Single-Cell Omics in Legumes: Research Trends and Applications
Abstract
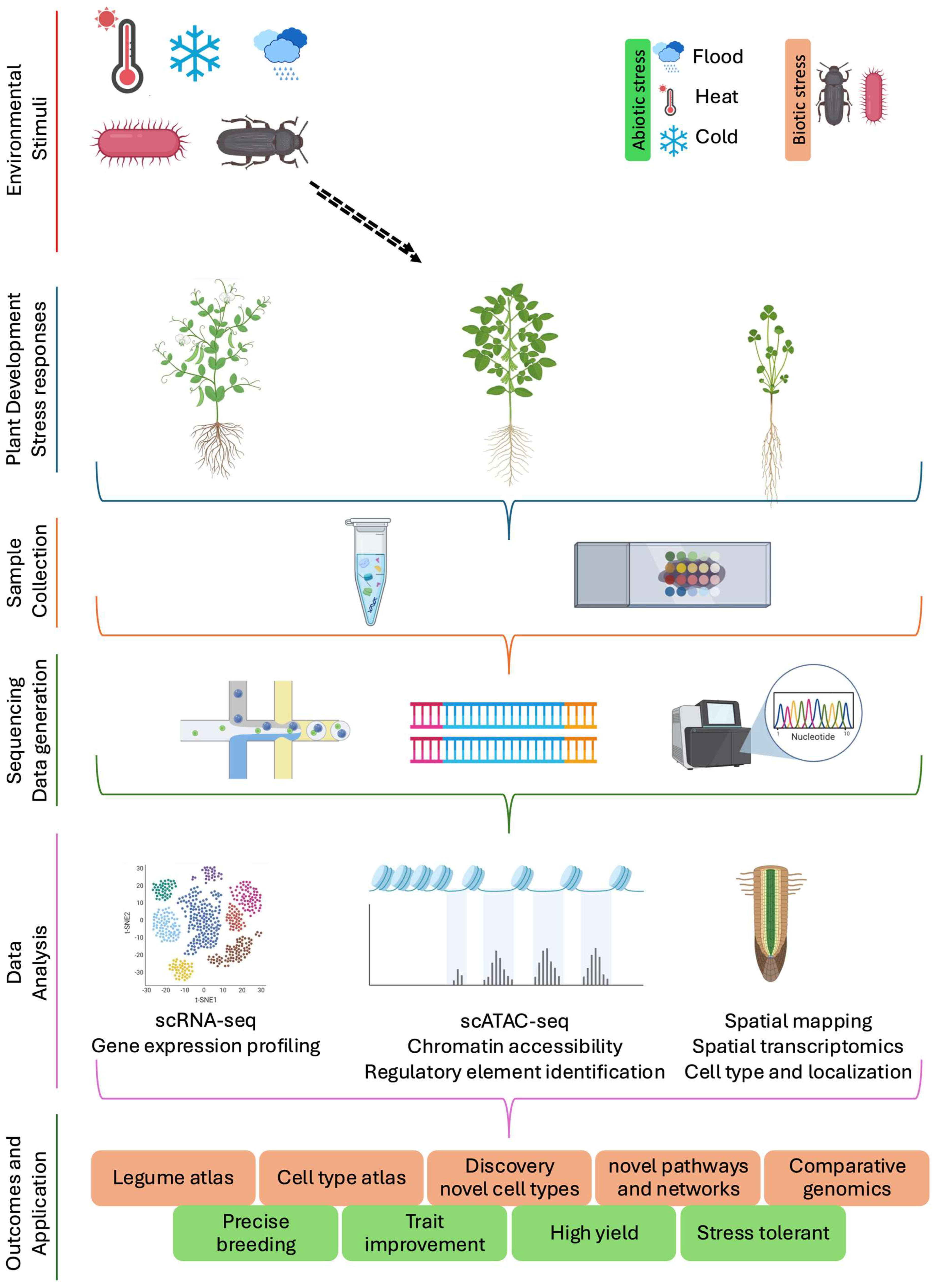
1. Introduction
2. From Bulk to Single-Cell Resolution: Building Cell-Level Insights
2.1. Findings Revealed by Bulk RNA-seq
2.2. Limitations of Bulk RNA-seq in Resolving Cellular Heterogeneity
2.3. Timeline of the Use of Single-Cell/Single-Nucleus RNA-seq in Legume Research
2.4. Implementation of Single-Cell Multi-omics Strategies to Predict Gene Regulatory Networks
2.5. Integration of Spatial and Single-Cell Data
3. Developing Single-Cell Legume Atlases to Capture Cellular Diversity and Plant Cell Environmental Responses
3.1. Developmental Atlases of Plant Organs: Single-Cell and Spatial Transcriptomes in Legumes
3.2. Stress and Immunity-Responsive Atlases
4. Symbiosis as a Model: Insights from Nodulation and Mycorrhizal Interactions
4.1. Root Nodule Symbiosis: As a Model for Organogenesis and Signaling
4.2. Root Nodule Development: Lineage Fate and Spatial Patterning
4.3. Early Signaling Genes Shared in Mycorrhizal and Rhizobia Symbiosis
5. Implementing Functional Genomic Solutions at Cellular Resolution
5.1. Deciphering Cell-Type-Specific Gene Expression Landscapes
5.2. Chromatin Accessibility and Protein–DNA Interaction Mapping
5.3. Regulatory Network Inference from Integrated Single-Cell Data
5.4. Perspectives and Future Directions
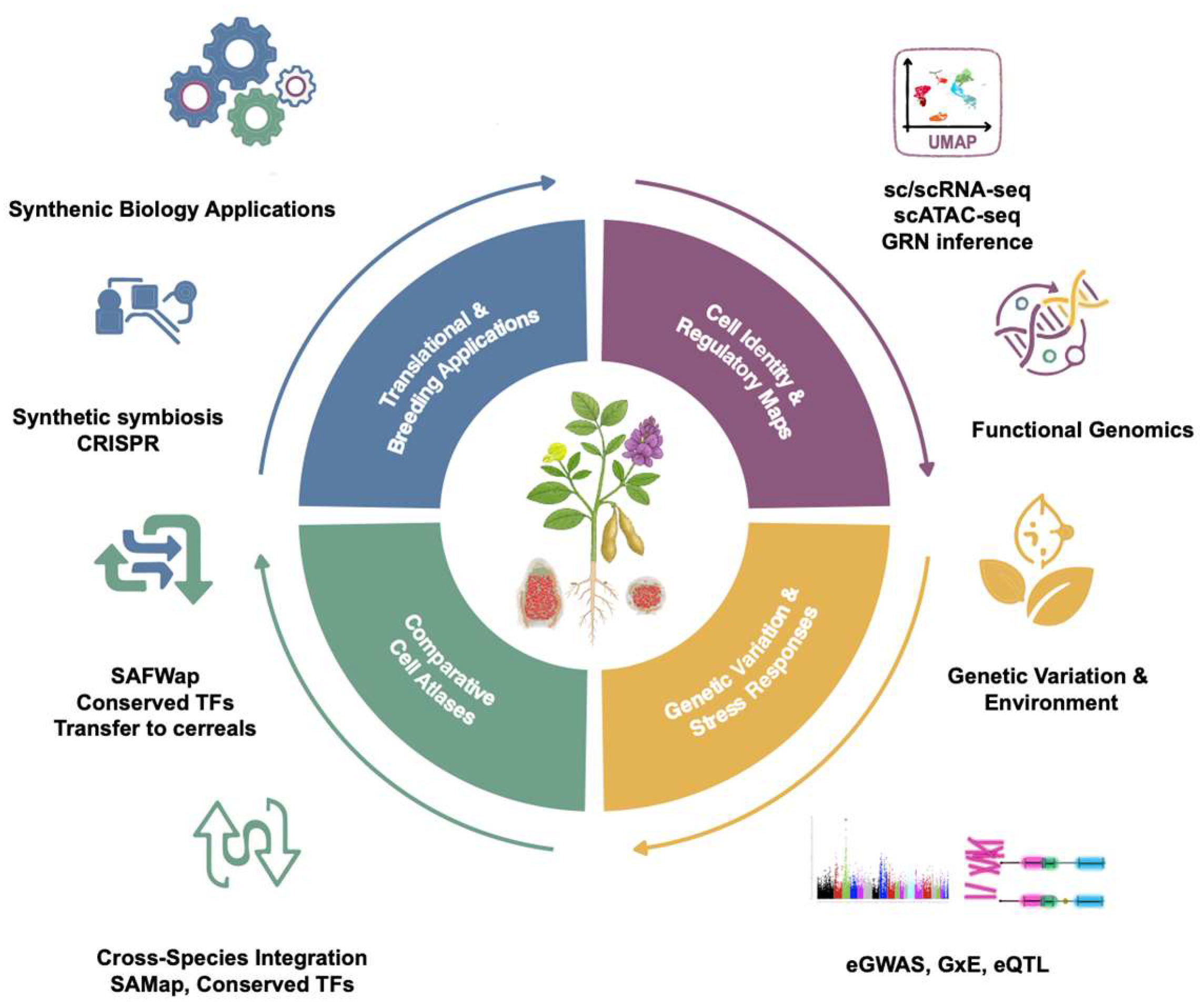
6. Integrating eGWAS and G×E with Cell Atlases: Toward Predictive Breeding
6.1. Cell-Type–Resolved eQTL and eGWAS Mapping
6.2. Single-Cell Resolution G×E Analysis
6.3. Future Directions and Public Platforms
7. Toward an Integrated Legume Single-Cell Biology: Cross-Species Integration and Synthetic Symbiosis
7.1. Standardizing Cell Ontologies and Annotation Systems
7.2. Cross-Species Atlas Integration and Conserved Regulatory Modules
7.3. Synthetic Symbiosis and Transferable Trait Modules
8. Challenges and Future Directions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABA | Abscisic acid |
| ACRs | Accessible chromatin regions |
| CSSP | Common Symbiotic Signaling Pathway |
| eGWAS | Expression genome-wide association studies |
| eQTL | Expression quantitative trait locus |
| FACS | Fluorescence-activated cell sorting |
| GRNs | Gene regulatory networks |
| GSTUs | Tau-class glutathione S-transferase |
| LCM | Laser capture microdissection |
| LCO | Lipo-chitooligosaccharides |
| MIRACL | Minimum information required to annotate a cell type |
| NF | Nod factor |
| NLR | Nucleotide-binding leucine-rich repeat |
| OMGs | Orthologous marker gene groups |
| PR | Pathogenesis-related |
| PRIMER | Pathogen-Responsive Immune Relay |
| PTI | Pattern-triggered immunity |
| QC | Quiescent center |
| RNS | Root nodule symbiosis |
| RNA-seq | RNA sequencing |
| ROS | Reactive oxygen species |
| SA | Salicylic acid |
| SAMap | Single-cell alignment mapping |
| SATURN | Species-agnostic transformers for universal cell embeddings and networks |
| sc/snRNA-seq | Single-cell/single-nucleus RNA sequencing |
| SMV | Soybean Mosaic Virus |
| snATAC-seq | Single-nucleus Assay for Transposase-Accessible Chromatin using sequencing |
| TFs | transcription factors |
References
- Oldroyd, G.E.D.; Leyser, O. A Plant’s Diet, Surviving in a Variable Nutrient Environment. Science 2020, 368, eaba0196. [Google Scholar] [CrossRef]
- Cervantes-Pérez, S.A.; Zogli, P.; Amini, S.; Thibivilliers, S.; Tennant, S.; Hossain, M.S.; Xu, H.; Meyer, I.; Nooka, A.; Ma, P.; et al. Single-Cell Transcriptome Atlases of Soybean Root and Mature Nodule Reveal New Regulatory Programs That Control the Nodulation Process. Plant Commun. 2024, 5, 100984. [Google Scholar] [CrossRef]
- Ye, K.; Bu, F.; Zhong, L.; Dong, Z.; Ma, Z.; Tang, Z.; Zhang, Y.; Yang, X.; Xu, X.; Wang, E.; et al. Mapping the Molecular Landscape of Lotus japonicus Nodule Organogenesis through Spatiotemporal Transcriptomics. Nat. Commun. 2024, 15, 6387. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Luo, Z.; Marand, A.P.; Yan, H.; Jang, H.; Bang, S.; Mendieta, J.P.; Minow, M.A.A.; Schmitz, R.J. A Spatially Resolved Multi-Omic Single-Cell Atlas of Soybean Development. Cell 2025, 188, 550–567.e19. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Shen, Y.; Chen, C.; Chen, X.; Yang, X.; Liu, H.; Chen, R.; Liu, S.; Zhang, B.; Zhang, M.; et al. A Large-Scale Integrated Transcriptomic Atlas for Soybean Organ Development. Mol. Plant 2025, 18, 669–689. [Google Scholar] [CrossRef]
- Gonzalez-Rizzo, S.; Crespi, M.; Frugier, F. The Medicago truncatula CRE1 Cytokinin Receptor Regulates Lateral Root Development and Early Symbiotic Interaction with Sinorhizobium meliloti. Plant Cell 2006, 18, 2680–2693. [Google Scholar] [CrossRef]
- Lin, J.; Li, X.; Luo, Z.; Mysore, K.S.; Wen, J.; Xie, F. NIN Interacts with NLPs to Mediate Nitrate Inhibition of Nodulation in Medicago truncatula. Nat. Plants 2018, 4, 942–952. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Jiang, S.; Wang, D.; Li, L.; Liu, B.; Ran, Q.; Hu, L.; Xiong, J.; Tang, Y.; Gu, X.; et al. Single-Cell RNA-Seq of Lotus japonicus Provide Insights into Identification and Function of Root Cell Types of Legume. J. Integr. Plant Biol. 2023, 65, 1147–1152. [Google Scholar] [CrossRef]
- Cervantes-Pérez, S.A.; Thibivilliers, S.; Amini, S.; Pelletier, J.M.; Meyer, I.; Xu, H.; Tennant, S.; Ma, P.; Sprueill, C.M.; Farmer, A.D.; et al. Tabula Glycine: The Whole-Soybean Single-Cell Resolution Transcriptome Atlas. bioRxiv 2024, 2024.07.08.602332. [Google Scholar] [CrossRef]
- Song, S.; Wang, J.; Zhou, J.; Cheng, X.; Hu, Y.; Wang, J.; Zou, J.; Zhao, Y.; Liu, C.; Hu, Z.; et al. Single-Cell RNA-Sequencing of Soybean Reveals Transcriptional Changes and Antiviral Functions of GmGSTU23 and GmGSTU24 in Response to Soybean Mosaic Virus. Plant Cell Environ. 2024. online ahead of print. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, Y.; Li, R.; Liang, J.; Tian, T.; Ji, J.; Chen, R.; Zhou, Y.; Fan, Q.; Ning, G.; et al. Single Cell-Type Transcriptome Profiling Reveals Genes That Promote Nitrogen Fixation in the Infected and Uninfected Cells of Legume Nodules. Plant Biotechnol. J. 2022, 20, 616–618. [Google Scholar] [CrossRef]
- Pereira, W.J.; Boyd, J.; Conde, D.; Triozzi, P.M.; Balmant, K.M.; Dervinis, C.; Schmidt, H.W.; Boaventura-Novaes, C.; Chakraborty, S.; Knaack, S.A.; et al. The Single-Cell Transcriptome Program of Nodule Development Cellular Lineages in Medicago truncatula. Cell Rep. 2024, 43, 113747. [Google Scholar] [CrossRef]
- Liu, Z.; Kong, X.; Long, Y.; Liu, S.; Zhang, H.; Jia, J.; Cui, W.; Zhang, Z.; Song, X.; Qiu, L.; et al. Integrated Single-Nucleus and Spatial Transcriptomics Captures Transitional States in Soybean Nodule Maturation. Nat. Plants 2023, 9, 515–524. [Google Scholar] [CrossRef]
- Frank, M.; Fechete, L.I.; Tedeschi, F.; Nadzieja, M.; Nørgaard, M.M.M.; Montiel, J.; Andersen, K.R.; Schierup, M.H.; Reid, D.; Andersen, S.U. Single-Cell Analysis Identifies Genes Facilitating Rhizobium Infection in Lotus japonicus. Nat. Commun. 2023, 14, 7171. [Google Scholar] [CrossRef]
- Guo, B.; Sun, L.; Jiang, S.; Ren, H.; Sun, R.; Wei, Z.; Hong, H.; Luan, X.; Wang, J.; Wang, X.; et al. Soybean Genetic Resources Contributing to Sustainable Protein Production. Theor. Appl. Genet. 2022, 135, 4095–4121. [Google Scholar] [CrossRef] [PubMed]
- Murphy, D.J. Agronomy and Environmental Sustainability of the Four Major Global Vegetable Oil Crops: Oil Palm, Soybean, Rapeseed, and Sunflower. Agronomy 2025, 15, 1465. [Google Scholar] [CrossRef]
- White, J.A.; Todd, J.; Newman, T.; Focks, N.; Girke, T.; de Ilárduya, O.M.; Jaworski, J.G.; Ohlrogge, J.B.; Benning, C. A New Set of Arabidopsis Expressed Sequence Tags from Developing Seeds. The Metabolic Pathway from Carbohydrates to Seed Oil. Plant Physiol. 2000, 124, 1582–1594. [Google Scholar] [CrossRef]
- Benedito, V.A.; Torres-Jerez, I.; Murray, J.D.; Andriankaja, A.; Allen, S.; Kakar, K.; Wandrey, M.; Verdier, J.; Zuber, H.; Ott, T.; et al. A Gene Expression Atlas of the Model Legume Medicago truncatula. Plant J. 2008, 55, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Lister, R.; O’Malley, R.C.; Tonti-Filippini, J.; Gregory, B.D.; Berry, C.C.; Millar, A.H.; Ecker, J.R. Highly Integrated Single-Base Resolution Maps of the Epigenome in Arabidopsis. Cell 2008, 133, 523–536. [Google Scholar] [CrossRef]
- Libault, M.; Farmer, A.; Joshi, T.; Takahashi, K.; Langley, R.J.; Franklin, L.D.; He, J.; Xu, D.; May, G.; Stacey, G. An Integrated Transcriptome Atlas of the Crop Model Glycine max, and Its Use in Comparative Analyses in Plants. Plant J. 2010, 63, 86–99. [Google Scholar] [CrossRef]
- Severin, A.J.; Woody, J.L.; Bolon, Y.-T.; Joseph, B.; Diers, B.W.; Farmer, A.D.; Muehlbauer, G.J.; Nelson, R.T.; Grant, D.; Specht, J.E.; et al. RNA-Seq Atlas of Glycine max: A Guide to the Soybean Transcriptome. BMC Plant Biol. 2010, 10, 160. [Google Scholar] [CrossRef]
- O’Rourke, J.A.; Iniguez, L.P.; Fu, F.; Bucciarelli, B.; Miller, S.S.; Jackson, S.A.; McClean, P.E.; Li, J.; Dai, X.; Zhao, P.X.; et al. An RNA-Seq Based Gene Expression Atlas of the Common Bean. BMC Genom. 2014, 15, 866. [Google Scholar] [CrossRef]
- Afzal, M.; Alghamdi, S.S.; Migdadi, H.H.; Khan, M.A.; Nurmansyah; Mirza, S.B.; El-Harty, E. Legume Genomics and Transcriptomics: From Classic Breeding to Modern Technologies. Saudi J. Biol. Sci. 2020, 27, 543–555. [Google Scholar] [CrossRef]
- Libault, M.; Farmer, A.; Brechenmacher, L.; Drnevich, J.; Langley, R.J.; Bilgin, D.D.; Radwan, O.; Neece, D.J.; Clough, S.J.; May, G.D.; et al. Complete Transcriptome of the Soybean Root Hair Cell, a Single-Cell Model, and Its Alteration in Response to Bradyrhizobium japonicum Infection. Plant Physiol. 2010, 152, 541–552. [Google Scholar] [CrossRef]
- Belamkar, V.; Weeks, N.T.; Bharti, A.K.; Farmer, A.D.; Graham, M.A.; Cannon, S.B. Comprehensive Characterization and RNA-Seq Profiling of the HD-Zip Transcription Factor Family in Soybean (Glycine max) during Dehydration and Salt Stress. BMC Genom. 2014, 15, 950. [Google Scholar] [CrossRef]
- Chen, W.; Yao, Q.; Patil, G.B.; Agarwal, G.; Deshmukh, R.K.; Lin, L.; Wang, B.; Wang, Y.; Prince, S.J.; Song, L.; et al. Identification and Comparative Analysis of Differential Gene Expression in Soybean Leaf Tissue under Drought and Flooding Stress Revealed by RNA-Seq. Front. Plant Sci. 2016, 7, 1044. [Google Scholar] [CrossRef]
- Du, J.; Wang, S.; He, C.; Zhou, B.; Ruan, Y.-L.; Shou, H. Identification of Regulatory Networks and Hub Genes Controlling Soybean Seed Set and Size Using RNA Sequencing Analysis. J. Exp. Bot. 2017, 68, 1955–1972. [Google Scholar] [CrossRef] [PubMed]
- Goettel, W.; Xia, E.; Upchurch, R.; Wang, M.-L.; Chen, P.; An, Y.-Q.C. Identification and Characterization of Transcript Polymorphisms in Soybean Lines Varying in Oil Composition and Content. BMC Genom. 2014, 15, 299. [Google Scholar] [CrossRef]
- Liu, A.; Xiao, Z.; Li, M.-W.; Wong, F.-L.; Yung, W.-S.; Ku, Y.-S.; Wang, Q.; Wang, X.; Xie, M.; Yim, A.K.-Y.; et al. Transcriptomic Reprogramming in Soybean Seedlings under Salt Stress. Plant Cell Environ. 2019, 42, 98–114. [Google Scholar] [CrossRef] [PubMed]
- Machado, F.B.; Moharana, K.C.; Almeida-Silva, F.; Gazara, R.K.; Pedrosa-Silva, F.; Coelho, F.S.; Grativol, C.; Venancio, T.M. Systematic Analysis of 1298 RNA-Seq Samples and Construction of a Comprehensive Soybean (Glycine max) Expression Atlas. Plant J. 2020, 103, 1894–1909. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Zhou, Z.; Wang, Z.; Li, W.; Fang, C.; Wu, M.; Ma, Y.; Liu, T.; Kong, L.-A.; Peng, D.-L.; et al. Global Dissection of Alternative Splicing in Paleopolyploid Soybean. Plant Cell 2014, 26, 996–1008. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, L.; Ma, Y.; Li, S.; Dong, S.; Zu, W. Transcriptome Profilling Analysis Characterized the Gene Expression Patterns Responded to Combined Drought and Heat Stresses in Soybean. Comput. Biol. Chem. 2018, 77, 413–429. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Xia, C.; Xia, Z.; Zhou, X.; Huang, J.; Huang, Z.; Liu, Y.; Jiang, Y.; Casteel, S.; Zhang, C. Physiological and Transcriptomic Responses of Reproductive Stage Soybean to Drought Stress. Plant Cell Rep. 2018, 37, 1611–1624. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.A.; Sharma, N.; Siddique, K.H.M.; Colmer, T.D.; Sutton, T.; Baumann, U. Comparative Transcriptome Analysis Reveals Molecular Regulation of Salt Tolerance in Two Contrasting Chickpea Genotypes. Front. Plant Sci. 2023, 14, 1191457. [Google Scholar] [CrossRef]
- Kudapa, H.; Barmukh, R.; Garg, V.; Chitikineni, A.; Samineni, S.; Agarwal, G.; Varshney, R.K. Comprehensive Transcriptome Profiling Uncovers Molecular Mechanisms and Potential Candidate Genes Associated with Heat Stress Response in Chickpea. Int. J. Mol. Sci. 2023, 24, 1369. [Google Scholar] [CrossRef]
- Malovichko, Y.V.; Shtark, O.Y.; Vasileva, E.N.; Nizhnikov, A.A.; Antonets, K.S. Transcriptomic Insights into Mechanisms of Early Seed Maturation in the Garden Pea (Pisum sativum L.). Cells 2020, 9, 779. [Google Scholar] [CrossRef]
- Alves-Carvalho, S.; Aubert, G.; Carrère, S.; Cruaud, C.; Brochot, A.-L.; Jacquin, F.; Klein, A.; Martin, C.; Boucherot, K.; Kreplak, J.; et al. Full-Length de Novo Assembly of RNA-Seq Data in Pea (Pisum sativum L.) Provides a Gene Expression Atlas and Gives Insights into Root Nodulation in This Species. Plant J. 2015, 84, 1–19. [Google Scholar] [CrossRef]
- Bhogireddy, S.; Xavier, A.; Garg, V.; Layland, N.; Arias, R.; Payton, P.; Nayak, S.N.; Pandey, M.K.; Puppala, N.; Varshney, R.K. Genome-Wide Transcriptome and Physiological Analyses Provide New Insights into Peanut Drought Response Mechanisms. Sci. Rep. 2020, 10, 4071. [Google Scholar] [CrossRef]
- Wan, L.; Li, B.; Lei, Y.; Yan, L.; Ren, X.; Chen, Y.; Dai, X.; Jiang, H.; Zhang, J.; Guo, W.; et al. Mutant Transcriptome Sequencing Provides Insights into Pod Development in Peanut (Arachis hypogaea L.). Front. Plant Sci. 2017, 8, 1900. [Google Scholar] [CrossRef]
- Rathod, V.; Hamid, R.; Tomar, R.S.; Patel, R.; Padhiyar, S.; Kheni, J.; Thirumalaisamy, P.P.; Munshi, N.S. Comparative RNA-Seq Profiling of a Resistant and Susceptible Peanut (Arachis hypogaea) Genotypes in Response to Leaf Rust Infection Caused by Puccinia Arachidis. 3 Biotech. 2020, 10, 284. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, M.L.; Dang, P.; Jiang, T.; Zhao, S.; Lamb, M.; Chen, C. Identification of Potential QTLs and Genes Associated with Seed Composition Traits in Peanut (Arachis hypogaea L.) Using GWAS and RNA-Seq Analysis. Gene 2021, 769, 145215. [Google Scholar] [CrossRef]
- Wang, L.; Rubio, M.C.; Xin, X.; Zhang, B.; Fan, Q.; Wang, Q.; Ning, G.; Becana, M.; Duanmu, D. CRISPR/Cas9 Knockout of Leghemoglobin Genes in Lotus japonicus Uncovers Their Synergistic Roles in Symbiotic Nitrogen Fixation. New Phytol. 2019, 224, 818–832. [Google Scholar] [CrossRef]
- Handa, Y.; Nishide, H.; Takeda, N.; Suzuki, Y.; Kawaguchi, M.; Saito, K. RNA-Seq Transcriptional Profiling of an Arbuscular Mycorrhiza Provides Insights into Regulated and Coordinated Gene Expression in Lotus japonicus and Rhizophagus irregularis. Plant Cell Physiol. 2015, 56, 1490–1511. [Google Scholar] [CrossRef]
- Luo, D.; Zhou, Q.; Wu, Y.; Chai, X.; Liu, W.; Wang, Y.; Yang, Q.; Wang, Z.; Liu, Z. Full-Length Transcript Sequencing and Comparative Transcriptomic Analysis to Evaluate the Contribution of Osmotic and Ionic Stress Components towards Salinity Tolerance in the Roots of Cultivated Alfalfa (Medicago sativa L.). BMC Plant Biol. 2019, 19, 32. [Google Scholar] [CrossRef]
- Zhao, M.; Wang, T.; Sun, T.; Yu, X.; Tian, R.; Zhang, W.-H. Identification of Tissue-Specific and Cold-Responsive LncRNAs in Medicago truncatula by High-Throughput RNA Sequencing. BMC Plant Biol. 2020, 20, 99. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Yang, X.; Zhang, F.; Yang, T.; Yang, C.; He, F.; Gao, T.; Wang, C.; Yang, Q.; Wang, Z.; et al. Combining QTL Mapping and RNA-Seq Unravels Candidate Genes for Alfalfa (Medicago sativa L.) Leaf Development. BMC Plant Biol. 2022, 22, 485. [Google Scholar] [CrossRef]
- Brown, A.V.; Conners, S.I.; Huang, W.; Wilkey, A.P.; Grant, D.; Weeks, N.T.; Cannon, S.B.; Graham, M.A.; Nelson, R.T. A New Decade and New Data at SoyBase, the USDA-ARS Soybean Genetics and Genomics Database. Nucleic Acids Res. 2021, 49, D1496–D1501. [Google Scholar] [CrossRef] [PubMed]
- Berendzen, J.; Brown, A.V.; Cameron, C.T.; Campbell, J.D.; Cleary, A.M.; Dash, S.; Hokin, S.; Huang, W.; Kalberer, S.R.; Nelson, R.T.; et al. The Legume Information System and Associated Online Genomic Resources. Legume Sci. 2021, 3, e74. [Google Scholar] [CrossRef]
- Mun, T.; Bachmann, A.; Gupta, V.; Stougaard, J.; Andersen, S.U. Lotus Base: An Integrated Information Portal for the Model Legume Lotus japonicus. Sci. Rep. 2016, 6, 39447. [Google Scholar] [CrossRef]
- Carrere, S.; Verdier, J.; Gamas, P. MtExpress, a Comprehensive and Curated RNAseq-Based Gene Expression Atlas for the Model Legume Medicago truncatula. Plant Cell Physiol. 2021, 62, 1494–1500. [Google Scholar] [CrossRef] [PubMed]
- Dash, S.; Cannon, E.K.S.; Kalberer, S.R.; Farmer, A.D.; Cannon, S.B. Chapter 8—PeanutBase and Other Bioinformatic Resources for Peanut. In Peanuts; Stalker, H.T., Wilson, R.F., Eds.; AOCS Press: Urbana, IL, USA, 2016; pp. 241–252. ISBN 978-1-63067-038-2. [Google Scholar]
- Takanashi, K.; Takahashi, H.; Sakurai, N.; Sugiyama, A.; Suzuki, H.; Shibata, D.; Nakazono, M.; Yazaki, K. Tissue-Specific Transcriptome Analysis in Nodules of Lotus japonicus. Mol. Plant Microbe Interact. 2012, 25, 869–876. [Google Scholar] [CrossRef] [PubMed]
- Limpens, E.; Moling, S.; Hooiveld, G.; Pereira, P.A.; Bisseling, T.; Becker, J.D.; Küster, H. Cell- and Tissue-Specific Transcriptome Analyses of Medicago truncatula Root Nodules. PLoS ONE 2013, 8, e64377. [Google Scholar] [CrossRef] [PubMed]
- Roux, B.; Rodde, N.; Jardinaud, M.-F.; Timmers, T.; Sauviac, L.; Cottret, L.; Carrère, S.; Sallet, E.; Courcelle, E.; Moreau, S.; et al. An Integrated Analysis of Plant and Bacterial Gene Expression in Symbiotic Root Nodules Using Laser-Capture Microdissection Coupled to RNA Sequencing. Plant J. 2014, 77, 817–837. [Google Scholar] [CrossRef] [PubMed]
- Ryu, K.H.; Huang, L.; Kang, H.M.; Schiefelbein, J. Single-Cell RNA Sequencing Resolves Molecular Relationships Among Individual Plant Cells. Plant Physiol. 2019, 179, 1444–1456. [Google Scholar] [CrossRef] [PubMed]
- Shulse, C.N.; Cole, B.J.; Ciobanu, D.; Lin, J.; Yoshinaga, Y.; Gouran, M.; Turco, G.M.; Zhu, Y.; O’Malley, R.C.; Brady, S.M.; et al. High-Throughput Single-Cell Transcriptome Profiling of Plant Cell Types. Cell Rep. 2019, 27, 2241–2247.e4. [Google Scholar] [CrossRef]
- Liu, H.; Hu, D.; Du, P.; Wang, L.; Liang, X.; Li, H.; Lu, Q.; Li, S.; Liu, H.; Chen, X.; et al. Single-Cell RNA-Seq Describes the Transcriptome Landscape and Identifies Critical Transcription Factors in the Leaf Blade of the Allotetraploid Peanut (Arachis hypogaea L.). Plant Biotechnol. J. 2021, 19, 2261–2276. [Google Scholar] [CrossRef]
- Cervantes-Pérez, S.A.; Thibivilliers, S.; Laffont, C.; Farmer, A.D.; Frugier, F.; Libault, M. Cell-Specific Pathways Recruited for Symbiotic Nodulation in the Medicago truncatula Legume. Mol. Plant 2022, 15, 1868–1888. [Google Scholar] [CrossRef]
- Ye, Q.; Zhu, F.; Sun, F.; Wang, T.-C.; Wu, J.; Liu, P.; Shen, C.; Dong, J.; Wang, T. Differentiation Trajectories and Biofunctions of Symbiotic and Un-Symbiotic Fate Cells in Root Nodules of Medicago truncatula. Mol. Plant 2022, 15, 1852–1867. [Google Scholar] [CrossRef]
- Sun, B.; Wang, Y.; Yang, Q.; Gao, H.; Niu, H.; Li, Y.; Ma, Q.; Huan, Q.; Qian, W.; Ren, B. A High-Resolution Transcriptomic Atlas Depicting Nitrogen Fixation and Nodule Development in Soybean. J. Integr. Plant Biol. 2023, 65, 1536–1552. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, J.; Long, Y.; Zhang, C.; Wang, D.; Zhang, X.; Dong, W.; Zhao, L.; Liu, C.; Zhai, J.; et al. Single-Nucleus Transcriptomes Reveal Spatiotemporal Symbiotic Perception and Early Response in Medicago. Nat. Plants 2023, 9, 1734–1748. [Google Scholar] [CrossRef]
- Cui, Y.; Su, Y.; Bian, J.; Han, X.; Guo, H.; Yang, Z.; Chen, Y.; Li, L.; Li, T.; Deng, X.W.; et al. Single-Nucleus RNA and ATAC Sequencing Analyses Provide Molecular Insights into Early Pod Development of Peanut Fruit. Plant Commun. 2024, 5, 100979. [Google Scholar] [CrossRef]
- Deng, Q.; Du, P.; Gangurde, S.S.; Hong, Y.; Xiao, Y.; Hu, D.; Li, H.; Lu, Q.; Li, S.; Liu, H.; et al. ScRNA-Seq Reveals Dark- and Light-Induced Differentially Expressed Gene Atlases of Seedling Leaves in Arachis hypogaea L. Plant Biotechnol. J. 2024, 22, 1848–1866. [Google Scholar] [CrossRef]
- Liu, H.; Guo, Z.; Gangurde, S.S.; Garg, V.; Deng, Q.; Du, P.; Lu, Q.; Chitikineni, A.; Xiao, Y.; Wang, W.; et al. A Single-Nucleus Resolution Atlas of Transcriptome and Chromatin Accessibility for Peanut (Arachis hypogaea L.) Leaves. Adv. Biol. 2024, 8, 2300410. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, R.; Huo, X.; Zhou, Y.; Umer, M.J.; Zheng, Z.; Jin, W.; Huang, L.; Li, H.; Yu, Q.; et al. Integration of Single-Nuclei Transcriptome and Bulk RNA-Seq to Unravel the Role of AhWRKY70 in Regulating Stem Cell Development in Arachis hypogaea L. Plant Biotechnol. J. 2025, 23, 1814–1831. [Google Scholar] [CrossRef]
- Lepetit, M.; Brouquisse, R. Control of the Rhizobium–Legume Symbiosis by the Plant Nitrogen Demand Is Tightly Integrated at the Whole Plant Level and Requires Inter-Organ Systemic Signaling. Front. Plant Sci. 2023, 14, 1114840. [Google Scholar] [CrossRef]
- Amini, S.; Doyle, J.J.; Libault, M. The Evolving Definition of Plant Cell Type. Front. Plant Sci. 2023, 14, 1271070. [Google Scholar] [CrossRef] [PubMed]
- Dorrity, M.W.; Alexandre, C.M.; Hamm, M.O.; Vigil, A.-L.; Fields, S.; Queitsch, C.; Cuperus, J.T. The Regulatory Landscape of Arabidopsis thaliana Roots at Single-Cell Resolution. Nat. Commun. 2021, 12, 3334. [Google Scholar] [CrossRef]
- Marand, A.P.; Chen, Z.; Gallavotti, A.; Schmitz, R.J. A Cis-Regulatory Atlas in Maize at Single-Cell Resolution. Cell 2021, 184, 3041–3055.e21. [Google Scholar] [CrossRef]
- Farmer, A.; Thibivilliers, S.; Ryu, K.H.; Schiefelbein, J.; Libault, M. Single-Nucleus RNA and ATAC Sequencing Reveals the Impact of Chromatin Accessibility on Gene Expression in Arabidopsis Roots at the Single-Cell Level. Mol. Plant 2021, 14, 372–383. [Google Scholar] [CrossRef]
- Yin, R.; Xia, K.; Xu, X. Spatial Transcriptomics Drives a New Era in Plant Research. Plant J. 2023, 116, 1571–1581. [Google Scholar] [CrossRef]
- Aida, M.; Beis, D.; Heidstra, R.; Willemsen, V.; Blilou, I.; Galinha, C.; Nussaume, L.; Noh, Y.-S.; Amasino, R.; Scheres, B. The PLETHORA Genes Mediate Patterning of the Arabidopsis Root Stem Cell Niche. Cell 2004, 119, 109–120. [Google Scholar] [CrossRef]
- Cao, Y.; Ma, J.; Han, S.; Hou, M.; Wei, X.; Zhang, X.; Zhang, Z.J.; Sun, S.; Ku, L.; Tang, J.; et al. Single-Cell RNA Sequencing Profiles Reveal Cell Type-Specific Transcriptional Regulation Networks Conditioning Fungal Invasion in Maize Roots. Plant Biotechnol. J. 2023, 21, 1839–1859. [Google Scholar] [CrossRef]
- Delannoy, E.; Batardiere, B.; Pateyron, S.; Soubigou-Taconnat, L.; Chiquet, J.; Colcombet, J.; Lang, J. Cell Specialization and Coordination in Arabidopsis Leaves upon Pathogenic Attack Revealed by ScRNA-Seq. Plant Commun. 2023, 4, 100676. [Google Scholar] [CrossRef]
- Li, R.; Wang, Z.; Wang, J.-W.; Li, L. Combining Single-Cell RNA Sequencing with Spatial Transcriptome Analysis Reveals Dynamic Molecular Maps of Cambium Differentiation in the Primary and Secondary Growth of Trees. Plant Commun. 2023, 4, 100665. [Google Scholar] [CrossRef]
- Nobori, T.; Oliva, M.; Lister, R.; Ecker, J.R. Multiplexed Single-Cell 3D Spatial Gene Expression Analysis in Plant Tissue Using PHYTOMap. Nat. Plants 2023, 9, 1026–1033. [Google Scholar] [CrossRef]
- DeFalco, T.A.; Zipfel, C. Molecular Mechanisms of Early Plant Pattern-Triggered Immune Signaling. Mol. Cell 2021, 81, 3449–3467. [Google Scholar] [CrossRef]
- Tang, B.; Feng, L.; Hulin, M.T.; Ding, P.; Ma, W. Cell-Type-Specific Responses to Fungal Infection in Plants Revealed by Single-Cell Transcriptomics. Cell Host Microbe 2023, 31, 1732–1747.e5. [Google Scholar] [CrossRef] [PubMed]
- Abdelaal, T.; Mourragui, S.; Mahfouz, A.; Reinders, M.J.T. SpaGE: Spatial Gene Enhancement Using ScRNA-Seq. Nucleic Acids Res. 2020, 48, e107. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Han, Y.; Sheng, K.; Yang, P.; Cao, Y.; Li, H.; Zhu, Q.-H.; Chen, J.; Zhu, S.; Zhao, T. Single-Cell Transcriptomic Analysis Reveals the Developmental Trajectory and Transcriptional Regulatory Networks of Pigment Glands in Gossypium bickii. Mol. Plant 2023, 16, 694–708. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Kadam, U.; Park, B.; Amariillis, S.; Nguyen, K.-N.T.; Can, M.-H.T.; Lee, K.O.; Park, S.J.; Chung, W.S.; Hong, J.C. Recent Progress in Single-Cell Transcriptomic Studies in Plants. Plant Biotechnol. Rep. 2025, 19, 91–103. [Google Scholar] [CrossRef]
- Nobori, T.; Monell, A.; Lee, T.A.; Sakata, Y.; Shirahama, S.; Zhou, J.; Nery, J.R.; Mine, A.; Ecker, J.R. A Rare PRIMER Cell State in Plant Immunity. Nature 2025, 638, 197–205. [Google Scholar] [CrossRef]
- Yang, Q.; Li, Z.; Guan, K.; Wang, Z.; Tang, X.; Hong, Y.; Liu, Z.; Zhai, J.; Huang, A.; Long, Y.; et al. Comparative Single-Nucleus RNA-Seq Analysis Revealed Localized and Cell Type-Specific Pathways Governing Root-Microbiome Interactions. Nat. Commun. 2025, 16, 3169. [Google Scholar] [CrossRef]
- Murray, J.D. Invasion by Invitation: Rhizobial Infection in Legumes. Mol. Plant Microbe Interact. 2011, 24, 631–639. [Google Scholar] [CrossRef]
- Schiessl, K.; Lilley, J.L.S.; Lee, T.; Tamvakis, I.; Kohlen, W.; Bailey, P.C.; Thomas, A.; Luptak, J.; Ramakrishnan, K.; Carpenter, M.D.; et al. NODULE INCEPTION Recruits the Lateral Root Developmental Program for Symbiotic Nodule Organogenesis in Medicago truncatula. Curr. Biol. 2019, 29, 3657–3668.e5. [Google Scholar] [CrossRef]
- Oldroyd, G.E.D.; Downie, J.A. Coordinating Nodule Morphogenesis with Rhizobial Infection in Legumes. Annu. Rev. Plant Biol. 2008, 59, 519–546. [Google Scholar] [CrossRef]
- Roy, S.; Liu, W.; Nandety, R.S.; Crook, A.; Mysore, K.S.; Pislariu, C.I.; Frugoli, J.; Dickstein, R.; Udvardi, M.K. Celebrating 20 Years of Genetic Discoveries in Legume Nodulation and Symbiotic Nitrogen Fixation. Plant Cell 2020, 32, 15–41. [Google Scholar] [CrossRef] [PubMed]
- Serrano, K.; Bezrutczyk, M.; Goudeau, D.; Dao, T.; O’Malley, R.; Malmstrom, R.R.; Visel, A.; Scheller, H.V.; Cole, B. Spatial Co-Transcriptomics Reveals Discrete Stages of the Arbuscular Mycorrhizal Symbiosis. Nat. Plants 2024, 10, 673–688. [Google Scholar] [CrossRef]
- Díaz, V.; Villalobos, M.; Arriaza, K.; Flores, K.; Hernández-Saravia, L.P.; Velásquez, A. Decoding the Dialog Between Plants and Arbuscular Mycorrhizal Fungi: A Molecular Genetic Perspective. Genes 2025, 16, 143. [Google Scholar] [CrossRef] [PubMed]
- Oldroyd, G.E.D. Speak, Friend, and Enter: Signalling Systems That Promote Beneficial Symbiotic Associations in Plants. Nat. Rev. Microbiol. 2013, 11, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Jean-Baptiste, K.; McFaline-Figueroa, J.L.; Alexandre, C.M.; Dorrity, M.W.; Saunders, L.; Bubb, K.L.; Trapnell, C.; Fields, S.; Queitsch, C.; Cuperus, J.T. Dynamics of Gene Expression in Single Root Cells of Arabidopsis thaliana. Plant Cell 2019, 31, 993–1011. [Google Scholar] [CrossRef]
- Soyano, T.; Shimoda, Y.; Kawaguchi, M.; Hayashi, M. A Shared Gene Drives Lateral Root Development and Root Nodule Symbiosis Pathways in Lotus. Science 2019, 366, 1021–1023. [Google Scholar] [CrossRef]
- Wijsman, T.; Mohd-Radzman, N.A.; Liu, J.; Oldroyd, G.E.D.; Kohlen, W.; Kulikova, O.; Heidstra, R.; Scheres, B.; Franssen, H.J. The Medicago truncatula LncRNA ENOD40 Is a Mediator of MicroRNA169-Controlled NF-YA Activity in Nodule Initiation. Proc. Natl. Acad. Sci. USA 2025, 122, e2424231122. [Google Scholar] [CrossRef]
- Fu, M.; Sun, J.; Li, X.; Guan, Y.; Xie, F. Asymmetric Redundancy of Soybean Nodule Inception (NIN) Genes in Root Nodule Symbiosis. Plant Physiol. 2022, 188, 477–489. [Google Scholar] [CrossRef]
- Li, X.; Zhou, H.; Cheng, L.; Ma, N.; Cui, B.; Wang, W.; Zhong, Y.; Liao, H. Shoot-to-Root Translocated GmNN1/FT2a Triggers Nodulation and Regulates Soybean Nitrogen Nutrition. PLoS Biol. 2022, 20, e3001739. [Google Scholar] [CrossRef] [PubMed]
- Jiao, W.; Wang, M.; Guan, Y.; Guo, W.; Zhang, C.; Wei, Y.; Zhao, Z.; Ma, H.; Wang, L.; Jiang, X.; et al. Transcriptional Regulatory Network Reveals Key Transcription Factors for Regulating Agronomic Traits in Soybean. Genome Biol. 2024, 25, 313. [Google Scholar] [CrossRef]
- Caballero, L.; Pasternak, T.; Riyazuddin, R.; Pérez-Pérez, J.M. Connecting High-Resolution 3D Chromatin Maps with Cell Division and Cell Differentiation at the Root Apical Meristem. Plant Cell Rep. 2024, 43, 232. [Google Scholar] [CrossRef] [PubMed]
- Aibar, S.; González-Blas, C.B.; Moerman, T.; Huynh-Thu, V.A.; Imrichova, H.; Hulselmans, G.; Rambow, F.; Marine, J.-C.; Geurts, P.; Aerts, J.; et al. SCENIC: Single-Cell Regulatory Network Inference and Clustering. Nat. Methods 2017, 14, 1083–1086. [Google Scholar] [CrossRef] [PubMed]
- Pérez, N.M.; Ferrari, C.; Engelhorn, J.; Depuydt, T.; Nelissen, H.; Hartwig, T.; Vandepoele, K. MINI-AC: Inference of Plant Gene Regulatory Networks Using Bulk or Single-Cell Accessible Chromatin Profiles. Plant J. 2024, 117, 280–301. [Google Scholar] [CrossRef]
- Knaack, S.A.; Conde, D.; Chakraborty, S.; Balmant, K.M.; Irving, T.B.; Maia, L.G.S.; Triozzi, P.M.; Dervinis, C.; Pereira, W.J.; Maeda, J.; et al. Temporal Change in Chromatin Accessibility Predicts Regulators of Nodulation in Medicago truncatula. BMC Biol. 2022, 20, 252. [Google Scholar] [CrossRef]
- Gao, J.-P.; Su, Y.; Jiang, S.; Liang, W.; Lou, Z.; Frugier, F.; Xu, P.; Murray, J.D. Applying Conventional and Cell-Type-Specific CRISPR/Cas9 Genome Editing in Legume Plants. aBIOTECH 2025, 6, 346–360. [Google Scholar] [CrossRef]
- Kremling, K.A.G.; Chen, S.-Y.; Su, M.-H.; Lepak, N.K.; Romay, M.C.; Swarts, K.L.; Lu, F.; Lorant, A.; Bradbury, P.J.; Buckler, E.S. Dysregulation of Expression Correlates with Rare-Allele Burden and Fitness Loss in Maize. Nature 2018, 555, 520–523. [Google Scholar] [CrossRef]
- van der Wijst, M.G.P.; de Vries, D.H.; Groot, H.E.; Trynka, G.; Hon, C.C.; Bonder, M.J.; Stegle, O.; Nawijn, M.C.; Idaghdour, Y.; van der Harst, P.; et al. The Single-Cell EQTLGen Consortium. Elife 2020, 9, e52155. [Google Scholar] [CrossRef]
- Xu, C.; Song, L.-Y.; Zhou, Y.; Ma, D.-N.; Ding, Q.-S.; Guo, Z.-J.; Li, J.; Song, S.-W.; Zhang, L.-D.; Zheng, H.-L. Integration of EQTL and GWAS Analysis Uncovers a Genetic Regulation of Natural Ionomic Variation in Arabidopsis. Plant Cell Rep. 2023, 42, 1473–1485. [Google Scholar] [CrossRef]
- Sun, Y.; Miller, C.; Rajurkar, A.B.; Lynch, R.C.; Alyward, A.; Zhang, L.; Shaner, M.; Copeland, C.D.; Ye, H.; Nguyen, H.T.; et al. Genome-Wide Association Study Reveals Influence of Cell-Specific Gene Networks on Soybean Root System Architecture. bioRxiv 2024, 2024.02.27.581071. [Google Scholar] [CrossRef]
- Seck, W.; Torkamaneh, D.; Belzile, F. Comprehensive Genome-Wide Association Analysis Reveals the Genetic Basis of Root System Architecture in Soybean. Front. Plant Sci. 2020, 11, 590740. [Google Scholar] [CrossRef] [PubMed]
- Kiegle, E.; Moore, C.A.; Haseloff, J.; Tester, M.A.; Knight, M.R. Cell-Type-Specific Calcium Responses to Drought, Salt and Cold in the Arabidopsis Root. Plant J. 2000, 23, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Xu, Y.; Song, Y.; Fan, D.; Li, J.; Zhang, Z.; Wang, L.; He, J.; Chen, C.; Ma, C. Hierarchical Regulatory Networks Reveal Conserved Drivers of Plant Drought Response at the Cell-Type Level. Adv. Sci. 2025, 12, 2415106. [Google Scholar] [CrossRef]
- Rosen, Y.; Brbić, M.; Roohani, Y.; Swanson, K.; Li, Z.; Leskovec, J. Toward Universal Cell Embeddings: Integrating Single-Cell RNA-Seq Datasets across Species with SATURN. Nat. Methods 2024, 21, 1492–1500. [Google Scholar] [CrossRef]
- Chau, T.N.; Timilsena, P.R.; Bathala, S.P.; Kundu, S.; Bargmann, B.O.R.; Li, S. Orthologous Marker Groups Reveal Broad Cell Identity Conservation across Plant Single-Cell Transcriptomes. Nat. Commun. 2025, 16, 201. [Google Scholar] [CrossRef]
- Tarashansky, A.J.; Musser, J.M.; Khariton, M.; Li, P.; Arendt, D.; Quake, S.R.; Wang, B. Mapping Single-Cell Atlases throughout Metazoa Unravels Cell Type Evolution. Elife 2021, 10, e66747. [Google Scholar] [CrossRef]
- Smith, B.; Ashburner, M.; Rosse, C.; Bard, J.; Bug, W.; Ceusters, W.; Goldberg, L.J.; Eilbeck, K.; Ireland, A.; Mungall, C.J.; et al. The OBO Foundry: Coordinated Evolution of Ontologies to Support Biomedical Data Integration. Nat. Biotechnol. 2007, 25, 1251–1255. [Google Scholar] [CrossRef] [PubMed]
- Diehl, A.D.; Meehan, T.F.; Bradford, Y.M.; Brush, M.H.; Dahdul, W.M.; Dougall, D.S.; He, Y.; Osumi-Sutherland, D.; Ruttenberg, A.; Sarntivijai, S.; et al. The Cell Ontology 2016: Enhanced Content, Modularization, and Ontology Interoperability. J. Biomed. Semant. 2016, 7, 44. [Google Scholar] [CrossRef] [PubMed]
- Lubiana, T.; Roncaglia, P.; Mungall, C.J.; Quardokus, E.M.; Fortriede, J.D.; Osumi-Sutherland, D.; Diehl, A.D. Guidelines for Reporting Cell Types: The MIRACL Standard. arXiv 2022, arXiv:2204.09673. [Google Scholar] [CrossRef]
- Bu, F.; Rutten, L.; Roswanjaya, Y.P.; Kulikova, O.; Rodriguez-Franco, M.; Ott, T.; Bisseling, T.; van Zeijl, A.; Geurts, R. Mutant Analysis in the Nonlegume Parasponia andersonii Identifies NIN and NF-YA1 Transcription Factors as a Core Genetic Network in Nitrogen-Fixing Nodule Symbioses. New Phytol. 2020, 226, 541–554. [Google Scholar] [CrossRef]
- Nishida, H.; Tanaka, S.; Handa, Y.; Ito, M.; Sakamoto, Y.; Matsunaga, S.; Betsuyaku, S.; Miura, K.; Soyano, T.; Kawaguchi, M.; et al. A NIN-LIKE PROTEIN Mediates Nitrate-Induced Control of Root Nodule Symbiosis in Lotus japonicus. Nat. Commun. 2018, 9, 499. [Google Scholar] [CrossRef]
- Misawa, F.; Ito, M.; Nosaki, S.; Nishida, H.; Watanabe, M.; Suzuki, T.; Miura, K.; Kawaguchi, M.; Suzaki, T. Nitrate Transport via NRT2.1 Mediates NIN-LIKE PROTEIN-Dependent Suppression of Root Nodulation in Lotus japonicus. Plant Cell 2022, 34, 1844–1862. [Google Scholar] [CrossRef]
- Boo, A.; Toth, T.; Yu, Q.; Pfotenhauer, A.; Fields, B.D.; Lenaghan, S.C.; Stewart, C.N.; Voigt, C.A. Synthetic Microbe-to-Plant Communication Channels. Nat. Commun. 2024, 15, 1817. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Hossain, M.S.; Libault, M. Single-Cell Omics in Legumes: Research Trends and Applications. Plants 2025, 14, 3615. https://doi.org/10.3390/plants14233615
Li Y, Hossain MS, Libault M. Single-Cell Omics in Legumes: Research Trends and Applications. Plants. 2025; 14(23):3615. https://doi.org/10.3390/plants14233615
Chicago/Turabian StyleLi, Yaohua, Md Sabbir Hossain, and Marc Libault. 2025. "Single-Cell Omics in Legumes: Research Trends and Applications" Plants 14, no. 23: 3615. https://doi.org/10.3390/plants14233615
APA StyleLi, Y., Hossain, M. S., & Libault, M. (2025). Single-Cell Omics in Legumes: Research Trends and Applications. Plants, 14(23), 3615. https://doi.org/10.3390/plants14233615





