Holobiome Structure and Microbial Core Assemblages of Deschampsia antarctica Across the South Shetland Islands
Abstract
1. Introduction
2. Results
2.1. Soil Physicochemical Analysis
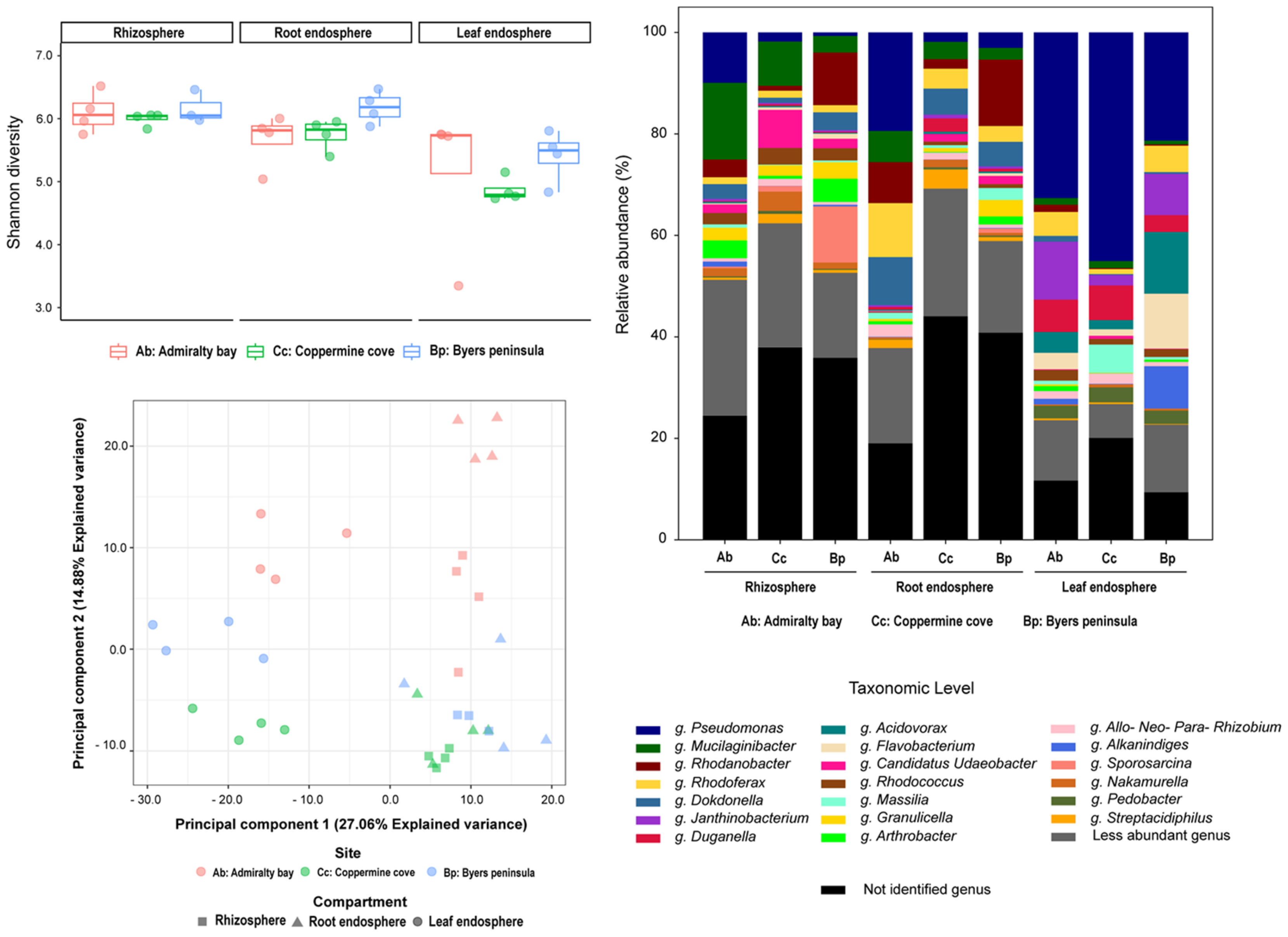
2.2. Bacterial Community Composition and Diversity
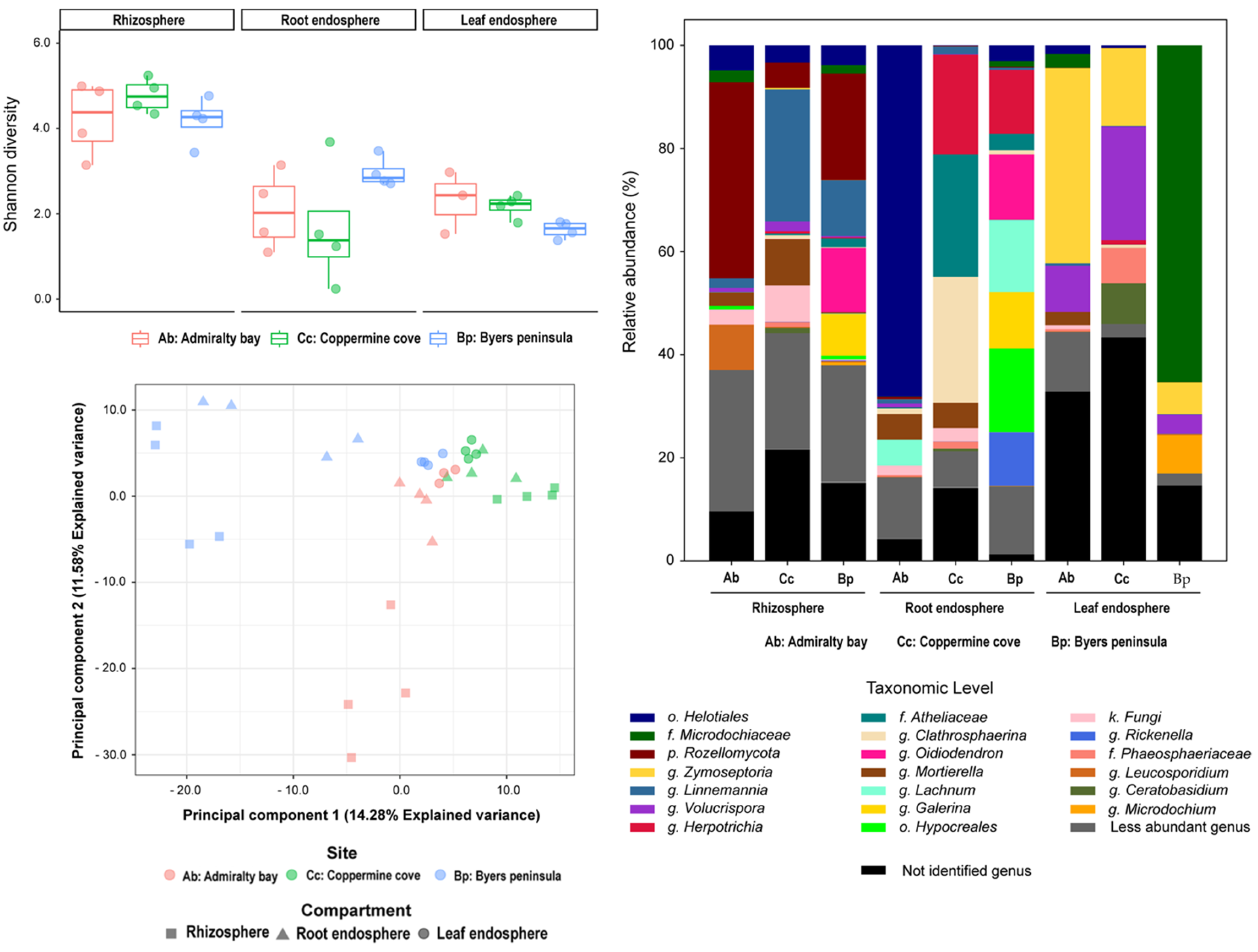
2.3. Fungal Community Composition and Diversity
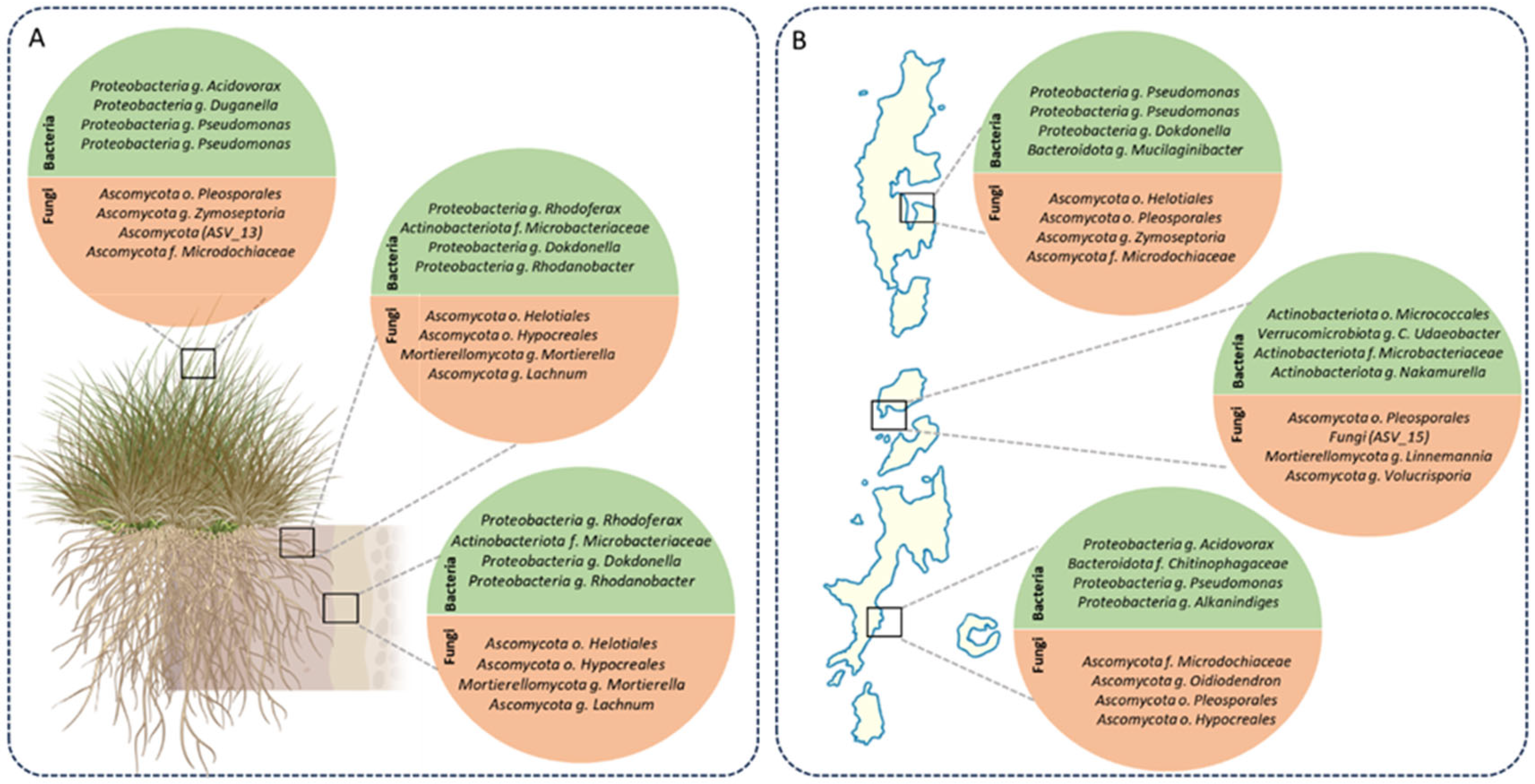
2.4. Core Microbiomes Across Compartments and Islands
2.5. Functional Guilds and Predicted Microbial Functions
2.6. Co-Occurrence Network Topology and Keystone Taxa Across Compartments
2.7. Distance-Decay Relationship
3. Discussion
3.1. Compartmentalization as the Dominant Driver of Microbiome Structure
3.2. Edaphic Filtering Shapes Fungal Communities and Site-Specific Assemblages
3.3. Geographic Heterogeneity and the Balance Between Conserved Cores and Local Exclusivity
3.4. Toward a Hierarchical Framework for Holobiont Resilience in Extreme Environments
4. Materials and Methods
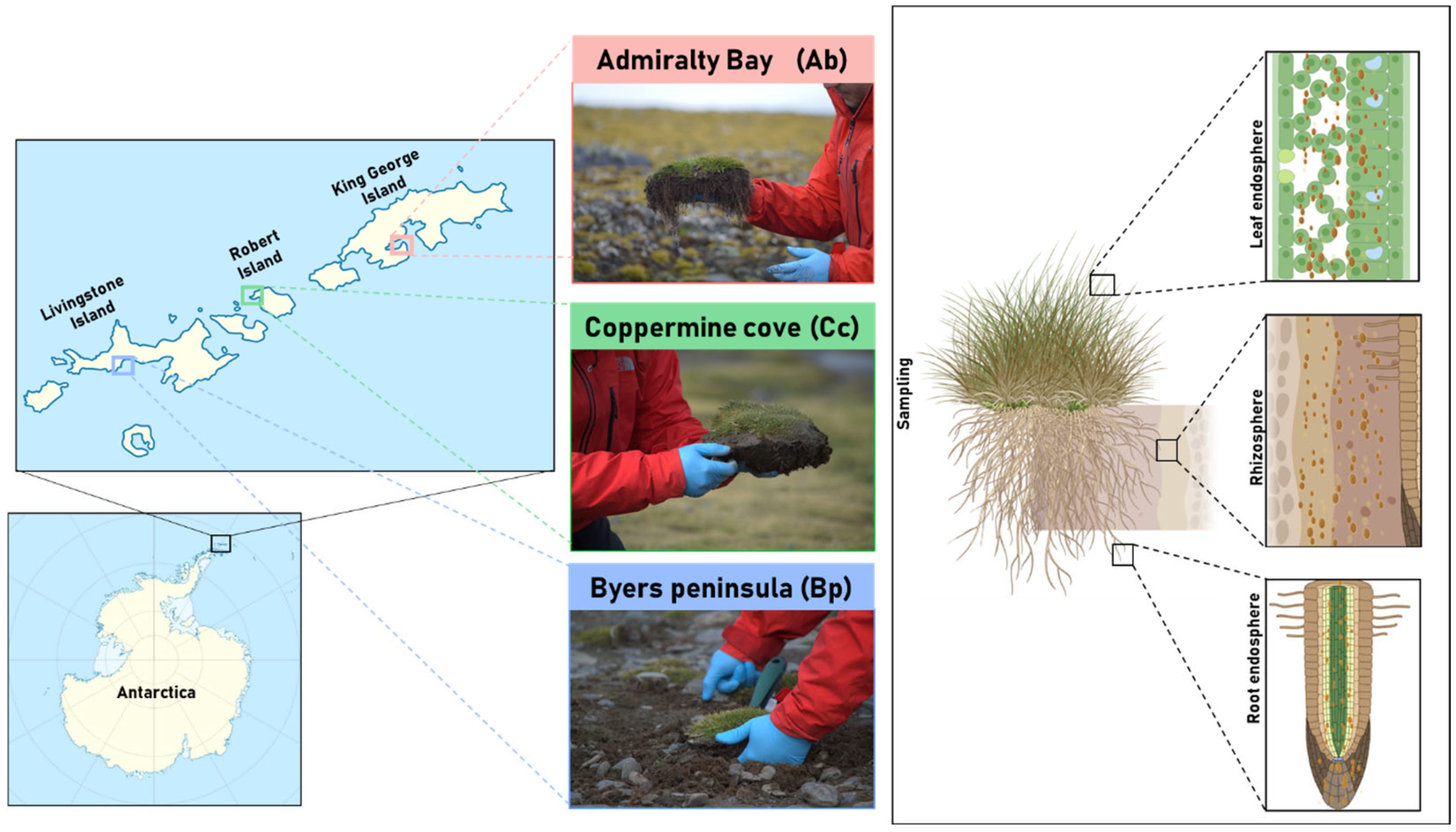
4.1. Sampling Site
4.2. Soil Physicochemical Analysis
4.3. DNA Isolation
4.4. Bioinformatics Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Ab | Admiralty Bay |
| Bp | Byers Peninsula |
| Cc | Coppermine Cove |
| CEC | Cation Exchange Capacity |
| DNA | Deoxyribonucleic Acid |
| ECA59 | 59th Chilean Antarctic Scientific Expedition |
| INACH | Instituto Antártico Chileno (Chilean Antarctic Institute) |
| ITS | Internal Transcribed Spacer |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KO | KEGG Orthology |
| OTU | Operational Taxonomic Unit |
| PCA | Principal Component Analysis |
| PCoA | Principal Coordinates Analysis |
| PNA | Peptide Nucleic Acid |
| QIIME2 | Quantitative Insights Into Microbial Ecology 2 |
| SOM | Soil Organic Matter |
| UV | Ultraviolet |
| ZOTU | Zero-radius Operational Taxonomic Unit |
References
- Convey, P.; Peck, L.S. Antarctic environmental change and biological responses. Sci. Adv. 2019, 5, eaaz0888. [Google Scholar] [CrossRef] [PubMed]
- Gutt, J.; Isla, E.; Xavier, J.; Adams, B.; Ahn, I.; Cheng, C.; Colesie, C.; Cummings, V.; Di Prisco, G.; Griffiths, H.; et al. Antarctic ecosystems in transition—Life between stresses and opportunities. Biol. Rev. 2020, 96, 798–821. [Google Scholar] [CrossRef] [PubMed]
- Fowbert, J.A.; Smith, R.I.L. Rapid population increases in native vascular plants in the Argentine Islands, Antarctic Peninsula. Arct. Alp. Res. 1994, 26, 290–296. [Google Scholar] [CrossRef]
- Parnikoza, I.; Kozeretska, I.; Kunakh, V. Vascular plants of the Maritime Antarctic: Origin and adaptation. Am. J. Plant Sci. 2011, 2, 381–395. [Google Scholar] [CrossRef]
- Marian, M.; Licciardello, G.; Vicelli, B.; Pertot, I.; Perazzolli, M. Ecology and potential functions of plant-associated microbial communities in cold environments. FEMS Microbiol. Ecol. 2021, 98, fiab161. [Google Scholar] [CrossRef]
- Znój, A.; Brej, T.; Chwedorzewska, K.J.; Giełwanowska, I.; Loro, M.; Matuła, J.; Nicia, P.; Zubek, S. Deschampsia antarctica and Colobanthus quitensis—A Microbiome Comparison. Microb. Ecol. 2022, 83, 1–14. [Google Scholar] [CrossRef]
- Wang, X.; Qu, Y.; Teng, X.; Xu, L.; Jin, L.; Xue, H.; Li, J.; Guo, Y.; Zhang, G.; Liu, Z.; et al. Meta-analysis of root-associated bacterial communities of widely distributed native and invasive Poaceae plants in Antarctica. Polar Biol. 2024, 47, 1023–1038. [Google Scholar] [CrossRef]
- Rodríguez, R.; Rabert, C.; Larama, G.; Fuentes-Lillo, I.; Corsini, G.; Morales-Quintana, L.; Ramos, P.; Tapia-Valdebenito, D.; González-Pastén, C.; Fuentes-Quiroz, A. Taxonomic and predicted functional profiling of coexisting rhizosphere microbiomes of Deschampsia antarctica and Colobanthus quitensis along an altitudinal transect in Admiralty Bay, Maritime Antarctica. J. Soil Sci. Plant Nutr. 2025, 25, 2201–2218. [Google Scholar] [CrossRef]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef]
- Berg, G.; Dorador, C.; Egamberdieva, D.; Kostka, J.; Ryu, C.; Wassermann, B. Shared governance in the plant holobiont and implications for one health. FEMS Microbiol. Ecol. 2024, 100, fiae004. [Google Scholar] [CrossRef]
- Rodríguez, R.; Durán, P. Natural holobiome engineering by using native extreme microbiome to counteract the climate change effects. Front. Bioeng. Biotechnol. 2020, 8, 568. [Google Scholar] [CrossRef] [PubMed]
- Aqeel, M.; Ran, J.; Hu, W.; Irshad, M.; Dong, L.; Akram, M.; Eldesoky, G.; Aljuwayid, A.; Chuah, L.; Deng, J. Plant–soil–microbe interactions in maintaining ecosystem stability and coordinated turnover under changing environmental conditions. Chemosphere 2023, 329, 137924. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, R.; Barra, P.J.; Larama, G.; Carrión, V.J.; de la Luz Mora, M.; Hale, L.; Durán, P. Microbiome engineering optimized by Antarctic microbiota to support a plant host under water deficit. Front. Plant Sci. 2023, 14, 1241612. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cha, Q.; Dang, Y.; Chen, X.; Wang, M.; McMinn, A.; Espina, G.; Zhang, Y.; Blamey, J.; Qin, Q. Reconstruction of the functional ecosystem in the high light, low temperature Union Glacier region, Antarctica. Front. Microbiol. 2019, 10, 2408. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef]
- Santoyo, G.; Guzmán-Guzmán, P.; Parra-Cota, F.; Santos-Villalobos, S.; Orozco-Mosqueda, M.; Glick, B. Plant growth stimulation by microbial consortia. Agronomy 2021, 11, 219. [Google Scholar] [CrossRef]
- Coelho, L.; de Carvalho, C.; Rosa, C.; Rosa, L. Diversity, distribution, and xerophilic tolerance of cultivable fungi associated with the Antarctic angiosperms. Polar Biol. 2021, 44, 379–388. [Google Scholar] [CrossRef]
- Compant, S.; Clément, C.; Sessitsch, A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 2010, 42, 669–678. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; Van der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef]
- Vorholt, J.A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828–840. [Google Scholar] [CrossRef]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef] [PubMed]
- Ball, B.A.; Convey, P.; Feeser, K.L.; Nielsen, U.N.; Van Horn, D. Habitat severity characteristics structure soil communities at regional and local spatial scales along the Antarctica Peninsula. Antarct. Sci. 2023, 35, 103–119. [Google Scholar] [CrossRef]
- Cavicchioli, R.; Ripple, W.J.; Timmis, K.N.; Azam, F.; Bakken, L.R.; Baylis, M.; Behrenfeld, M.J.; Boetius, A.; Boyd, P.W.; Classen, A.T.; et al. Scientists’ warning to humanity: Microorganisms and climate change. Nat. Rev. Microbiol. 2019, 17, 569–586. [Google Scholar] [CrossRef]
- Yergeau, E.; Bokhorst, S.; Huiskes, A.H.L.; Boschker, H.T.S.; Aerts, R.; Kowalchuk, G.A. Size and structure of bacterial, fungal and nematode communities along an Antarctic environmental gradient. FEMS Microbiol. Ecol. 2007, 59, 436–451. [Google Scholar] [CrossRef]
- Turner, T.R.; James, E.K.; Poole, P.S. The plant microbiome. Genome Biol. 2013, 14, 209. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; van Themaat, E.V.L.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef]
- Mapelli, F.; Marasco, R.; Rolli, E.; Barbato, M.; Cherif, H.; Guesmi, A.; Ouzari, H.I.; Daffonchio, D.; Borin, S. Potential for plant growth promotion of rhizobacteria associated with Salicornia growing in Tunisian hypersaline soils. Biomed Res. Int. 2013, 2013, 248078. [Google Scholar] [CrossRef]
- D’Amico, S.; Collins, T.; Marx, J.C.; Feller, G.; Gerday, C. Psychrophilic microorganisms: Challenges for life. EMBO Rep. 2006, 7, 385–389. [Google Scholar] [CrossRef]
- Pearce, D.A.; Bridge, P.D.; Hughes, K.A.; Sattler, B.; Psenner, R.; Russell, N.J. Microorganisms in the atmosphere over Antarctica. FEMS Microbiol. Ecol. 2009, 69, 143–157. [Google Scholar] [CrossRef]
- Newsham, K.K. A meta-analysis of plant responses to dark septate root endophytes. New Phytol. 2011, 190, 783–793. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Põlme, S.; Abarenkov, K.; Henrik Nilsson, R.; Lindahl, B.D.; Clemmensen, K.E.; Kauserud, H.; Nguyen, N.; Kjøller, R.; Bates, S.T.; Baldrian, P.; et al. FungalTraits: A user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Divers. 2020, 105, 1–16. [Google Scholar] [CrossRef]
- Esperschuetz, J.; Baisden, W.T.; Balks, M.R.; Schipper, L.A. The role of soil chemistry and microbial communities in soil carbon stabilization and turnover in Antarctica. Soil Biol. Biochem. 2017, 109, 76–86. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef]
- Andreev, I.O.; Parnikoza, I.Y.; Konvalyuk, I.I.; Metcheva, R.; Kozeretska, I.A.; Kunakh, V.A. Genetic Divergence of Deschampsia antarctica (Poaceae) Population Groups in the Maritime Antarctic. Biol. J. Linn. Soc. 2022, 135, 223–234. [Google Scholar] [CrossRef]
- Peay, K.G.; Kennedy, P.G.; Talbot, J.M. Dimensions of biodiversity in the Earth mycobiome. Nat. Rev. Microbiol. 2016, 14, 434–447. [Google Scholar] [CrossRef]
- Schulz, B.; Wanke, U.; Draeger, S.; Aust, H.J. Endophytes from herbaceous plants and shrubs: Effectiveness of surface sterilization methods. Mycol. Res. 1993, 97, 1447–1450. [Google Scholar] [CrossRef]
- Sadzawka, A.; Carrasco, M.A.; Grez, R.; Mora, M.L.; Flores, H.; Neaman, A. Métodos de análisis recomendados para los suelos de Chile. In Instituto de Investigaciones Agropecuarias; Centro Regional de Investigación La Platina: Santiago, Chile, 2006; 164p. [Google Scholar]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Edgar, R.C. UNOISE2: Improved error-correction for Illumina 16S and ITS amplicon sequencing. bioRxiv 2016. [Google Scholar] [CrossRef]
- Rivers, A.R.; Weber, K.C.; Gardner, T.G.; Liu, S.; Armstrong, S.D. ITSxpress: Software to rapidly trim internally transcribed spacer sequences with quality scores for marker gene analysis. F1000Research 2018, 7, 1418. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Abarenkov, K.; Nilsson, R.H.; Kirk, P.M.; Larsson, K.-H.; Põldmaa, K.; Zhou, Q.; Sjökvist, E.; May, T.; Tedersoo, L.; Hibbet, D.S.; et al. The UNITE database for molecular identification of fungi: Latest updates and future directions. Nucleic Acids Res. 2024, 52, D766–D772. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.H.; Kristiansson, E.; Ryberg, M.; Hallenberg, N.; Larsson, K.-H. Intraspecific ITS variability in the kingdom Fungi as expressed in the international sequence databases and its implications for molecular species identification. Evol. Bioinform. 2008, 4, 193–201. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package, R package Version 2.5-7. 2020. Available online: https://cran.r-project.org/package=vegan (accessed on 25 September 2025).
- Wei, Z.; Yang, T.; Friman, V.-P.; Xu, Y.; Shen, Q.; Jousset, A. Trophic network architecture of root-associated bacterial communities determines pathogen invasion and plant health. Nat. Commun. 2015, 6, 8413. [Google Scholar] [CrossRef]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2: An improved and extensible approach for metagenome inference. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. InterJ. Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Pedersen, T.L. ggraph: An Implementation of Grammar of Graphics for Graphs and Networks, R Package Version 2.0.6. 2022. Available online: https://cran.r-project.org/package=ggraph (accessed on 25 July 2025).
- Hijmans, R.J. Geosphere: Spherical Trigonometry, R Package Version 1.5-14. 2023. Available online: https://CRAN.R-project.org/package=geosphere (accessed on 5 July 2025).

| Parameters | Coppermine Cove | Admiralty Bay | Byers Peninsula |
|---|---|---|---|
| Al (cmol+ kg−1) | 5.83 ± 0.06 | 4.47 ± 0.18 | 3.85 ± 0.07 |
| Ca (cmol+ kg−1) | 5.22 ± 0.1 | 0.32 ± 0.06 | 0.68 ± 0.01 |
| Cu (mg kg−1) | 1.79 ± 0.01 | 2.37 ± 0.11 | 1.23 ± 0.08 |
| Fe (mg kg−1) | 48.88 ± 0.67 | 111.89 ± 5.16 | 239.0 ± 9.9 |
| K (cmol+ kg−1) | 1.17 ± 0.03 | 0.91 ± 0.06 | 0.64 ± 0.05 |
| MO (%) | 2.0 ± 0.0 | 2.0 ± 0.0 | 2.0 ± 0.0 |
| Mg (cmol+ kg−1) | 6.66 ± 0.68 | 0.17 ± 0.07 | 0.22 ± 0.01 |
| Mn (mg kg−1) | 3.13 ± 0.01 | 0.85 ± 0.01 | 1.63 ± 0.07 |
| Na (cmol+ kg−1) | 1.88 ± 0.11 | 0.57 ± 0.06 | 0.66 ± 0.01 |
| P (mg kg−1) | 132.0 ± 11.31 | 315.5 ± 10.61 | 117.8 ± 0.28 |
| Zn (mg kg−1) | 7.92 ± 0.06 | 0.06 ± 0.01 | 0.16 ± 0.01 |
| pH | 5.5 ± 0.01 | 4.34 ± 0.02 | 4.34 ± 0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodriguez, R.; Barra, P.J.; Saldivar-Diaz, M.; Larama, G.; Alvarado, R.; López, D.; Delgado, M.; Orlando, J.; Oses, R.; Merino, C.; et al. Holobiome Structure and Microbial Core Assemblages of Deschampsia antarctica Across the South Shetland Islands. Plants 2025, 14, 3657. https://doi.org/10.3390/plants14233657
Rodriguez R, Barra PJ, Saldivar-Diaz M, Larama G, Alvarado R, López D, Delgado M, Orlando J, Oses R, Merino C, et al. Holobiome Structure and Microbial Core Assemblages of Deschampsia antarctica Across the South Shetland Islands. Plants. 2025; 14(23):3657. https://doi.org/10.3390/plants14233657
Chicago/Turabian StyleRodriguez, Rodrigo, Patricio Javier Barra, Manuel Saldivar-Diaz, Giovanni Larama, Roxana Alvarado, Dariel López, Mabel Delgado, Julieta Orlando, Rómulo Oses, Carolina Merino, and et al. 2025. "Holobiome Structure and Microbial Core Assemblages of Deschampsia antarctica Across the South Shetland Islands" Plants 14, no. 23: 3657. https://doi.org/10.3390/plants14233657
APA StyleRodriguez, R., Barra, P. J., Saldivar-Diaz, M., Larama, G., Alvarado, R., López, D., Delgado, M., Orlando, J., Oses, R., Merino, C., Tortella, G., & Duran, P. (2025). Holobiome Structure and Microbial Core Assemblages of Deschampsia antarctica Across the South Shetland Islands. Plants, 14(23), 3657. https://doi.org/10.3390/plants14233657







