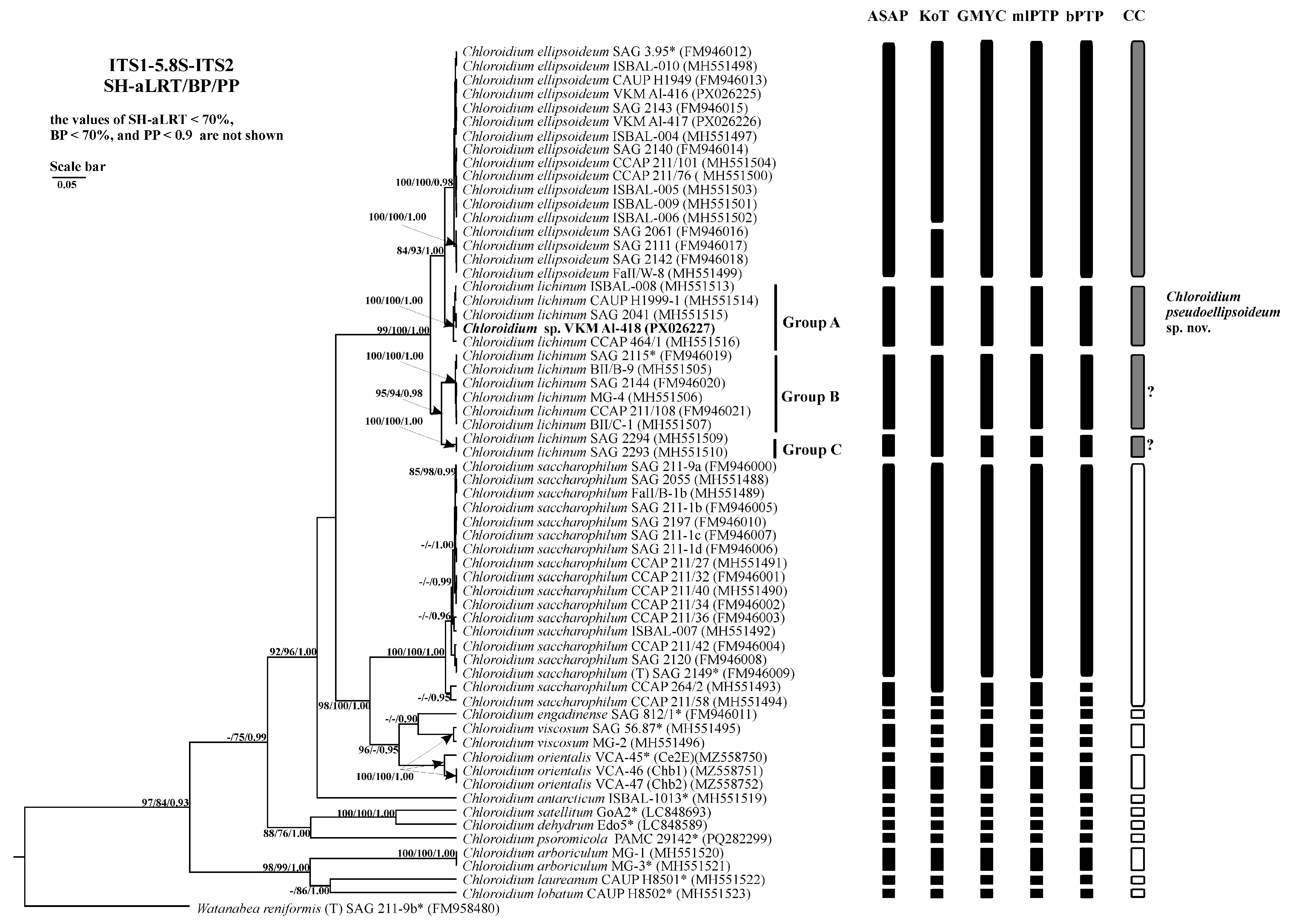
2.3. Phylogenetic Analysis
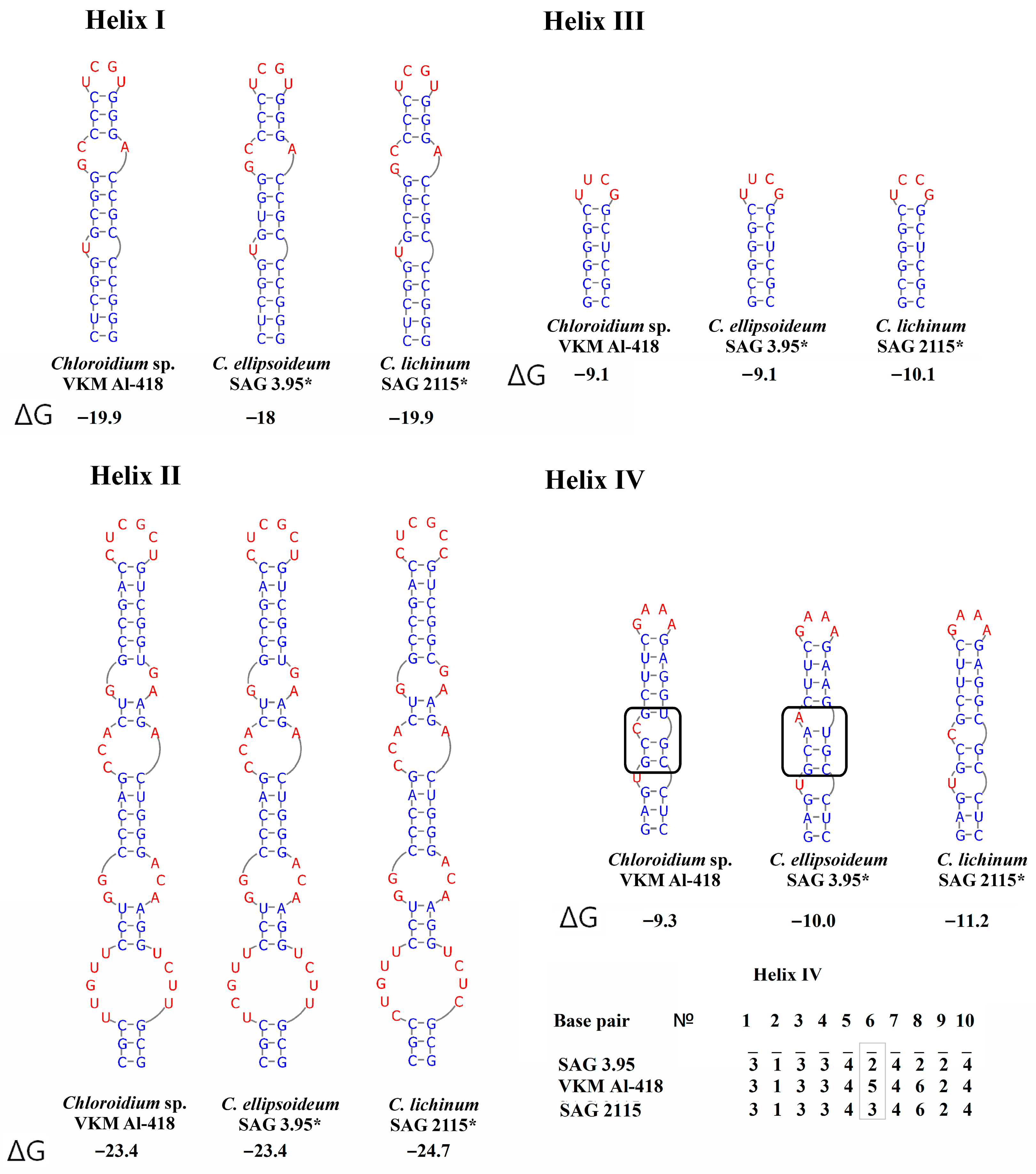
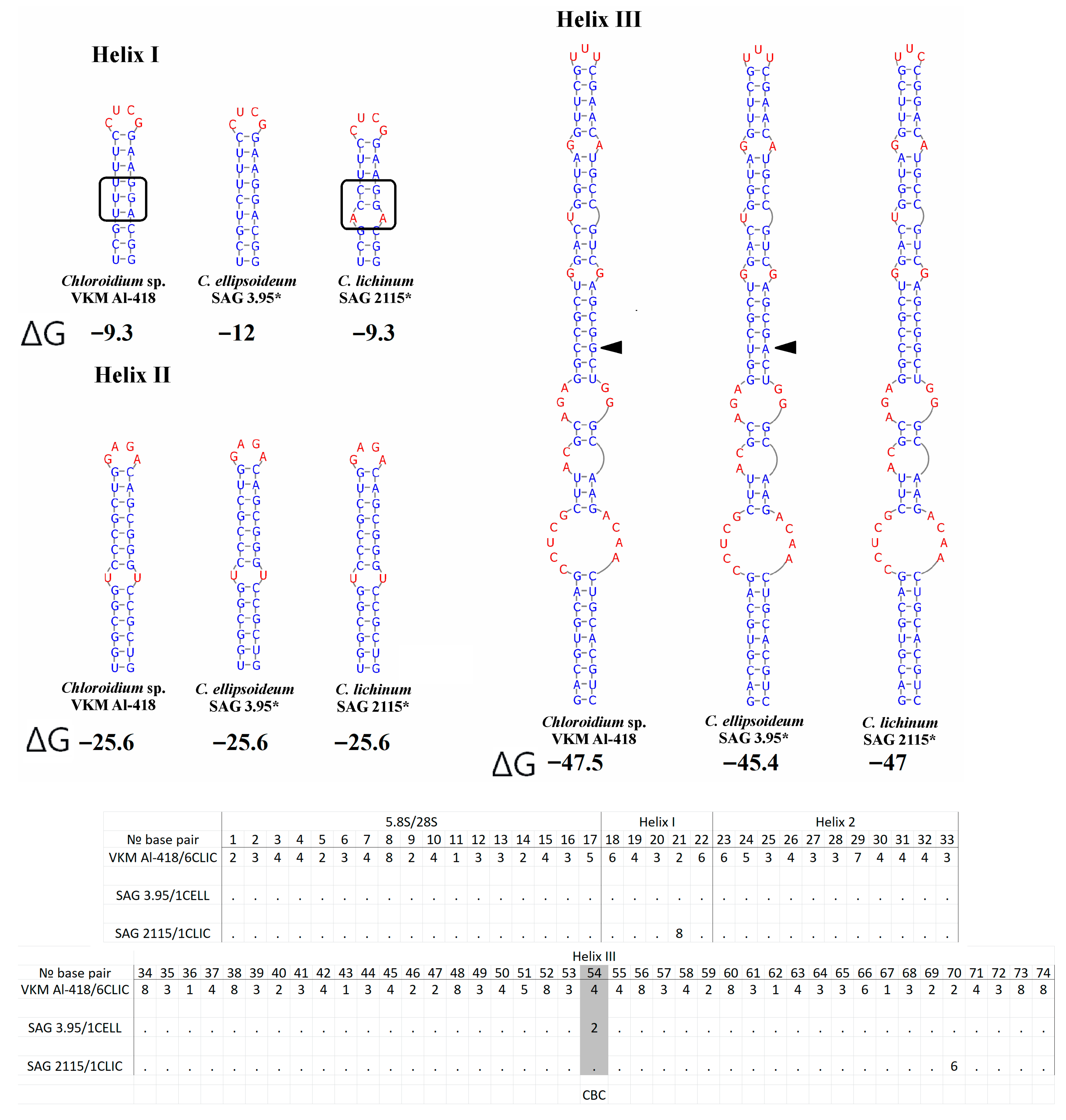
2.4. ITS1 Secondary Structure
2.5. ITS2 Secondary Structure
2.6. Analysis of Various DNA Barcodes to Distinguish Species
2.7. Species Delimitation
2.8. Habitat
2.9. Fatty Acid Composition
3. Discussion
4. Materials and Methods
4.1. Isolation and Cultivation of Algal Strains
4.2. Light Microscopy (LM)
4.3. Statistical Analysis of Morphological Traits and Their Congruence with Phylogeny
4.4. DNA Isolation, Amplification, Purification, and Sequencing
4.5. Phylogenetic Analysis
4.6. Folding of ITS1 and ITS2 Secondary Structures
4.7. Species Delimitation
4.8. Determination of the Total Lipid (TL) Content and the Fatty Acid (FA) Composition of TL
5. Conclusions
6. Taxonomic Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| C. dehydrum | C. satellitum | C. lichinum Group B | C. lichinum Group C | C. pseudoellipsoideum sp. nov. Group A | C. ellipsoideum | C. engadinense | C. viscosum | C. psoromicola | C. orientale | C. saccharophilum | C. antarcticum | C. lobatum | C. laureanum | C. arboricola | |
| C. dehydrum | 0 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| C. satellitum | 2.8 | 0 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| C. lichinum Group B | 2.9–3 | 4 | 0 | - | - | - | - | - | - | - | - | - | - | - | - |
| C. lichinum Group C | 3 | 3.8 | 0.5–0.6 | 0 | - | - | - | - | - | - | - | - | - | - | - |
| C. pseudoellipsoideum sp. nov. Group A | 3–3.1 | 4.2–4.3 | 0.8–0.9 | 0.7–0.8 | 0.1 | - | - | - | - | - | - | - | - | - | - |
| C. ellipsoideum | 3.4–3.5 | 4.4–4.5 | 1.1–1.2 | 0.9 | 0.7–0.8 | 0–0.1 | - | - | - | - | - | - | - | - | - |
| C. engadinense | 4.7 | 5.7 | 3.1–3.2 | 3 | 3–3.1 | 3.2 | 0 | - | - | - | - | - | - | - | - |
| C. viscosum | 4.7–4.8 | 5.8 | 3.1–3.2 | 3.3 | 3–3.2 | 3.1–3.2 | 1.8–1.9 | 0.1 | - | - | - | - | - | - | - |
| C. psoromicola | 4 | 5.5 | 4.3–4.4 | 4.4 | 4.1–4.2 | 4.3–4.4 | 3.1 | 3.2 | 0 | - | - | - | - | - | - |
| C. orientale | 4.4–4.5 | 5.3 | 3 | 2.9 | 3.1–3.2 | 3.4–3.5 | 2.1 | 2.6 | 4–4.1 | 0–0.4 | - | - | - | - | - |
| C. saccharophilum | 4.4–4.6 | 5.8–6 | 3.5–3.8 | 3.6–3.8 | 3.5–3.8 | 4.1–4.3 | 2.7–2.9 | 2.4–2.7 | 3.5–3.9 | 3.2–3.6 | 0–0.6 | - | - | - | - |
| C. antarcticum | 4.6 | 5.6 | 3.4 | 3.4 | 3.1–3.2 | 3.4–3.5 | 3.2 | 3.3 | 3.7 | 3.2–3.5 | 3.6–4 | 0 | - | - | - |
| C. lobatum | 7.4 | 8.1 | 6.8 | 6.9 | 6.9 | 6.9–7 | 7 | 6.7 | 5.9 | 7 | 6.5–6.9 | 6.5 | 0 | - | - |
| C. laureanum | 6.8 | 7.1 | 6.1–6.2 | 6 | 6.1–6.2 | 6.4–6.5 | 6.4 | 6.4 | 6 | 6.6–6.8 | 6–6.2 | 6.1 | 4.1 | 0 | - |
| C. arboricola | 7.1 | 7.3 | 6.3 | 6.1 | 6.3 | 6.6–6.7 | 6.1 | 6–6.1 | 6.1 | 6.1–6.3 | 6–6.2 | 6.3 | 4.4 | 4 | 0 |
Appendix B
| Species | Number of Introns | |||||||
|---|---|---|---|---|---|---|---|---|
| No | 1 | Length, n. | 2 | Length, n. | 3 | Length, n. | ||
| C. ellipsoideum | - | SAG 3.95 | 387 | SAG 2061 SAG 2142 SAG 2111 Fall/W-8 | 387; 379 | - | - | |
| CAUP H1949 | ||||||||
| SAG 2143 | ||||||||
| ISBAL-004 | ||||||||
| VKM Al-416 | ||||||||
| ISBAL-010 | ||||||||
| VKM Al-417 | ||||||||
| SAG 2140 | ||||||||
| ISBAL-005 | ||||||||
| CCAP 211/76 | ||||||||
| ISBAL-006 | ||||||||
| ISBAL-009 | ||||||||
| CCAP 211/101 | ||||||||
| C. pseudoellipsoideum | ISBAL-008 | CCAP 464/1 | 381 | CAUP H1999-1 | 339; 381 | - | - | |
| VKM Al-418 | ||||||||
| SAG 2041 | ||||||||
| C. lichinum Group B | SAG 2115 | - | - | - | - | - | - | |
| CCAP 211/108 | ||||||||
| BII/B-9 | ||||||||
| MG-4 | ||||||||
| BII/C-1 | ||||||||
| SAG 2144 | ||||||||
| C. lichinum Group C | - | - | - | - | - | SAG 2294 | 374; 335; 378 | |
| SAG 2293 | ||||||||
| C. dehydrum | - | - | - | Edo5 | 342; 399 | - | - | |
| C. satellitum | - | - | - | GoA2 | 374; 416 | - | - | |
| C. saccharophilum | SAG 211-9a SAG 2055 Fall/B-1b CCAP211/42 CCAP 264/2 CCAP 211/58 | SAG 211-1c | 400 | ISBAL-007 | 377; 400 | - | - | |
| CCAP 211/32 | ||||||||
| CCAP 211/40 | ||||||||
| CCAP 211/31 | ||||||||
| SAG 211-1b | ||||||||
| SAG 211-1d | ||||||||
| CCAP 211/27 | ||||||||
| SAG 2120 | ||||||||
| SAG 2140 | ||||||||
| CCAP 211/36 | 377 | |||||||
| C. engadinense | - | - | - | - | - | SAG 812/1 | 394; 368; 404 | |
| C. orientale | VCA-45 | - | - | - | - | - | - | |
| VCA-46 | ||||||||
| VCA-47 | ||||||||
| C. viscosum | - | - | - | MG-2 SAG 56.87 | 363; 401 | - | - | |
| C. antarcticum | - | ISBAL-1013 | 422 | - | - | - | - | |
| C. psoromicola | PAMC 29142 | - | - | - | - | - | - | |
| C. arboriculum | - | - | - | MG-1 | 484; 463 | - | - | |
| MG-3 | ||||||||
| C. laureanum | - | - | - | CAUP H8501 | 1280; 434 | - | - | |
| C. lobatum | - | - | - | CAUP H8502 | 412; 466 | - | - | |
Appendix C
| C. dehydrum | C. satellitum | C. lichinum Group B | C. lichinum Group C | C. pseudoellipsoideum sp. nov. Group A | C. ellipsoideum | C. engadinense | C. viscosum | C. psoromicola | C. orientale | C. saccharophilum | C. antarcticum | C. lobatum | C. laureanum | C. arboricola | |
| C. dehydrum | 0 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| C. satellitum | 13.4 | 0 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| C. lichinum Group B | 23.7 | 24.4 | 0 | - | - | - | - | - | - | - | - | - | - | - | - |
| C. lichinum Group C | 27.1 | 26.5 | 5.2 | 0 | - | - | - | - | - | - | - | - | - | - | - |
| C. pseudoellipsoideum sp. nov. Group A | 26.2–27.6 | 27.1–28.6 | 7.4–8.4 | 8.4–9.8 | 0–2.5 | - | - | - | - | - | - | - | - | - | - |
| C. ellipsoideum | 29.1 | 27.9 | 8.8–9.8 | 9.8–10.7 | 4.2–6.9 | 0–0.8 | - | - | - | - | - | - | - | - | - |
| C. engadinense | 35.4 | 36.4 | 25 | 23.9 | 21.8–23 | 23.5–23.6 | 0 | - | - | - | - | - | - | - | - |
| C. viscosum | 33.2–34 | 33.9–34.7 | 22–23.2 | 23.7–25 | 18.3–20.6 | 20–21.2 | 9.1–9.9 | 0.7 | - | - | - | - | - | - | - |
| C. psoromicola | 34.5 | 37.5 | 31.2 | 29.9 | 25.7–27.6 | 28.8 | 23.7 | 21.9 | 0 | - | - | - | - | - | - |
| C. orientale | 30.8–31.1 | 29.2–29.5 | 20.7–22.2 | 19.6–21.2 | 21.1–23.9 | 23.5–26.3 | 20.8–23.5 | 20.4–23.2 | 33.4–34.3 | 0–3.9 | - | - | - | - | - |
| C. saccharophilum | 27.3–30.8 | 30.7–33 | 18.9–21.8 | 21.9–24.9 | 17.6–21.1 | 21.8–23.6 | 12.7–14.9 | 13–15.7 | 25.6–27.8 | 24.4–27 | 0–5.1 | - | - | - | - |
| C. antarcticum | 33.3 | 38.1 | 18.9 | 19.5 | 17.6–18.8 | 20.4–21.5 | 17.9 | 17.8–18.9 | 21.7–23.2 | 20.5–21.7 | 22.1–24.5 | 0 | - | - | - |
| C. lobatum | 59.5 | 58.9 | 54.3 | 53 | 51.3 | 49.5 | 53.2 | 49.9 | 45 | 51.9–52.4 | 48–51 | 53.6 | 0 | - | - |
| C. laureanum | 46.7 | 46.7 | 39.1 | 40 | 40.4–42.7 | 42–43.5 | 47.8 | 45.4 | 46.9 | 44.5–45 | 42.6–44.8 | 47 | 43.6 | 0 | - |
| C. arboricola | 55 | 54.6 | 42.6 | 40 | 42–43.5 | 46–46.8 | 49.6 | 46.6–48.3 | 49.9 | 40.1–43.7 | 38.4–42 | 44.7 | 44.7 | 41.8 | 0 |
Appendix D
| C. dehydrum | C. satellitum | C. lichinum Group B | C. lichinum Group C | C. pseudoellipsoideum sp. nov. Group A | C. ellipsoideum | C. engadinense | C. viscosum | C. psoromicola | C. orientale | C. saccharophilum | C. antarcticum | C. lobatum | C. laureanum | C. arboricola | |
| C. dehydrum | 0 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| C. satellitum | 27.3 | 0 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| C. lichinum Group B | 28.5 | 48 | 0 | - | - | - | - | - | - | - | - | - | - | - | - |
| C. lichinum Group C | 31 | 47.7 | 4.4 | 0 | - | - | - | - | - | - | - | - | - | - | - |
| C. pseudoellipsoideum sp. nov. Group A | 28.1 | 44 | 3.4 | 5.1 | 0–0.2 | - | - | - | - | - | - | - | - | - | - |
| C. ellipsoideum | 30.9–31.8 | 45.2–46.5 | 4.4–4.9 | 6.2–6.7 | 1.9–2.4 | 0–0.5 | - | - | - | - | - | - | - | - | - |
| C. engadinense | 53 | 62.5 | 33.3 | 32.1 | 31.8 | 33.5–34.4 | 0 | - | - | - | - | - | - | - | - |
| C. viscosum | 48.4–49.6 | 59.7–61.2 | 28.9–29 | 30–30.1 | 26.8 | 28.3–29.1 | 15.8 | 1.7 | - | - | - | - | - | - | - |
| C. psoromicola | 32.3 | 42.6 | 44.8 | 47.1 | 46.5 | 47.7–49 | 36.5 | 39.3–40.4 | 0 | - | - | - | - | - | - |
| C. orientale | 48.6–49.8 | 56.9–63.2 | 24.5–26 | 23.7–25.1 | 23.5–24.7 | 24.6–26.9 | 15.1–16.1 | 18.9–19.5 | 41.1–43.3 | 0–5.9 | - | - | - | - | - |
| C. saccharophilum | 42.8–45 | 61.6–64.7 | 33.3–36 | 32.2–35 | 30.8–33.5 | 32.6–38.1 | 37.7–41.9 | 32.7–37.9 | 42.6–44.9 | 38.3–41.3 | 0–4 | - | - | - | - |
| C. antarcticum | 47.1 | 60.2 | 35.6 | 33.8 | 38.6 | 36.7–37.6 | 49.5 | 47.5–47.6 | 55.3 | 36.9–43.4 | 40.3–42.2 | 0 | - | - | - |
| C. lobatum | 57.3 | 67.8 | 52.2 | 52.7 | 54 | 54–55.2 | 83.7 | 72.7 | 59.8 | 70.7–71 | 64.1–68 | 57.9 | 0 | - | - |
| C. laureanum | 41.5 | 55.9 | 39.9 | 39.7 | 38.3 | 42.1 | 61.1 | 59.5–60.9 | 52.2 | 53.1–56.4 | 45.1–48.1 | 46 | 30.4 | 0 | - |
| C. arboricola | 64.1 | 68.1 | 54.2 | 51 | 56.5 | 59.1 | 58.5 | 57.4–60.8 | 60.6 | 52.8–57.7 | 55.2–56.4 | 57 | 43 | 28.3 | 0 |
Appendix E
| C. dehydrum | C. satellitum | C. lichinum Group B | C. lichinum Group C | C. pseudoellipsoideum sp. nov. Group A | C. ellipsoideum | C. engadinense | C. viscosum | C. psoromicola | C. orientale | C. saccharophilum | C. antarcticum | C. lobatum | C. laureanum | C. arboricola | |
| C. dehydrum | 0 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| C. satellitum | 14.1 | 0 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| C. lichinum Group B | 17.8 | 22.6 | 0 | - | - | - | - | - | - | - | - | - | - | - | - |
| C. lichinum Group C | 20 | 23.1 | 3.7 | 0 | - | - | - | - | - | - | - | - | - | - | - |
| C. pseudoellipsoideum sp. nov. Group A | 18.6–19.1 | 23.1–23.7 | 4.4–4.8 | 5.6–6.1 | 0–1 | – | – | – | – | – | – | – | – | – | – |
| C. ellipsoideum | 20.4–20.6 | 23.7–24 | 5.3–5.7 | 6.5–6.8 | 2.3–3.4 | 0–0.5 | – | – | – | – | – | – | – | – | – |
| C. engadinense | 29 | 31 | 20.8 | 20.2 | 19.1–19.6 | 20.3–20.5 | 0 | – | – | – | – | – | – | – | – |
| C. viscosum | 27.2–27.8 | 30 | 18.4–18.8 | 19.6–20 | 16.4–17.2 | 17.5–18.1 | 9.4–9.7 | 0.9 | – | – | – | – | – | – | – |
| C. psoromicola | 24.1 | 27.8 | 25.4 | 25.8 | 23.6–24.3 | 25.1–25.4 | 20 | 20.4–20.6 | 0 | – | – | – | – | – | – |
| C. orientale | 26–26.3 | 27–28.1 | 16.4–17.4 | 16.3–16.5 | 16.6–18.1 | 17.9–19.4 | 14.3–14.9 | 14.8–15.7 | 25.9–26 | 0–3.6 | – | – | – | – | – |
| C. saccharophilum | 23.4–25.2 | 27.9–29.6 | 18.1–19.3 | 19.2–20.4 | 17.6–18.8 | 20–21 | 17.6–18.8 | 16.1–17.8 | 23–24.2 | 21.9–22.8 | 0–3.2 | – | – | – | – |
| C. antarcticum | 26.3 | 31.3 | 18.9 | 18.9 | 18.5–19 | 19.2–19.9 | 21.9 | 21.5–21.9 | 25.4 | 20.7–21.9 | 21.8–22.7 | 0 | – | – | – |
| C. lobatum | 38.6 | 41.3 | 36.4 | 36.5 | 35.6–36.1 | 35.2–35.5 | 44.5 | 41.1 | 36.8 | 41–41.2 | 37.2–38.9 | 39.1 | 0 | – | – |
| C. laureanum | 30.4 | 33.4 | 27.5 | 27.5 | 27.4–28.1 | 29–29.5 | 36.5 | 35.3–35.6 | 34.9 | 33.8–34.4 | 30.6–31.9 | 33.3 | 29 | 0 | – |
| C. arboricola | 38.2 | 37.9 | 31.9 | 30.1 | 32.8–33.3 | 34.7–35 | 36.7 | 35.4–36.8 | 37.7 | 32.7–32.9 | 31.3–32.6 | 35 | 32.5 | 26.4 | 0 |
Appendix F
| Parameter | 18S-ITS1-5.8S-ITS2 | ITS1 | ITS2 | ITS1-5.8S-ITS2 |
|---|---|---|---|---|
| Maximum intraspecific distance, % | 0.6 | 5.1 | 5.9 | 3.6 |
| Minimum interspecific distance, % | 0.70 | 4.2 | 1.9 | 2.3 |
| Overlap minimizing threshold, % | 0.65 | 4.65 | 3.9 | 2.95 ≈ 3.0 |
| Area of distribution overlap, % | ≈2–3 | ≈10 | ≈40–45 | ≈15–20 |
| Optimal ROC Threshold, % | 0.65 | 4.6 | 3.8 | 3.0 |
| Area Under Curve (AUC) | 0.982 | 0.992 | 0.973 | 0.987 |
| 95% CI (AUC) | 0.94–1.00 | 0.96–1.00 | 0.93–1.00 | 0.95–1.00 |
| Sensitivity | 1.00 | 0.99 | 0.96 | 0.98 |
| Specificity | 0.98 | 0.99 | 0.95 | 0.97 |
| Accuracy | 0.99 | 0.99 | 0.96 | 0.98 |
References
- Darienko, T.; Lukešová, A.; Pröschold, T. The polyphasic approach revealed new species of Chloroidium (Trebouxiophyceae, Chlorophyta). Phytotaxa 2018, 372, 51–66. [Google Scholar] [CrossRef]
- Nadson, G.A. K morfologii nizshykh vodorosley (Predvaritelnoe soobshchenie) [To the morphology of inferior algae (Preliminary communication)]. Izv. Imp. St. Peterbg. Bot. Sada 1906, 6, 184–194. [Google Scholar]
- Guiry, M.D.; Guiry, G.M. AlgaeBase, World-Wide Electronic Publication. University of Galway. Available online: https://www.AlgaeBase.org/ (accessed on 8 October 2025).
- Darienko, T.; Gustavs, L.; Mudimu, O.; Menendez, C.R.; Schumann, R.; Karsten, U.; Friedl, T.; Pröschold, T. Chloroidium, a common terrestrial coccoid green alga previously assigned to Chlorella (Trebouxiophyceae, Chlorophyta). Eur. J. Phycol. 2010, 45, 79–95. [Google Scholar] [CrossRef]
- Neustupa, J.; Nĕmcová, Y.; Veselá, J.; Steinová, J.; Škaloud, P. Parachloroidium gen. nov. (Trebouxiophyceae, Chlorophyta), a novel genus of coccoid green algae from subaerial corticolous biofilms. Phycologia 2013, 52, 411–421. [Google Scholar] [CrossRef]
- Chodat, R. Monographies d‘Algues en Culture Pure. In Matériaux Pour la Flore Cryptogamique Suisse; K.J. Wyss: Berne, Switzerland, 1906; Volume 4, pp. Fasc. 2–Fasc. 266. [Google Scholar]
- Gontcharov, A.A.; Nikulin, A.Y.; Nikulin, V.Y.; Bagmet, V.B.; Allaguvatova, R.Z.; Abdullin, S.R. New species of Chloroidium (Trebouxiophyceae, Chlorophyta) from East Asia. Plants 2021, 10, 2560. [Google Scholar] [CrossRef] [PubMed]
- Chae, H.; Lee, Y.M.; Lee, H.; So, J.E.; So, J.; Choi, H.-G.; Kim, S.; Kim, J.H. Morphology and molecular characterization of a new Chloroidium (Trebouxiophyceae, Chlorophyta) species isolated from lichen in Antarctica. Phycol. Res. 2025, 73, 125–133. [Google Scholar] [CrossRef]
- Motomatsu, H.; Kawaguchi, R.; Toyoshima, H.; Kawasaki, S. Two microalgal strains, Chloroidium dehydrum sp. nov. and Chloroidium satellitum sp. nov (Trebouxiophyceae, Chlorophyta), isolated under high light exposure conditions and exhibiting stress-induced morphological changes. Phycol. Res. 2025, 73, 280–293. [Google Scholar] [CrossRef]
- Sanders, W.B.; de los Rios, A.; Pérez-Ortega, S. Chloroidium phycobionts (Watanabeales, Trebouxiophyceae) partner with lecanoralean mycobionts in foliicolous lichen communities of Tenerife (Canary Islands) and Navarra (Iberian Peninsula), Spain. Lichenologist 2024, 56, 107–119. [Google Scholar] [CrossRef]
- Wheeler, Q.D. Taxonomic triage and the poverty of phylogeny. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2004, 359, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Malavasi, V.; Škaloud, P.; Rindi, F.; Tempesta, S.; Paoletti, M.; Pasqualetti, M. DNA-based taxonomy in ecologically versatile microalgae: A re-evaluation of the species concept within the coccoid green algal genus Coccomyxa (Trebouxiophyceae, Chlorophyta). PLoS ONE 2016, 11, e0151137. [Google Scholar] [CrossRef]
- Zou, S.; Fei, C.; Wang, C.; Gao, Z.; Bao, Y.; He, M.; Wang, C. How DNA barcoding can be more effective in microalgae identification: A case of cryptic diversity revelation in Scenedesmus (Chlorophyceae). Sci. Rep. 2016, 9, 36822. [Google Scholar] [CrossRef] [PubMed]
- Luo, A.; Ho, S.Y.W. The molecular clock and evolutionary timescales. Biochem. Soc. Trans. 2018, 46, 1183–1190. [Google Scholar] [CrossRef] [PubMed]
- Karabanov, D.P.; Kotov, A.A.; Borovikova, E.A.; Kodukhova, Y.V.; Zhang, X. Comparison of the efficiency of single-locus species delimitation methods: A case study of a single lake fish population in comparison against the barcodes from international databases. Water 2023, 15, 1851. [Google Scholar] [CrossRef]
- Krivina, E.; Sinetova, M.; Zadneprovskaya, E.; Ivanova, M.; Starikov, A.; Shibzukhova, K.; Lobakova, E.; Budkin, Y.; Portnov, A.; Temraleeva, A. The genus Coelastrella (Chlorophyceae, Chlorophyta): Molecular species delimitation, biotechnological potential, and description of a new species Coelastrella affinis sp. nov., based on an integrative taxonomic approach. Antonie Van. Leeuwenhoek 2024, 117, 113. [Google Scholar] [CrossRef]
- Leliaert, F.; Smith, D.R.; Moreau, H.; Herron, M.D.; Verbruggen, H.; Delwiche, C.F.; De Clerck, O. Phylogeny and molecular evolution of the green algae. Crit. Rev. Plant Sci. 2012, 31, 1–46. [Google Scholar] [CrossRef]
- Vorobyev, K.; Andronov, E.; Rautian, M.; Skoblo, I.; Migunova, A.; Kvitko, K. An atypical Chlorella symbiont from Paramecium bursaria. Protistology 2009, 6, 39–44. [Google Scholar]
- Gaonkar, C.C.; Piredda, R.; Minucci, C.; Mann, D.G.; Montresor, M.; Sarno, D.; Kooistra, W.H. Annotated 18S and 28S rDNA reference sequences of taxa in the planktonic diatom family Chaetocerotaceae. PLoS ONE 2018, 13, e0208929. [Google Scholar] [CrossRef] [PubMed]
- Hoshina, R.; Iwataki, M.; Imamura, N. Chlorella variabilis and Micractinium reisseri sp. nov. (Chlorellaceae, Trebouxiophyceae): Redescription of the endosymbiotic green algae of Paramecium bursaria (Peniculia, Oligohymenophorea) in the 120th year. Phycol. Res. 2010, 58, 188–201. [Google Scholar] [CrossRef]
- Coleman, A.W. Nuclear rRNA transcript processing versus internal transcribed spacer secondary structure. Trends Genet. 2015, 31, 157–163. [Google Scholar] [CrossRef]
- Chae, H.; Lim, S.; Kim, H.; Choi, H.-G.; Kim, J.H. Morphology and phylogenetic relationships of Micractinium (Chlorellaceae, Trebouxiophyceae) taxa, including three new species from Antarctica. Algae 2019, 34, 267–275. [Google Scholar] [CrossRef]
- Krivina, E.S.; Boldina, O.N.; Bukin, Y.S.; Bykova, S.V.; Temraleeva, A.D. Species delimitation polyphasic approach reveals Meyerella similis sp. nov.: A new species of “small green balls” within the Chlorella-clade (Trebouxiophyceae, Chlorophyta). Org. Divers. Evol. 2023, 23, 25–40. [Google Scholar] [CrossRef]
- Nishida, I.; Murata, N. Chilling sensitivity in plants and cyanobacteria: The crucial contribution of membrane lipids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 541–568. [Google Scholar] [CrossRef]
- Morgan-Kiss, R.M.; Priscu, J.C.; Pocock, T.; Gudynaite-Savitch, L.; Huner, N.P. Adaptation and acclimation of photosynthetic microorganisms to permanently cold environments. Microbiol. Mol. Biol. Rev. 2006, 70, 222–252. [Google Scholar] [CrossRef]
- Hempel, N.; Petrick, I.; Behrendt, F. Biomass productivity and productivity of fatty acids and amino acids of microalgae strains as key characteristics of suitability for biodiesel production. J. Appl. Phycol. 2012, 24, 1407–1418. [Google Scholar] [CrossRef] [PubMed]
- Lang, I.; Hodac, L.; Friedl, T.; Feussner, I. Fatty acid profiles and their distribution patterns in microalgae: A comprehensive analysis of more than 2000 strains from the SAG culture collection. BMC Plant. Biol. 2011, 11, 124. [Google Scholar] [CrossRef] [PubMed]
- González, M.A.; Pröschold, T.; Palacios, Y.; Aguayo, P.; Inostroza, I.; Gómez, P.I. Taxonomic identification and lipid production of two Chilean Chlorella-like strains isolated from a marine and an estuarine coastal environment. AoB Plants 2013, 5, plt020. [Google Scholar] [CrossRef]
- Takeshita, T.; Ota, S.; Yamazaki, T.; Hirata, A.; Zachleder, V.; Kawano, S. Starch and lipid accumulation in eight strains of six Chlorella species under comparatively high light intensity and aeration culture conditions. Bioresour. Technol. 2014, 158, 127–134. [Google Scholar] [CrossRef]
- Nelson, D.R.; Khraiwesh, B.; Fu, W.; Alseekh, S.; Jaiswal, A.; Chaiboonchoe, A.; Hazzouri, K.M.; O’Connor, M.J.; Butterfoss, G.L.; Drou, N.; et al. The genome and phenome of the green alga Chloroidium sp. UTEX 3007 reveal adaptive traits for desert acclimatization. Elife 2017, 6, e25783. [Google Scholar] [CrossRef]
- Khozin-Goldberg, I. Lipid metabolism in microalgae. In The Physiology of Microalgae Developments in Applied Phycology; Borowitzka, M., Beardall, J., Raven, J., Eds.; Springer: Cham, Switzerlands, 2016; Volume 6, pp. 414–484. [Google Scholar] [CrossRef]
- Guschina, I.A.; Harwood, J.L. Lipids and lipid metabolism in eukaryotic algae. Prog. Lipid. Res. 2006, 45, 160–186. [Google Scholar] [CrossRef]
- Stanier, R.Y.; Kunisawa, R.; Mandel, M.; Cohen-Bazire, G. Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev. 1971, 35, 171–205. [Google Scholar] [CrossRef]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar]
- Legendre, P.; Legendre, L. Numerical Ecology, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2012; pp. 1–969. [Google Scholar]
- Oksanen, J.; Simpson, G.; Blanchet, F.; Kindt, R.; Legendre, P.; Minchin, P.; O’Hara, R.; Solymos, P.; Stevens, M.; Szoecs, E.; et al. Vegan: Community Ecology Package. R Package Version 2.6-4. Available online: https://CRAN.R-project.org/package=vegan (accessed on 4 October 2025).
- Sokal, R.R.; Michener, C.D. A statistical method for evaluating systematic relationships. Univ. Kans. Sci. Bull. 1958, 38, 1409–1438. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023; Available online: https://www.R-project.org (accessed on 4 October 2025).
- Paradis, E.; Schliep, K. Ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef] [PubMed]
- Katana, A.; Kwiatowski, J.; Spalik, K.; Zakryś, B.; Szalacha, E.; Szymańska, H. Phylogenetic position of Koliella (Chlorophyta) as inferred from nuclear and chloroplast small subunit rDNA. J. Phycol. 2001, 37, 443–451. [Google Scholar] [CrossRef]
- Johnson, J.L.; Fawley, M.W.; Fawley, K.P. The diversity of Scenedesmus and Desmodesmus (Chlorophyceae) in Itasca State Park, Minnesota, USA. Phycologia 2007, 46(2), 214–229. [Google Scholar] [CrossRef]
- Kuraku, S.; Zmasek, C.M.; Nishimura, O.; Katoh, K. Leaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 2013, 41, W22–W28. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Le Vinh, S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Shimodaira, H. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 2002, 51, 492–508. [Google Scholar] [CrossRef]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchene, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kuhnert, D.; de Maio, N.; et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Drummond, A.J.; Bouckaert, R.R. Bayesian Evolutionary Analysis with BEAST2; Cambridge University Press: Cambridge, UK, 2015; ISBN 978-1-107-01965-2. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Pastore, M. Overlapping: A R package for Estimating Overlapping in Empirical Distributions. J. Open Source Softw. 2018, 3, 1023. [Google Scholar] [CrossRef]
- Hosmer, D.W.; Lemeshow, S. Applied Logistic Regression, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2000. [Google Scholar]
- Caisová, L.; Marin, B.; Melkonian, M. A consensus secondary structure of ITS2 in the Chlorophyta identified by phylogenetic reconstruction. Protist 2013, 164, 482–496. [Google Scholar] [CrossRef] [PubMed]
- Seibel, P.N.; Müller, T.; Dandekar, T.; Schultz, J.; Wolf, M. 4SALE: A tool for synchronous RNA sequence and secondary structure alignment and editing. BMC Bioinform. 2006, 7, 498. [Google Scholar] [CrossRef] [PubMed]
- Seibel, P.N.; Müller, T.; Dandekar, T.; Wolf, M. Synchronous visual analysis and editing of RNA sequence and secondary structure alignments using 4SALE. BMC Res. Notes 2008, 1, 91. [Google Scholar] [CrossRef] [PubMed]
- Coleman, A.W. The significance of a coincidence between evolutionary landmarks found in mating affinity and a DNA sequence. Protist 2000, 151, 1–9. [Google Scholar] [CrossRef]
- Demchenko, E.; Mikhailyuk, T.; Coleman, A.; Pröschold, T. Generic and species concepts in Microglena (previously the Chlamydomonas monadina group) revised using an integrative approach. Eur. J. Phycol. 2012, 47, 264–290. [Google Scholar] [CrossRef]
- Spöri, Y.; Stoch, F.; Dellicour, S.; Birky, C.W.; Flot, J.-F. KoT: An automatic implementation of the K/θ method for species delimitation. bioRxiv 2021. [Google Scholar] [CrossRef]
- Puillandre, N.; Brouillet, S.; Achaz, G. ASAP: Assemble species by automatic partitioning. Mol. Ecol. Resour. 2021, 21, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Vences, M.; Miralles, A.; Brouillet, S.; Ducasse, J.; Fedosov, A.; Kharchev, V.; Kostdinov, I.; Kumari, S.; Patmanidis, S.; Scherz, M.D.; et al. iTaxoTools 0.1: Kickstarting a specimen-based software toolkit for taxonomists. Megataxa 2021, 6, 77–92. [Google Scholar] [CrossRef]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef]
- Magoga, G.; Fontaneto, D.; Montagna, M. Factors affecting the efficiency of molecular species delimitation in a species-rich insect family. Mol. Ecol. Resour. 2021, 21, 1475–1489. [Google Scholar] [CrossRef]
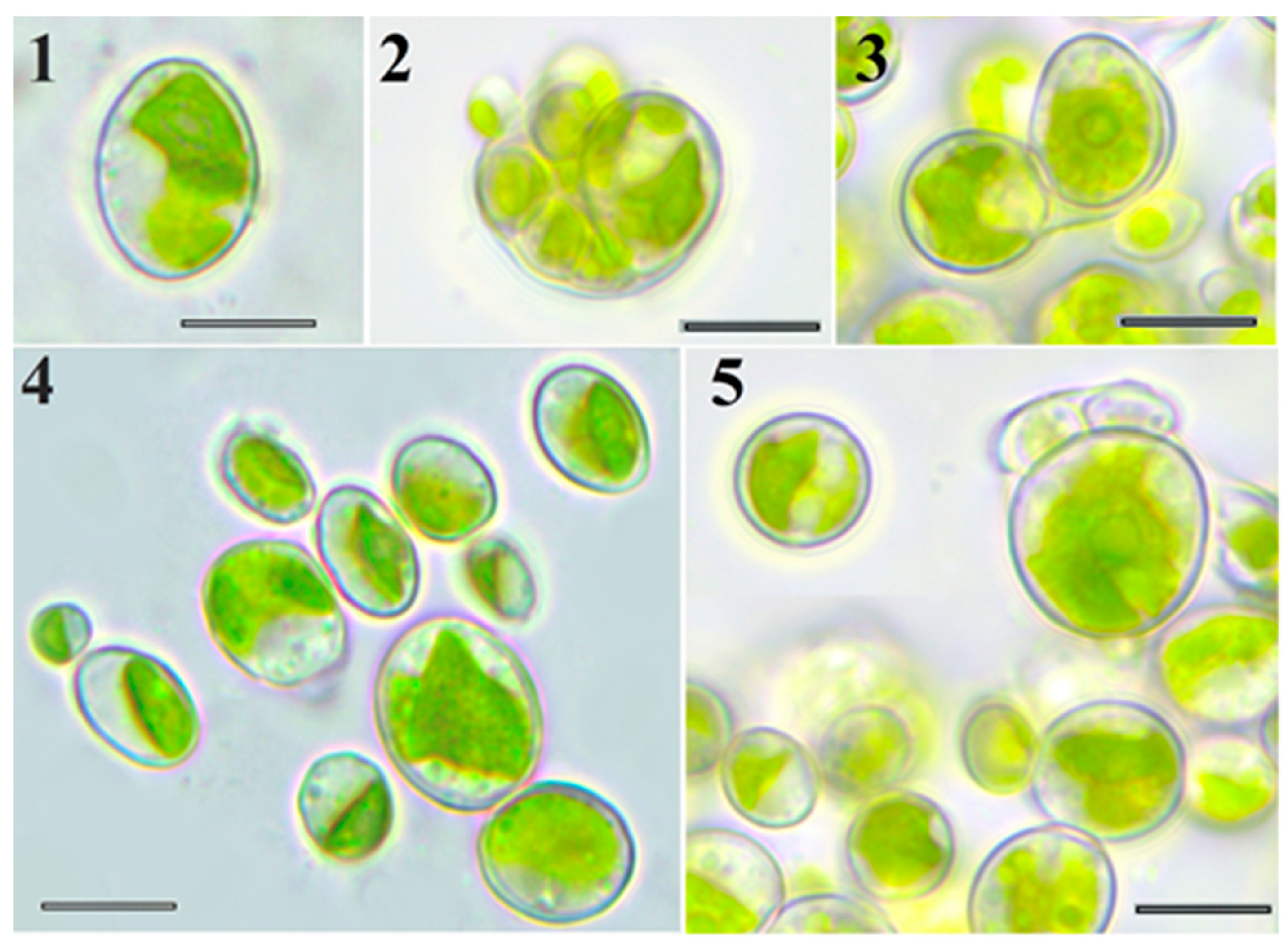
| Species | Young Cell | Adult Cell | Pyrenoid | Reproduction by Autospores | ||||
|---|---|---|---|---|---|---|---|---|
| Cell Shape | Cell Size, μm | Chloroplast Shape | Cell Shape | Cell Size, μm | Chloroplast Shape | |||
| Chloroidium psoromicola | Ellipsoidal, broadly ellipsoidal, almost spherical | 4.6 × 2.0– 10.5 × 6.4 (5.8 × 5.2– 9.2 × 8.5) | Parietal, band- to cup-shaped | Broadly ellipsoidal, almost spherical | 6.8 × 6.7– 18.2 × 18.1 | Parietal, band- to cup-shaped, deeply lobed | Naked | 2–16 |
| Chloroidium saccharophilum | Narrowly ellipsoidal, ovoid, almost cylindrical | - | Parietal, band-shaped | Ellipsoidal, almost spherical, slightly pyriform | 6.9 × 5.3– 13.6 × 9.4 | Parietal, band-shaped, slightly lobed | Naked | 2–16 |
| Chloroidium antarcticum | Ellipsoidal, broadly ellipsoidal, almost spherical | 13.6 × 10.9– 16.4 × 11.8 | Parietal, band-shaped, deeply lobed | Ellipsoidal, broadly ellipsoidal | 19.1 × 16.4– 24.5 × 20.0 | Parietal, deeply lobed | Naked | 4–64 |
| Chloroidium arboricola | Broadly ellipsoidal, spherical | 4.1 × 2.8– 5.6 × 5.0 | Parietal, band-shaped | Ellipsoidal, almost spherical | 5.3 × 5.0–8.2 × 8.5 (9.2–10.0) | Parietal, band-shaped | Naked | 4–8 |
| Chloroidium ellipsoideum | Ellipsoidal, ovoid, irregular | 4.4 × 2.7– 6.9 × 4.4 | Parietal, plate-like, band-shaped | Ellipsoidal, broadly ellipsoidal, almost spherical | 6.0 × 5.2– 10.4 × 9.2 (14.0 × 9.0) | Parietal, band-shaped, deep lobes | Present | 2–8 |
| Chloroidium engadinense | Narrowly ellipsoidal, almost cylindrical, pointed on one side | 4.4 × 2.7– 7.8 × 4.2 | Parietal, band-shaped | Narrowly ellipsoidal, ellipsoidal | 7.6 × 4.0– 10.6 × 7.5 | Parietal, band-shaped | Absent | 2–8 |
| Chloroidium laureanum | - | - | - | Spherical | (2.5–)3.0–7.5 (–9.8) | Parietal, cup-shaped, lobes | Absent | 2–8 |
| Chloroidium lichinum | Narrowly ellipsoidal, ovoid | 6.0 × 3.0– 7.5 × 4.0 | Ellipsoidal, ovoid, spherical | Ellipsoidal, ovoid, spherical | 10.1 × 7.1– 20.8 × 9.0 (×11.5) | Parietal, deeply lobed | Present | 2–8 (16) |
| Chloroidium lobatum | - | - | Spherical | Spherical | (3.5–)4.0–10.5 (–13.5) | Parietal, cup-shaped, often with two lobes | Absent | 2–8 |
| Chloroidium orientale | Ellipsoidal, broadly ellipsoidal | 4.1 × 1.8– 7.4 × 5.1 | Ellipsoidal, broadly ellipsoidal, almost spherical | Ellipsoidal, broadly ellipsoidal, almost spherical | 3.8 × 2.1– 16.7 × 9.0 (5.4– 10.8) | Parietal, band- to cup-shaped | Present | 2–16 |
| Chloroidium viscosum | Ellipsoidal, broadly ellipsoidal | 4.1 × 2.8– 5.6 × 5.0 | Ellipsoidal, almost spherical | Ellipsoidal, almost spherical | 6.3 × 5.0–8.8 × 6.9 (9.4–10.0) | Parietal, band-shaped | Naked | 2–32 |
| Chloroidium pseudoellipsoideum sp. nov. | Ellipsoidal or broadly ellipsoidal | 4.5–9 × 2.1–8 | Parietal, band-shaped to plate-shaped, containing a single pyrenoid | Ellipsoidal, broadly ellipsoidal or sometimes spherical | 9–16 × 5.1–16 | Parietal, band-, plate- and lobed- shaped | Present | 2–16 |
| Chloroidium dehydrum | Ellipsoidal, broadly ellipsoidal, narrowly ellipsoidal, ovoid, irregular | 4.3 × 3.0– 11.3 × 7.2 | Parietal, ellipsoidal, ovoid, spherical | Ellipsoidal, broadly ellipsoidal, spherical | 7.3 × 5.4– 18.2 × 15.3 | Parietal, deeply lobed | Present | 2–16 |
| Chloroidium satellitum | Ellipsoidal, narrowly ellipsoidal, ovoid | 4.1 × 3.3– 9.6 × 5.1 | Parietal, ellipsoidal, broadly ellipsoidal, almost spherical | Broadly ellipsoidal, almost spherical | 5.5 × 5.4– 10.7 × 10.2 | Parietal, slightly lobed | Present | 2–32 |
| Morphological Characteristics | Pearson Correlation Coefficient | p-Value |
|---|---|---|
| Shape of young cells | 0.69 | 0.0001 |
| Maximum size of young cells | 0.65 | 0.0012 |
| Shape of adult cells | 0.62 | 0.0002 |
| Pyrenoid presence | 0.52 | 0.0001 |
| Chloroplast type of young cells | 0.52 | 0.0048 |
| Number of autospores | 0.35 | 0.0038 |
| Chloroplast type of adult cells | 0.3 | 0.0092 |
| Maximum size of adult cells | 0.29 | 0.0053 |
| V4 Region | V9 Region | ITS1 | ITS2 | ITS1-5.8S-ITS2 | |
|---|---|---|---|---|---|
| Barcode length, bp | 430–432 | 175–176 | 244–360 | 211–247 | 591–617 |
| Proportion of variable sites, % | 4.3 | 15.3 | 62.3 | 59.6 | 52.7 |
| Range of intraspecific distances | 0–0.9 | 0–2.9 | 0–5.1 | 0–5.9 | 0–3.6 |
| Range of interspecific distances | 0–2.2 | 0–8.6 | 4.2–59.6 | 1.9–83.7 | 2.3–44.5 |
| Percentage of overlapping distances from the total range of genetic differences, % | 40.9 | 33.4 | 1.5 | 4.8 | 2.9 |
| Efficiency of species identification, % | 21.4 | 28.6 | 78.6 | 78.6 | 78.6 |
| 18S-ITS1-5.8S-ITS2 | ITS1 | ITS2 | ITS1-5.8S-ITS2 | ||
|---|---|---|---|---|---|
| ASAP | matches | 90.9 | 90.9 | 81.8 | 81.8 |
| splits | 9.1 | 9.1 | 18.2 | 18.2 | |
| KoT | matches | 81.8 | 81.8 | 72.7 | 72.7 |
| splits | 18.2 | 18.2 | 27.3 | 27.3 | |
| GMYC | matches | 81.8 | 90.9 | 81.8 | 81.8 |
| splits | 18.2 | 9.1 | 18.2 | 18.2 | |
| mlPTP | matches | 81.8 | 72.7 | 81.8 | 72.7 |
| splits | 18.2 | 27.3 | 18.2 | 27.3 | |
| bPTP | matches | 81.8 | 81.8 | 72.7 | 72.7 |
| splits | 18.2 | 18.2 | 27.3 | 27.3 |
| FAs (Mass % of Total) | VKM Al-418 | η2 | ||
|---|---|---|---|---|
| 9 °C | 22 °C | 27 °C | ||
| 14:0 | 1.0 ± 0.1 | 1.2 ± 0.3 | 1.1 ± 0.1 | |
| 16:0 | 20.6 ± 1.5 a | 23.0 ± 1.9 a | 41.8 ± 3.5 b* | 0.958 |
| 16:1 Δ7 | 1.9 ± 0.6 | 0.4 ± 0.1 | 0.2 ± 0.2 | |
| 16:1 Δ9 | n.d. | 1.0 ± 0.1 | 0.3 ± 0.3 | |
| 16:2 Δ7,10 | 1.8 ± 0.1 | 2.7 ± 0.1 | 0.4 ± 0.2 | |
| 16:3 Δ7,10,13 | 6.0 ± 0.9 a* | 1.7 ± 0.4 b | 0.1 ± 0.1 c | 0.966 |
| 18:0 | 2.4 ± 0.7 | 0.8 ± 0.6 | 3.4 ± 1.3 | |
| 18:1 Δ9 | 5.7 ± 1.3 a | 7.3 ± 0.1 a | 25.4 ± 3.0 b* | 0.971 |
| 18:1 Δ11 | 1.3 ± 0.1 | 2.6 ± 0.3 | 1.4 ± 0.1 | |
| 18:2 Δ9,12 | 17.1 ± 0.9 a | 36.1 ± 2.2 b* | 21.2 ± 4.0 a | 0.933 |
| 18:3 Δ9,12,15 | 41.6 ± 2.0 a* | 22.8 ± 1.4 b* | 3.0 ± 1.1 c* | 0.993 |
| UI | 1.892 ± 0.075 a | 1.623 ± 0.066 b | 0.802 ± 0.095 c* | 0.981 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krivina, E.; Sinetova, M.; Starikov, A.; Portnov, A.; Temraleeva, A. Evaluating Species Delimitation Methods in Chloroidium (Trebouxiophyceae, Chlorophyta): Efficacy of DNA Barcodes and Description of Chloroidium pseudoellipsoideum sp. nov. from Arctic Soils. Plants 2025, 14, 3739. https://doi.org/10.3390/plants14243739
Krivina E, Sinetova M, Starikov A, Portnov A, Temraleeva A. Evaluating Species Delimitation Methods in Chloroidium (Trebouxiophyceae, Chlorophyta): Efficacy of DNA Barcodes and Description of Chloroidium pseudoellipsoideum sp. nov. from Arctic Soils. Plants. 2025; 14(24):3739. https://doi.org/10.3390/plants14243739
Chicago/Turabian StyleKrivina, Elena, Maria Sinetova, Alexander Starikov, Aleksey Portnov, and Anna Temraleeva. 2025. "Evaluating Species Delimitation Methods in Chloroidium (Trebouxiophyceae, Chlorophyta): Efficacy of DNA Barcodes and Description of Chloroidium pseudoellipsoideum sp. nov. from Arctic Soils" Plants 14, no. 24: 3739. https://doi.org/10.3390/plants14243739
APA StyleKrivina, E., Sinetova, M., Starikov, A., Portnov, A., & Temraleeva, A. (2025). Evaluating Species Delimitation Methods in Chloroidium (Trebouxiophyceae, Chlorophyta): Efficacy of DNA Barcodes and Description of Chloroidium pseudoellipsoideum sp. nov. from Arctic Soils. Plants, 14(24), 3739. https://doi.org/10.3390/plants14243739






