Development of a Multiplex RT-PCR Assay for Simultaneous Detection of Velarivirus arecae, Arepavirus arecae and Arepavirus arecamaculatum
Abstract
1. Background
2. Methods
2.1. Plant Materials
2.2. RNA Extraction and Reverse Transcription
2.3. Design of Virus-Specific Primers
2.4. Evaluation of Primer Specificity
2.5. Optimization of Multiplex RT-PCR Assay
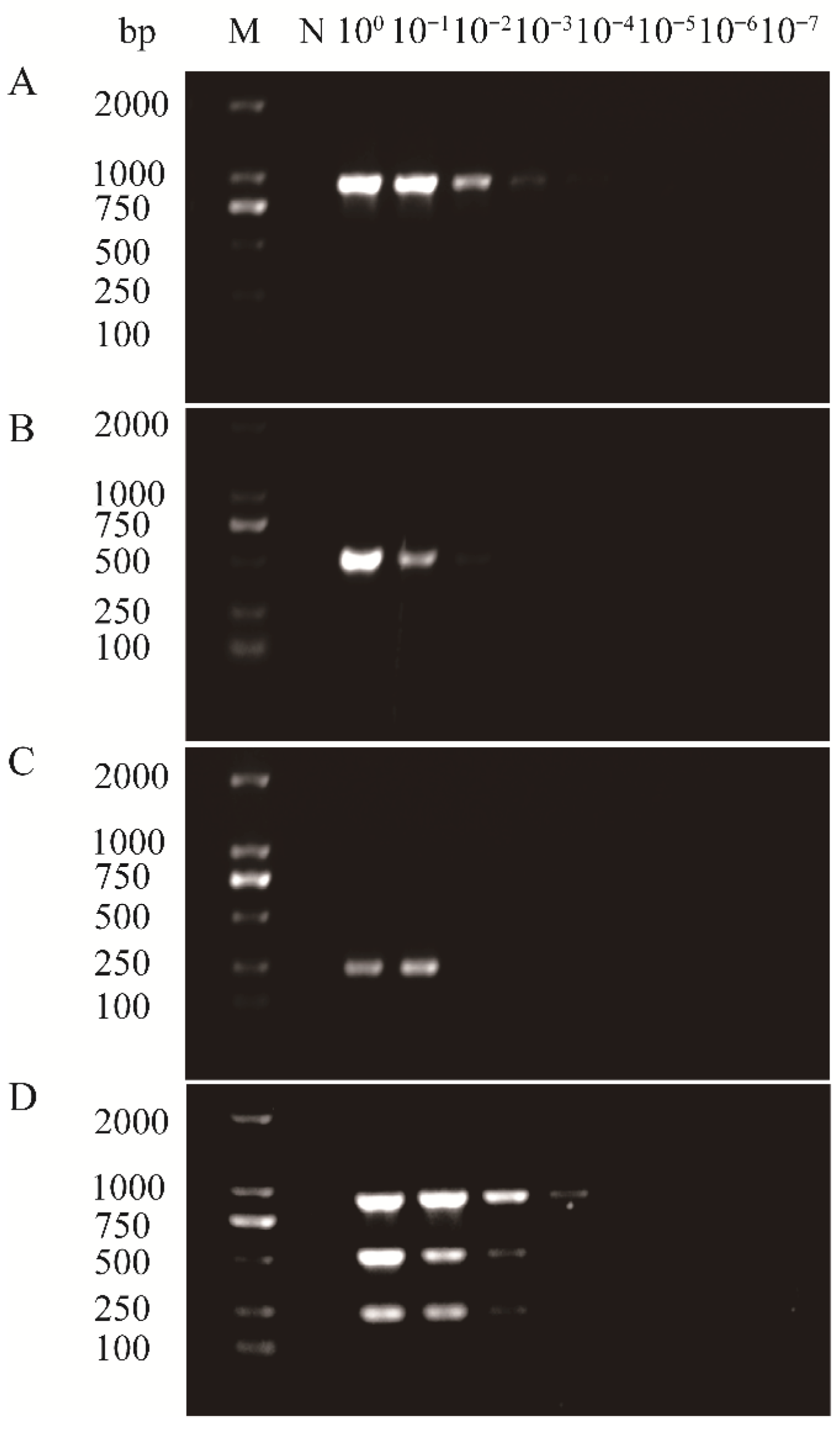
2.6. Sensitivity of Multiplex PCR Assay
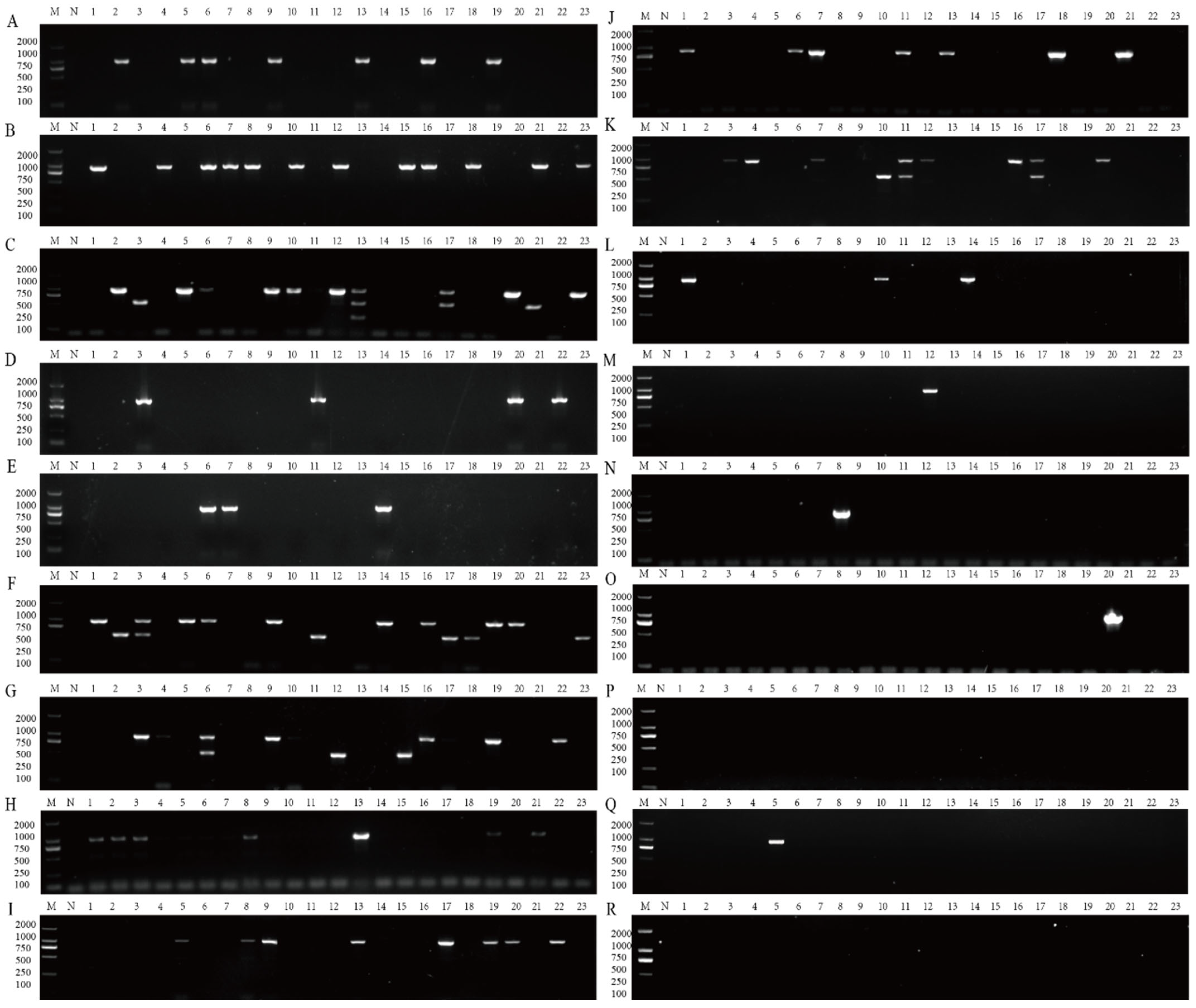
2.7. Survey of Areca Viruses by Multiplex RT-PCR Assay
3. Results
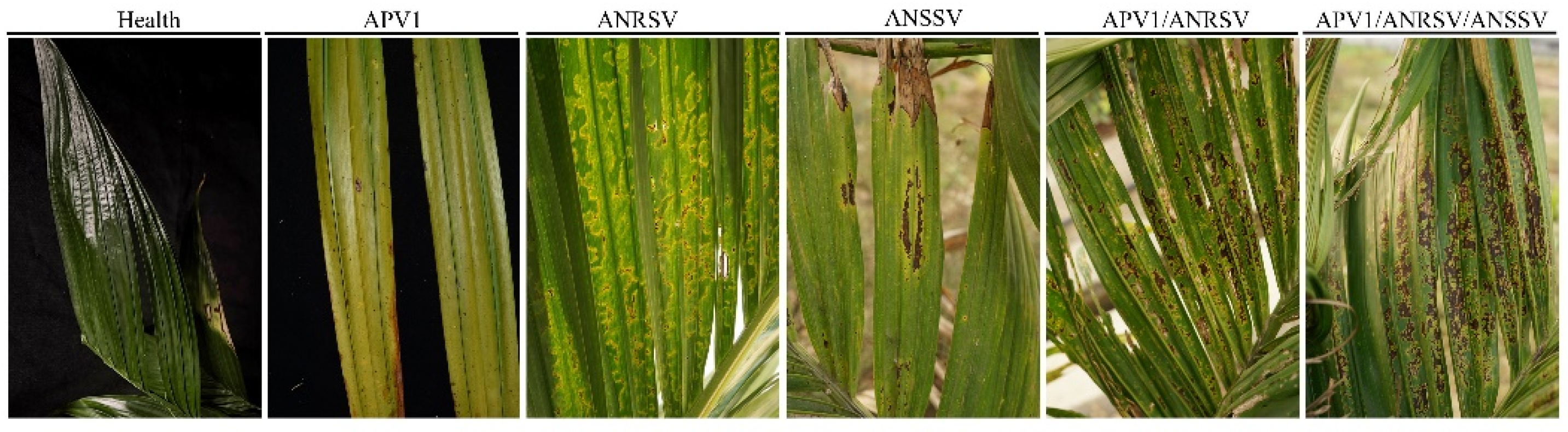
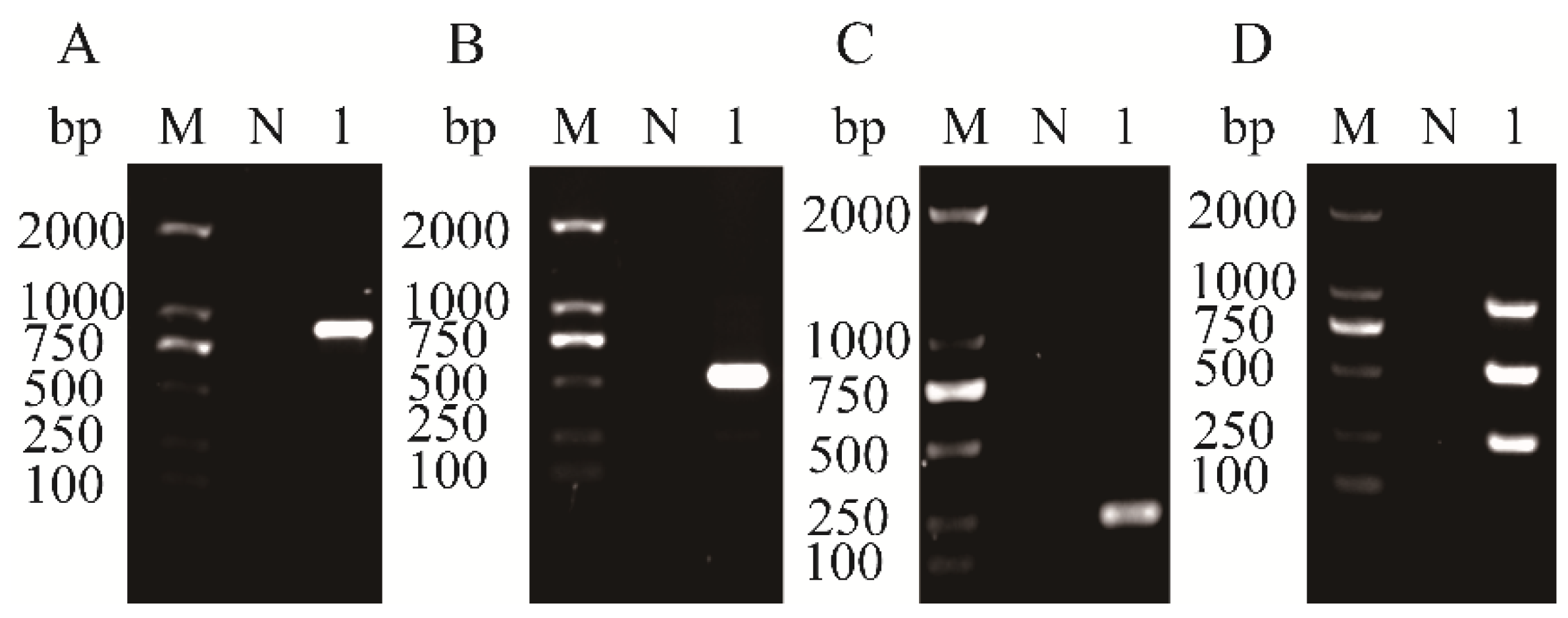
3.1. Specificity and Compatibility of Primer Pairs
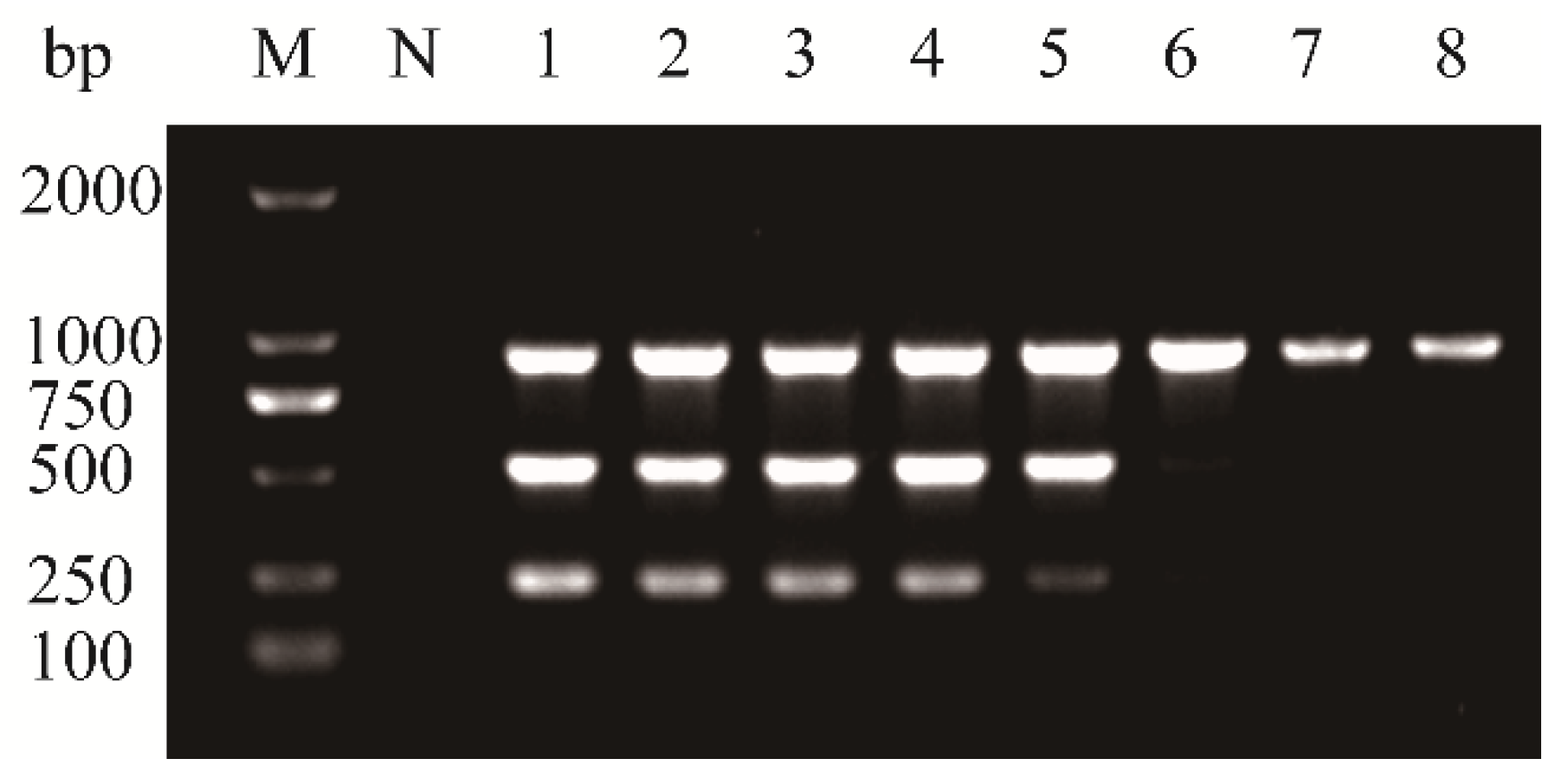
3.2. Optimization of Multiplex RT-PCR Conditions and Evaluation of Primer Concentrations
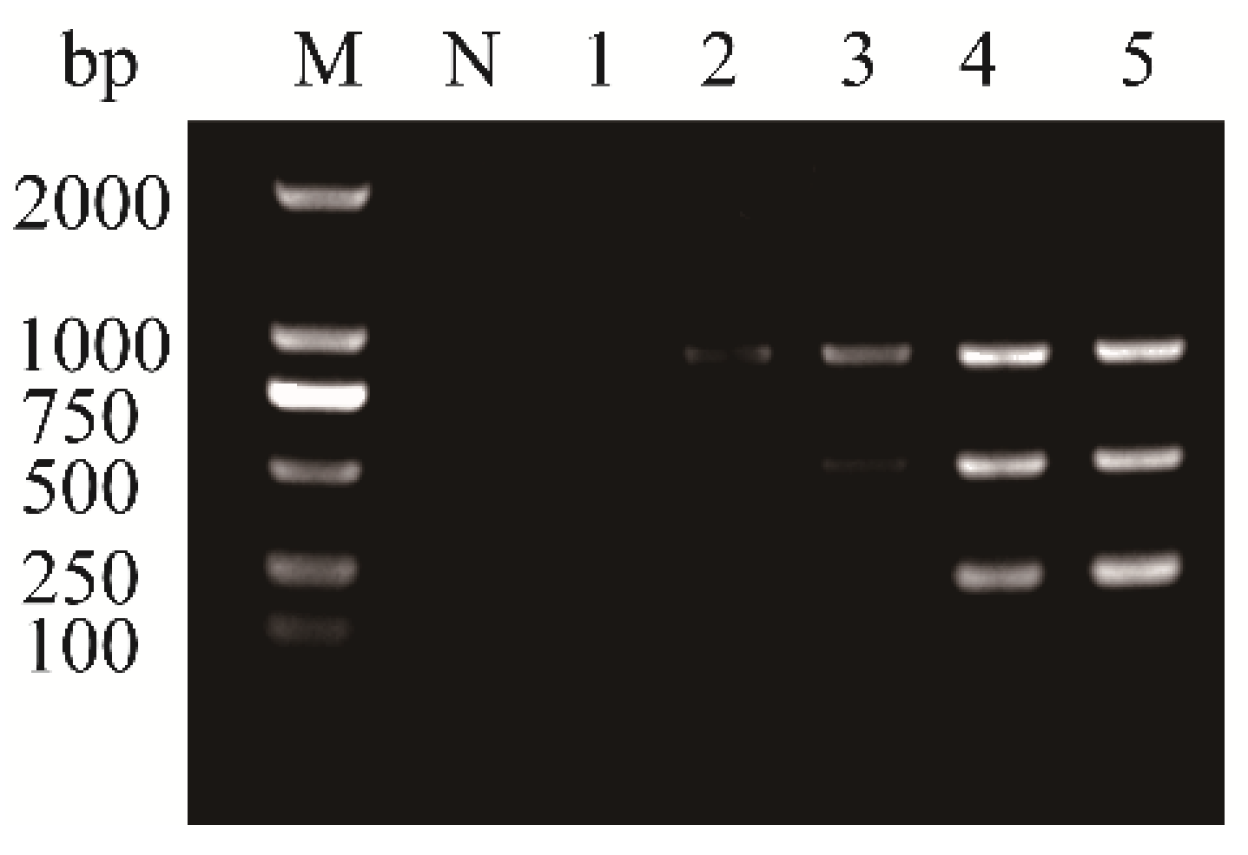
3.3. Sensitivity of the Multiplex RT-PCR Assay
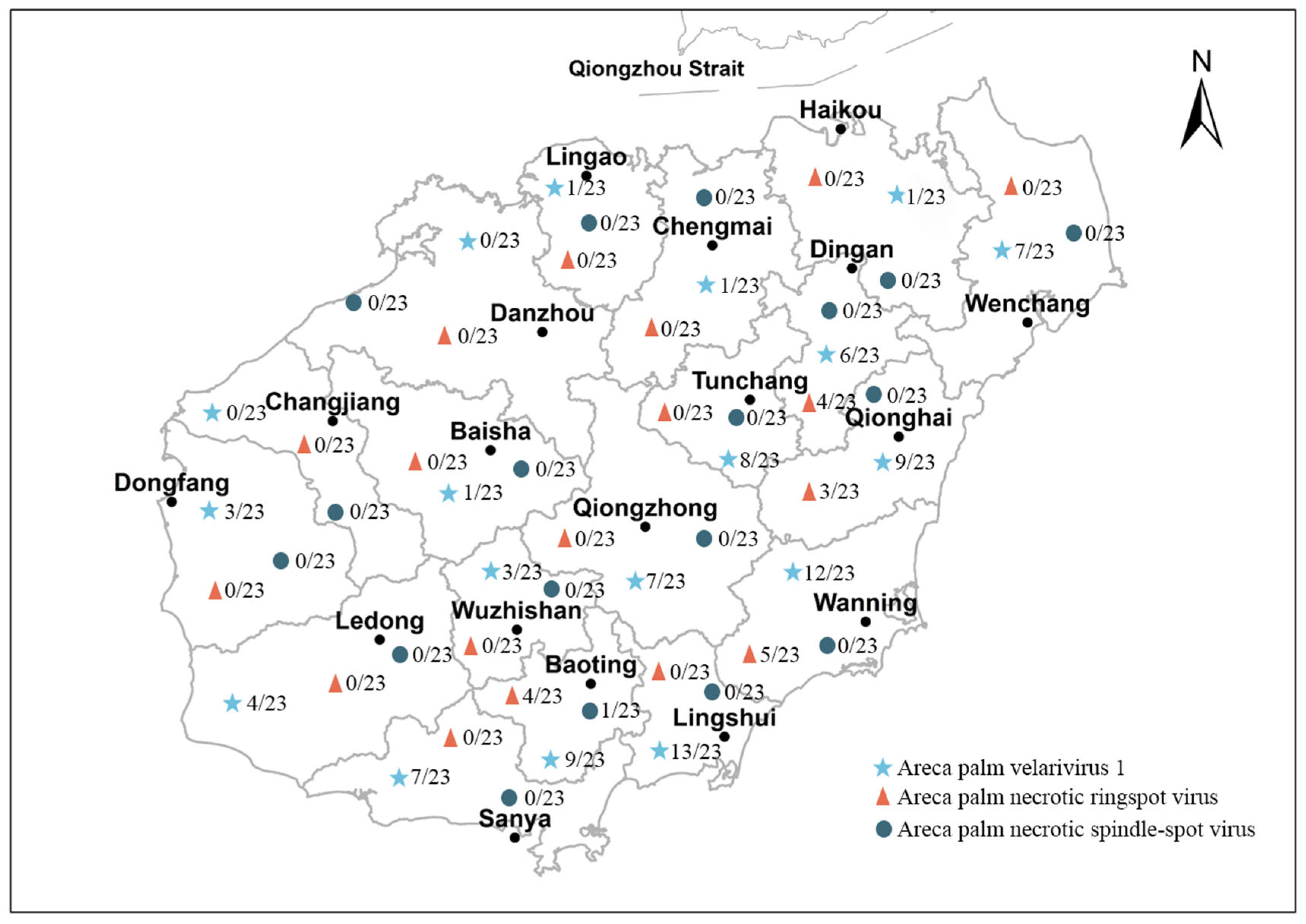
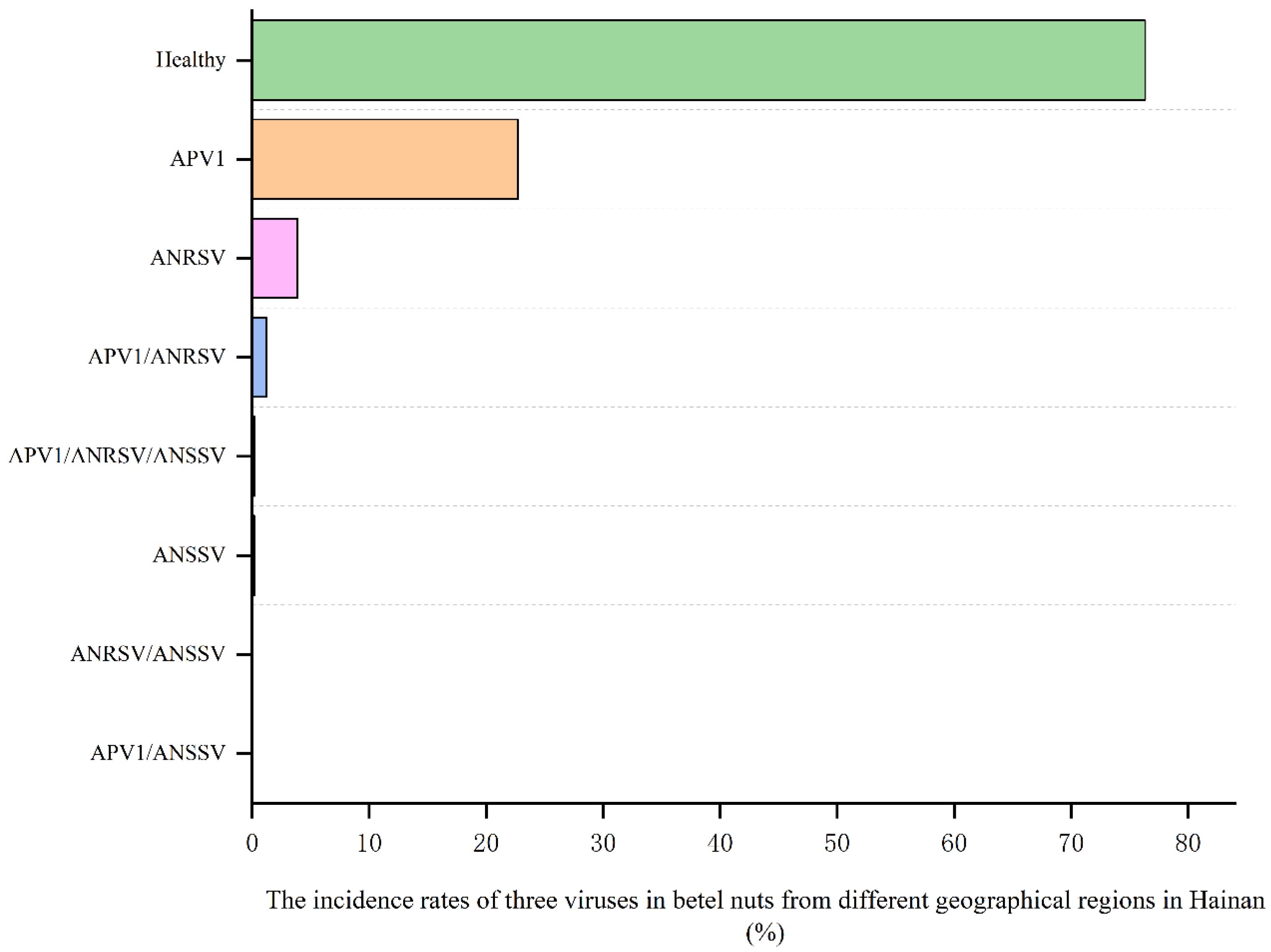
3.4. Application of Multiplex RT-PCR Assay in Survey of Areca Viruses
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Heatubun, C.D.; Dransfield, J.; Flynn, T.; Tjitrosoedirdjo, S.S.; Mogea, J.P.; Baker, W.J. A monograph of the betel nut palms (Areca: Arecaceae) of East Malesia. Bot. J. Linn. Soc. 2012, 168, 147–173. [Google Scholar] [CrossRef]
- Salama, H.S.; Zaki, F.N.; Abdel-Razek, A.S. Ecological and biological studies on the red palm weevil Rhynchophorus ferrugineus (Olivier). Arch. Phytopathol. Plant Prot. 2009, 42, 392–399. [Google Scholar] [CrossRef]
- Yu, H.; Qi, S.; Chang, Z.; Rong, Q.; Akinyemi, I.A.; Wu, Q. Complete genome sequence of a novel velarivirus infecting areca palm in China. Arch. Virol. 2015, 160, 2367–2370. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Zhao, R.; Wang, H.; Zhang, H.; Zhao, X.; Khan, L.U.; Huang, X. Genomic diversity of Areca Palm Velarivirus 1 (APV1) in Areca palm (Areca catechu) plantations in Hainan, China. BMC Genom. 2021, 22, 725. [Google Scholar] [CrossRef]
- Cao, X.; Gao, B.; Lu, J.; Wang, H.; Zhao, R.; Huang, X. Areca palm velarivirus 1 infection caused disassembly of chloroplast and reduction of photosynthesis in areca palm. Front. Microbiol. 2024, 15, 1424489. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, X.; Cao, X.; Khan, L.U.; Zhao, R.; Wang, H.; Huang, X. Transmission of Areca Palm Velarivirus 1 by Mealybugs Causes Yellow Leaf Disease in Betel Palm (Areca catechu). Phytopathology 2022, 112, 700–707. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, H.; Huang, X. Preparation of Polyclonal Antibody against Areca Palm Virus APV1 and Its Detection by Enzyme-Linked Immunosorbent Assay. Mol. Plant Breed. 2022, 20, 518–523. [Google Scholar] [CrossRef]
- Yang, K.; Ran, M.; Li, Z.; Hu, M.; Zheng, L.; Liu, W.; Jin, P.; Miao, W.; Zhou, P.; Shen, W.; et al. Analysis of the complete genomic sequence of a novel virus, areca palm necrotic spindle-spot virus, reveals the existence of a new genus in the family Potyviridae. Arch. Virol. 2018, 163, 3471–3475. [Google Scholar] [CrossRef]
- Khan, L.U.; Zhao, R.; Wang, H.; Huang, X. Recent advances of the causal agent of yellow leaf disease (YLD) on areca palm (Areca catechu L.). Trop. Plants 2023, 2, 7. [Google Scholar] [CrossRef]
- Yang, K.; Shen, W.; Li, Y.; Li, Z.; Miao, W.; Wang, A.; Cui, H. Areca Palm Necrotic Ringspot Virus, Classified Within a Recently Proposed Genus Arepavirus of the Family Potyviridae, Is Associated with Necrotic Ringspot Disease in Areca Palm. Phytopathology 2019, 109, 887–894. [Google Scholar] [CrossRef]
- Boonham, N.; Kreuze, J.; Winter, S.; van der Vlugt, R.; Bergervoet, J.; Tomlinson, J.; Mumford, R. Methods in virus diagnostics: From ELISA to next generation sequencing. Virus Res. 2014, 186, 20–31. [Google Scholar] [CrossRef]
- Santiago, G.A.; Vázquez, J.; Courtney, S.; Matías, K.Y.; Andersen, L.E.; Colón, C.; Butler, A.E.; Roulo, R.; Bowzard, J.; Villanueva, J.M.; et al. Performance of the Trioplex real-time RT-PCR assay for detection of Zika, dengue, and chikungunya viruses. Nat. Commun. 2018, 9, 1391. [Google Scholar] [CrossRef]
- Rubio, L.; Galipienso, L.; Ferriol, I. Detection of Plant Viruses and Disease Management: Relevance of Genetic Diversity and Evolution. Front. Plant Sci. 2020, 11, 1092. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; Liu, Y.J.; Wu, N.; Sun, T.; He, X.Y.; Gao, Y.X.; Wu, C.J. Areca catechu L. (Arecaceae): A review of its traditional uses, botany, phytochemistry, pharmacology and toxicology. J. Ethnopharmacol. 2015, 164, 340–356. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Shen, W.; Tuo, D.; Cui, H.; Yan, P.; Tang, Q.; Zhu, G.; Li, X.; Zhou, P.; Zhang, Y. Rapid detection of two emerging viruses associated with necrotic symptoms in Areca catechu L. by reverse transcription loop-mediated isothermal amplification (RT-LAMP). J. Virol. Methods 2020, 281, 113795. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.Y.; Jeong, R.D.; Yoon, Y.N.; Lee, S.H.; Shin, D.B.; Kang, H.W.; Lee, B.C. One-step multiplex reverse transcription-polymerase chain reaction for the simultaneous detection of three rice viruses. J. Virol. Methods 2013, 193, 674–678. [Google Scholar] [CrossRef]
- Zhang, P.; Liu, Y.; Liu, W.; Massart, S.; Wang, X. Simultaneous detection of wheat dwarf virus, northern cereal mosaic virus, barley yellow striate mosaic virus and rice black-streaked dwarf virus in wheat by multiplex RT-PCR. J. Virol. Methods 2017, 249, 170–174. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; Hu, W.; Li, Y.; Li, Y.; Chen, S.; Wang, J. Simultaneous multiplex RT-PCR detection of four viruses associated with maize lethal necrosis disease. J. Virol. Methods 2021, 298, 114286. [Google Scholar] [CrossRef]
- Xue, B.; Shang, J.; Yang, J.; Zhang, L.; Du, J.; Yu, L.; Yang, W.; Naeem, M. Development of a multiplex RT-PCR assay for the detection of soybean mosaic virus, bean common mosaic virus and cucumber mosaic virus in field samples of soybean. J. Virol. Methods 2021, 298, 114278. [Google Scholar] [CrossRef]
- Xu, Y.; Yang, L.; Zhou, J.; Yang, Y.; Lu, M.; Li, S. Multiplex RT-PCR to simultaneously detect three viruses that infect peach. Lett. Appl. Microbiol. 2019, 69, 318–324. [Google Scholar] [CrossRef]
- Faggioli, F.; Luigi, M. Multiplex RT-PCR. Methods Mol. Biol. 2022, 2316, 163–179. [Google Scholar] [CrossRef] [PubMed]
- Baggio, F.; Hetzel, U.; Prähauser, B.; Dervas, E.; Michalopoulou, E.; Thiele, T.; Kipar, A.; Hepojoki, J. A Multiplex RT-PCR Method for the Detection of Reptarenavirus Infection. Viruses 2023, 15, 2313. [Google Scholar] [CrossRef] [PubMed]
- Goto, Y.; Fukunari, K.; Tada, S.; Ichimura, S.; Chiba, Y.; Suzuki, T. A multiplex real-time RT-PCR system to simultaneously diagnose 16 pathogens associated with swine respiratory disease. J. Appl. Microbiol. 2023, 134, lxad263. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Ye, T.; Hao, L.; Guo, L.; Fan, Z.; Li, S.; Zhou, T. Genetic variation analysis of apple chlorotic leaf spot virus coat protein reveals a new phylogenetic type and two recombinants in China. Arch. Virol. 2014, 159, 1431–1438. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Peterson, D.; Tamura, K. MEGA-CC: Computing core of molecular evolutionary genetics analysis program for automated and iterative data analysis. Bioinformatics 2012, 28, 2685–2686. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Di Serio, F.; Ambrós, S.; Sano, T.; Flores, R.; Navarro, B. Viroid Diseases in Pome and Stone Fruit Trees and Koch’s Postulates: A Critical Assessment. Viruses 2018, 10, 612. [Google Scholar] [CrossRef]
- Yao, B.; Wang, G.; Ma, X.; Liu, W.; Tang, H.; Zhu, H.; Hong, N. Simultaneous detection and differentiation of three viruses in pear plants by a multiplex RT-PCR. J. Virol. Methods 2014, 196, 113–119. [Google Scholar] [CrossRef]
- Hao, L.; Xie, J.; Chen, S.; Wang, S.; Gong, Z.; Ling, K.S.; Guo, L.; Fan, Z.; Zhou, T. A multiple RT-PCR assay for simultaneous detection and differentiation of latent viruses and apscarviroids in apple trees. J. Virol. Methods 2016, 234, 16–21. [Google Scholar] [CrossRef]
- Wang, Y.; Shen, W.; Dai, Z.; Gou, B.; Liu, H.; Hu, W.; Qin, L.; Li, Z.; Tuo, D.; Cui, H. Biological and Molecular Characterization of Two Closely Related Arepaviruses and Their Antagonistic Interaction in Nicotiana benthamiana. Front. Microbiol. 2021, 12, 755156. [Google Scholar] [CrossRef]
- Lin, Z.; Tang, Q.; Meng, X.; Song, W.; Yu, F.; Huang, S.; Niu, X.; Qin, W. Occurrence, Distribution and Phytoplasma Detection of Areca Palm Pathological Yellowing in Hainan Province. Chin. J. Trop. Crops 2022, 43, 2106–2113. [Google Scholar]
| Virus | Primer | Primer Sequence (5′–3′) | Tm (°C) | GC% | Position (nt) | Products (bp) | Target Gene |
|---|---|---|---|---|---|---|---|
| APV1 | APV1-F | ATCGCTAAATATTATGGATAGACTT | 52 | 28 | 12,768–12,792 | 938 | CP |
| APV1-R | TATTCAGAAGCATAAGATTGTGACA | 54 | 31 | 13,681–13,705 | |||
| ANRSV | ANRSV-F | TCGCTGACATTGAAAAGG | 52 | 44 | 5709–5726 | 527 | Nla-VPg/Nla-Pro |
| ANRSV-R | CTTAGCTGCTAGTCGCG | 57 | 59 | 6219–6235 | |||
| ANSSV | ANSSV-F | CTCTAAAGGACAACAAACCA | 50 | 50 | 8691–8708 | 250 | Nla-VPg/Nla-Pro |
| ANSSV-R | GAAAATTTGCGAAATCTGCATTGTC | 54 | 42 | 8922–8940 |
| Location | No. of Samples | No. of Positive Samples and Positive Rate (%) | ||
|---|---|---|---|---|
| APV1 | ANRSV | ANSSV | ||
| Sanya | 23 | 7(30.00) | 0(00.00) | 0(00.00) |
| Lingshui | 23 | 12(52.17) | 0(0.00) | 0(00.00) |
| Baoting | 23 | 12(52.17) | 4(17.39) | 1(04.34) |
| Ledong | 23 | 4(17.39) | 0(00.00) | 0(0.00) |
| Dongfang | 23 | 3(13.04) | 0(00.00) | 0(00.00) |
| Wanning | 23 | 9(39.13) | 6(26.09) | 0(00.00) |
| Qionghai | 23 | 6(26.09) | 3(13.00) | 0(00.00) |
| Qiongzhong | 23 | 7(30.00) | 0(00.00) | 0(00.00) |
| Tunchang | 23 | 8(34.78) | 0(00.00) | 0(00.00) |
| Wenchang | 23 | 7(30.00) | 0(00.00) | 0(00.00) |
| Dingan | 23 | 8(34.78) | 3(13.04) | 0(00.00) |
| Wuzhishan | 23 | 3(13.04) | 0(00.00) | 0(00.00) |
| Haikou | 23 | 1(08.69) | 0(00.00) | 0(00.00) |
| Chengmai | 23 | 1(04.34) | 0(00.00) | 0(00.00) |
| Lingao | 23 | 1(04.34) | 0(00.00) | 0(00.00) |
| Danzhou | 23 | 0(00.00) | 0(00.00) | 0(00.00) |
| Baisha | 23 | 1(04.34) | 0(00.00) | 0(00.00) |
| Changjiang | 23 | 0(00.00) | 0(00.00) | 0(00.00) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, K.; Zhang, L.; Li, Z.; Zhao, P.; Wan, S. Development of a Multiplex RT-PCR Assay for Simultaneous Detection of Velarivirus arecae, Arepavirus arecae and Arepavirus arecamaculatum. Plants 2025, 14, 3683. https://doi.org/10.3390/plants14233683
Sun K, Zhang L, Li Z, Zhao P, Wan S. Development of a Multiplex RT-PCR Assay for Simultaneous Detection of Velarivirus arecae, Arepavirus arecae and Arepavirus arecamaculatum. Plants. 2025; 14(23):3683. https://doi.org/10.3390/plants14233683
Chicago/Turabian StyleSun, Kexin, Li Zhang, Zemu Li, Peng Zhao, and Siyu Wan. 2025. "Development of a Multiplex RT-PCR Assay for Simultaneous Detection of Velarivirus arecae, Arepavirus arecae and Arepavirus arecamaculatum" Plants 14, no. 23: 3683. https://doi.org/10.3390/plants14233683
APA StyleSun, K., Zhang, L., Li, Z., Zhao, P., & Wan, S. (2025). Development of a Multiplex RT-PCR Assay for Simultaneous Detection of Velarivirus arecae, Arepavirus arecae and Arepavirus arecamaculatum. Plants, 14(23), 3683. https://doi.org/10.3390/plants14233683





