Overexpression of Hevea brasiliensis HbCDS2 Gene Enhances Cold Tolerance in Transgenic Arabidopsis
Abstract
1. Introduction
2. Results
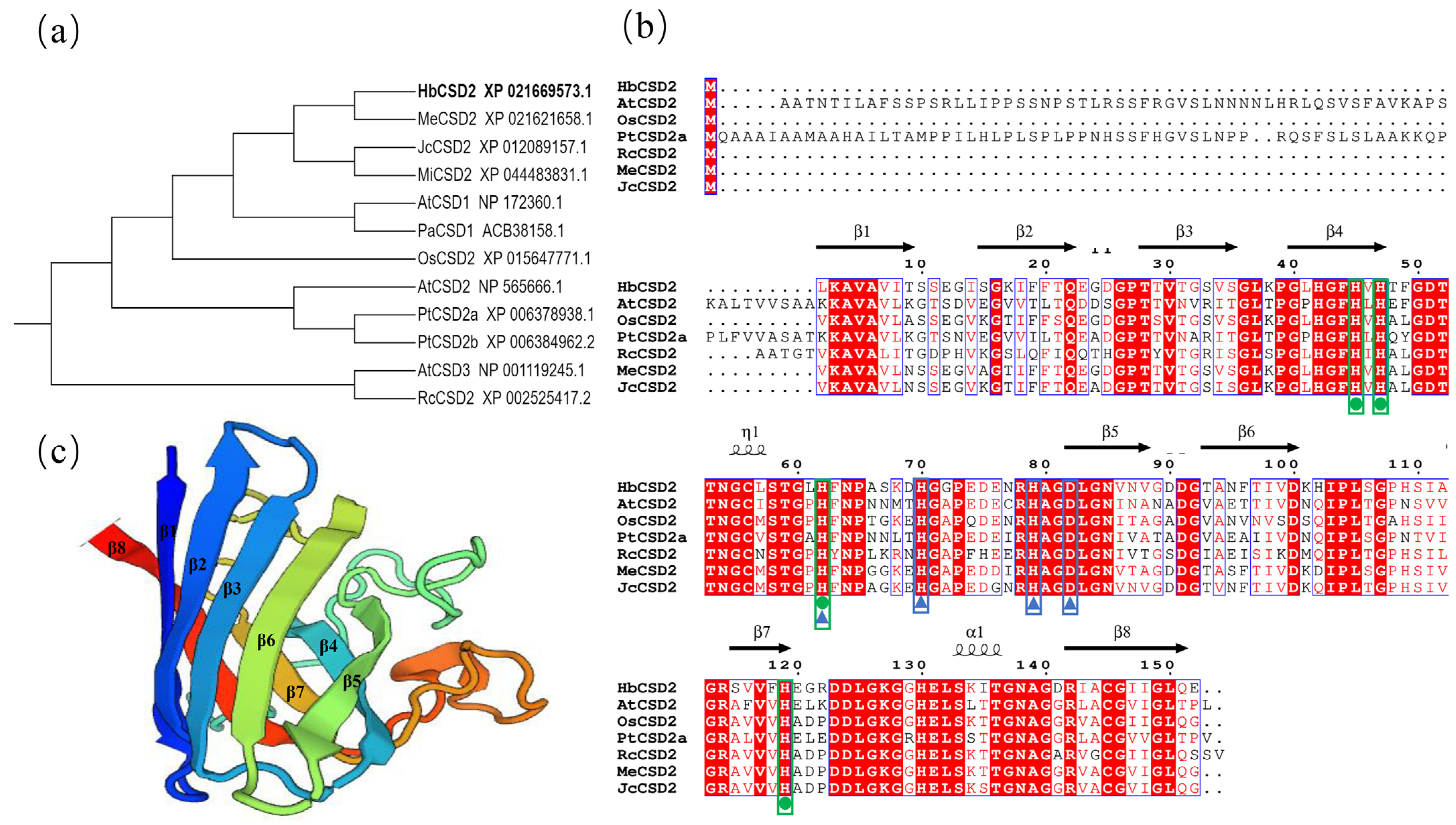
2.1. Gene Cloning and Sequence Analysis of HbCSD2
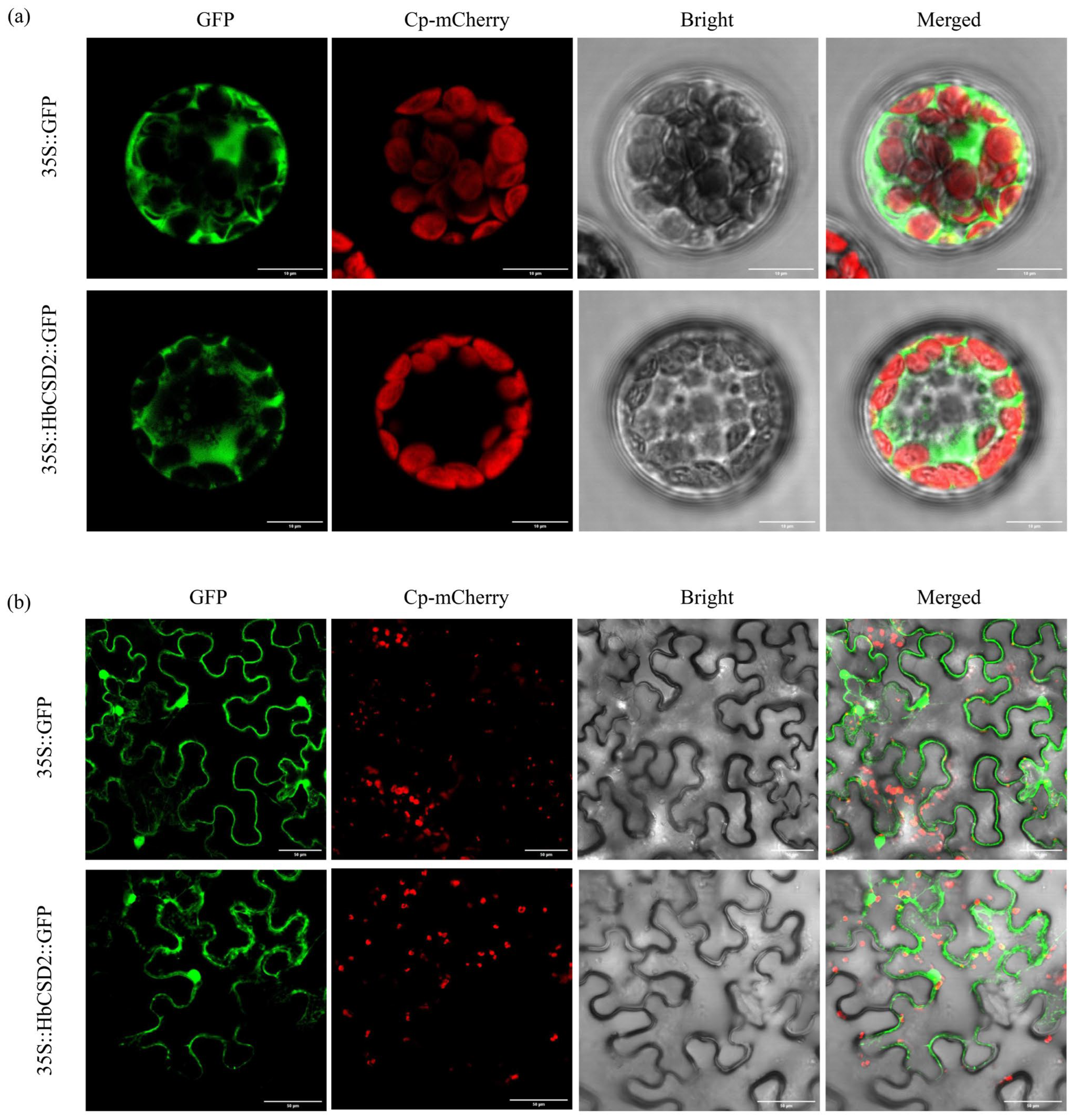
2.2. Subcellular Localization Analysis
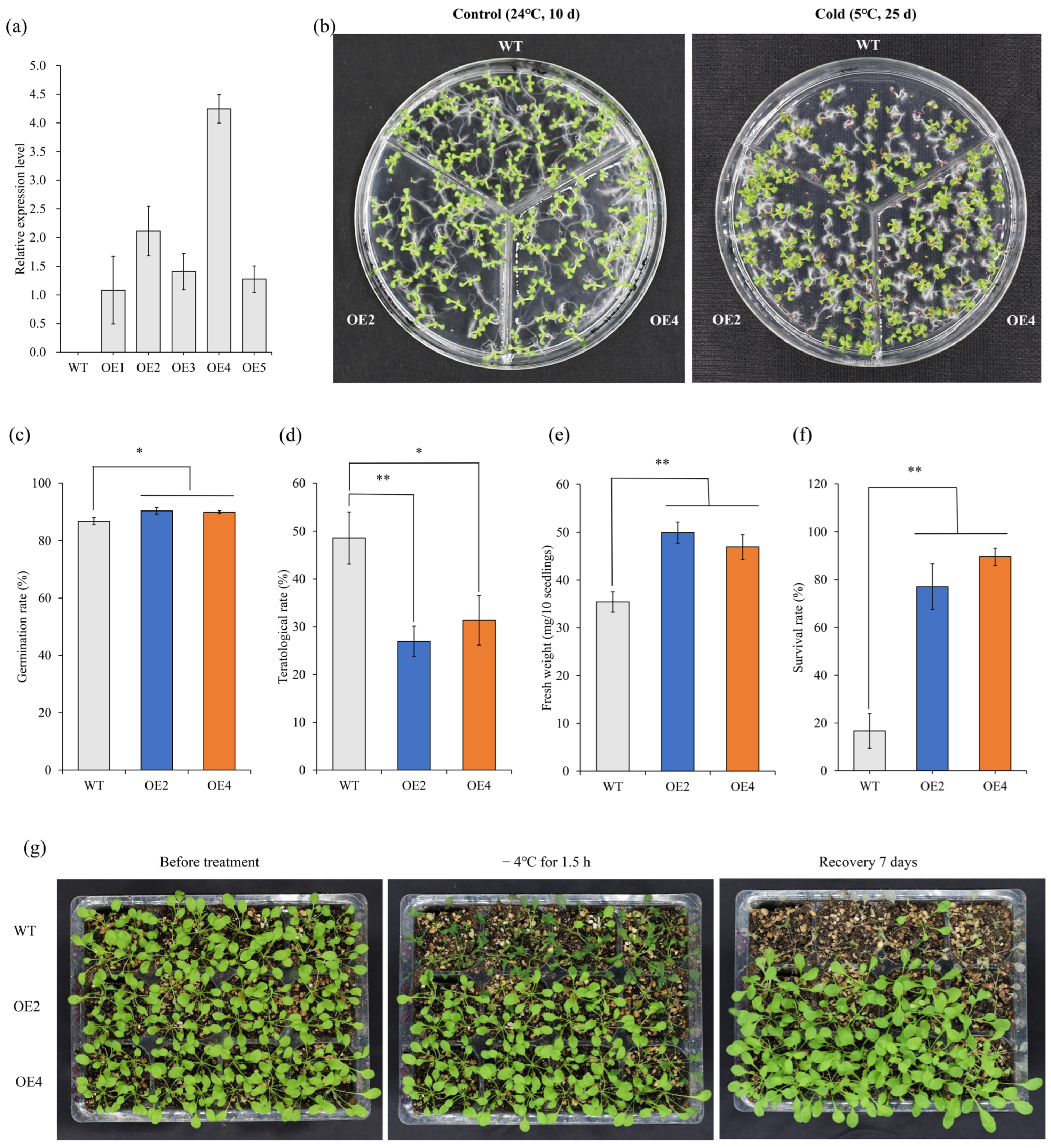
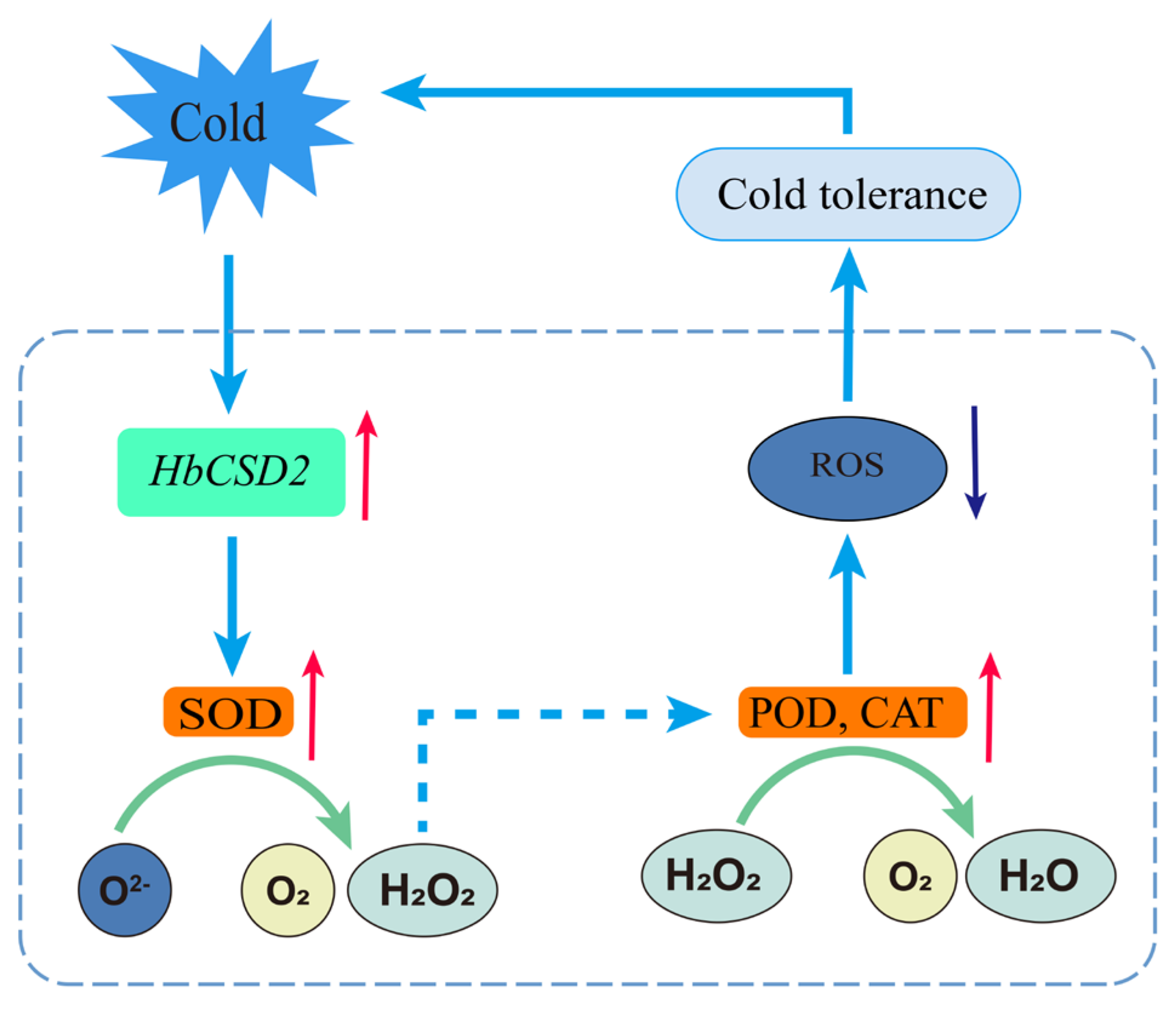
2.3. Overexpression of HbCSD2 Enhances Cold Tolerance in Transgenic Arabidopsis
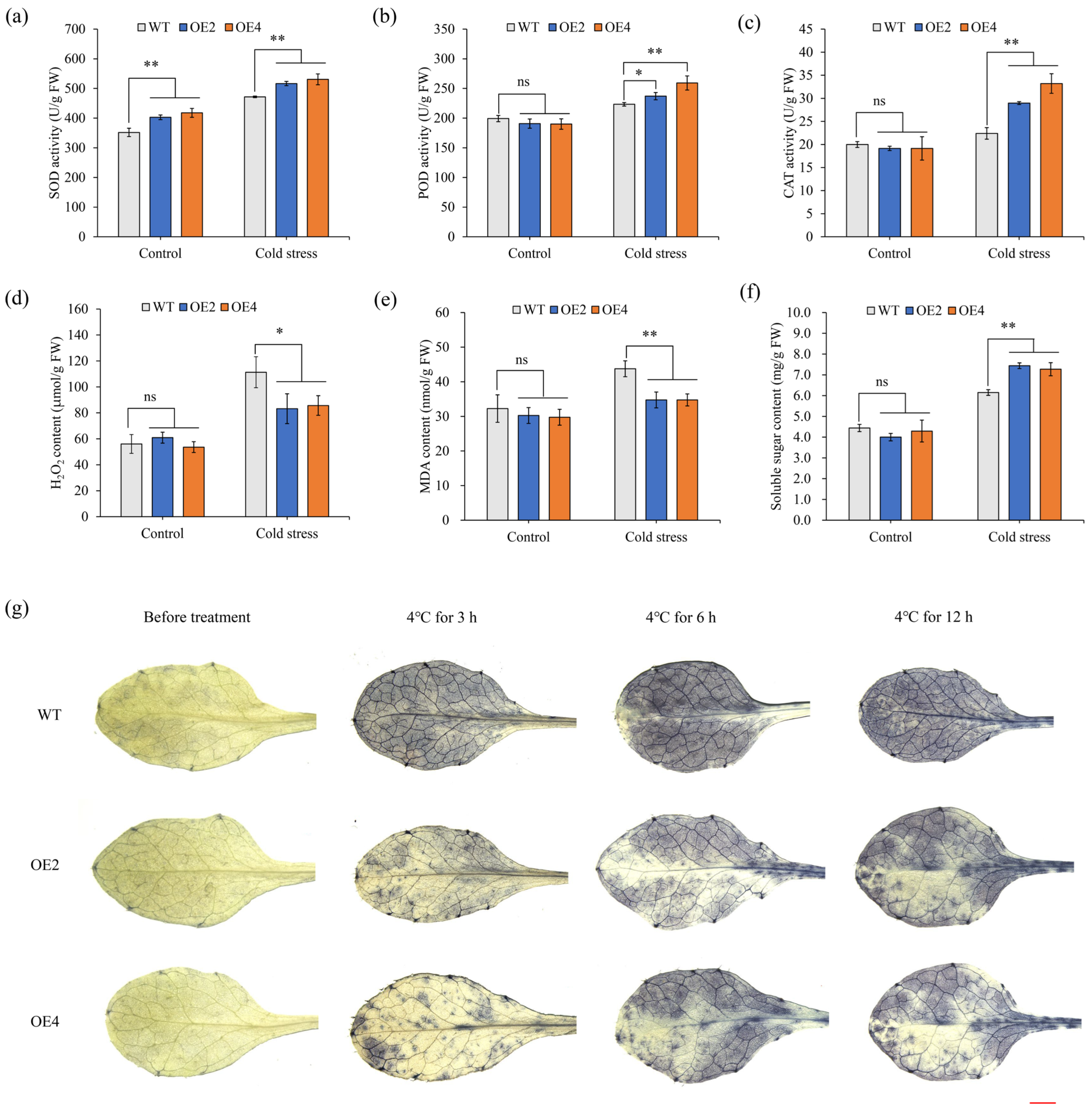
2.4. Physiological Response of HbCSD2 Transgenic Arabidopsis Plants Under Cold Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. RNA Extraction and cDNA Synthesis
4.3. HbCSD2 Cloning and Sequence Analysis
4.4. Subcellular Localization of the HbCSD2 Protein
4.5. Transformation of HbCSD2 into Arabidopsis thaliana
4.6. HbCSD2 Enhanced the Cold Tolerance of Transgenic Arabidopsis
4.7. Physiological Responses of Transgenic Arabidopsis Under Cold Stress
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhu, J. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.A.; Hsu, P.K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef]
- del Río, L.A. ROS and RNS in plant physiology: An overview. J. Exp. Bot. 2015, 66, 2827–2837. [Google Scholar] [CrossRef]
- Dmitrieva, V.A.; Tyutereva, E.V.; Voitsekhovskaja, O.V. Singlet oxygen in plants: Generation, detection, and signaling roles. Int. J. Mol. Sci. 2020, 21, 3237. [Google Scholar] [CrossRef] [PubMed]
- Rajput, V.D.; Harish; Singh, R.K.; Verma, K.K.; Sharma, L.; Quiroz-Figueroa, F.R.; Meena, M.; Gour, V.S.; Minkina, T.; Sushkova, S.; et al. Recent developments in enzymatic antioxidant defence mechanism in plants with special reference to abiotic stress. Biology 2021, 10, 267. [Google Scholar] [CrossRef]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2016, 90, 856–867. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.H.M.; Zulfiqar, F.; Raza, A.; Mohsin, S.; Mahmud, J.; Fujita, M.; Fotopoulos, V. Reactive oxygen species and antioxidant defense in plants under abiotic stress: Revisiting the crucial role of a universal defense regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Raihan, M.R.H.; Masud, A.A.C.; Rahman, K.; Nowroz, F.; Rahman, M.; Nahar, K.; Fujita, M. Regulation of reactive oxygen species and antioxidant defense in plants under salinity. Int. J. Mol. Sci. 2021, 22, 9326. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Y.; Abreu, I.A.; Cabelli, D.E.; Maroney, M.J.; Miller, A.-F.; Teixeira, M.; Valentine, J.S. Superoxide dismutases and superoxide reductases. Chem. Rev. 2014, 114, 3854–3918. [Google Scholar] [CrossRef]
- Gill, S.S.; Anjum, N.A.; Gill, R.; Yadav, S.; Hasanuzzaman, M.; Fujita, M.; Mishra, P.; Sabat, S.C.; Tuteja, N. Superoxide dismutase—Mentor of abiotic stress tolerance in crop plants. Environ. Sci. Pollut. Res. 2015, 22, 10375–10394. [Google Scholar] [CrossRef] [PubMed]
- Fink, R.C.; Scandalios, J.G. Molecular evolution and structure–function relationships of the superoxide dismutase gene families in angiosperms and their relationship to other eukaryotic and prokaryotic superoxide dismutases. Arch. Biochem. Biophys. 2002, 399, 19–36. [Google Scholar] [CrossRef]
- Abreu, I.A.; Cabelli, D.E. Superoxide dismutases—A review of the metal-associated mechanistic variations. Biochim. Biophys. Acta 2010, 1804, 263–274. [Google Scholar] [CrossRef]
- Negi, N.P.; Shrivastava, D.C.; Sharma, V.; Sarin, N.B. Overexpression of Cu/ZnSOD from Arachis hypogaea alleviates salinity and drought stress in tobacco. Plant Cell Rep. 2015, 34, 1109–1126. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Han, X.; Song, X.; Zhang, Y.; Jiang, J.; Han, Q.; Liu, M.; Qiao, G.; Zhuo, R. Overexpressing the Sedum alfredii Cu/Zn superoxide dismutase increased resistance to oxidative stress in transgenic Arabidopsis. Front. Plant Sci. 2017, 8, 1010. [Google Scholar] [CrossRef]
- Zhou, L.; Yu, C.; Nan, S.; Li, Y.; Hu, J.; Zhao, K.; Guo, J.; Wang, S. PagSOD2a improves poplar salt tolerance by elevating superoxide dismutase activity and decreasing malondialdehyde contents. Front. Plant Sci. 2024, 15, 1456249. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.B.; Zhang, W.J.; Gong, X.D.; Zhang, Q.; Zhou, L.R. A Cu/Zn superoxide dismutase from Jatropha curcas enhances salt tolerance of Arabidopsis thaliana. Genet Mol Res. 2015, 14, 2086–2098. [Google Scholar] [CrossRef]
- Lin, K.; Sei, S.; Su, Y.; Chiang, C. Overexpression of the Arabidopsis and winter squash superoxide dismutase genes enhances chilling tolerance via ABA-sensitive transcriptional regulation in transgenic Arabidopsis. Plant Signaling Behav. 2019, 14, 1685728. [Google Scholar] [CrossRef]
- Onokpise, O.U. Natural rubber, Hevea brasiliensis (Willd. ex A. Juss.) Müll. Arg., germplasm collection in the amazon basin, brazil: A retrospective. Econ. Bot. 2004, 58, 544–555. [Google Scholar] [CrossRef]
- Chao, J.; Wu, S.; Shi, M.; Xu, X.; Gao, Q.; Du, H.; Gao, B.; Guo, D.; Yang, S.; Zhang, S.; et al. Genomic insight into domestication of rubber tree. Nat. Commun. 2023, 14, 4651. [Google Scholar] [CrossRef]
- Gao, J.; Cheng, H. The rubber tree that changed the world in 150 years. Sci. Sin. Vitae 2024, 54, 1744–1751. [Google Scholar] [CrossRef]
- Carr, M.K.V. The water relations of rubber (Hevea brasiliensis): A review. Exp. Agric. 2011, 48, 176–193. [Google Scholar] [CrossRef]
- Tang, X.; Zhang, Y.; Yuan, H.-M.; Zhai, J.; Huang, X. Reprogramming of the Hevea brasiliensis epigenome and transcriptome in response to cold stress. Front. Plant Sci. 2022, 13, 831839. [Google Scholar] [CrossRef]
- Mao, C.; Li, L.; Yang, T.; Gui, M.; Li, X.; Zhang, F.; Zhao, Q.; Wu, Y. Transcriptomics integrated with widely targeted metabolomics reveals the cold resistance mechanism in Hevea brasiliensis. Front. Plant Sci. 2023, 13, 1092411. [Google Scholar] [CrossRef]
- Yu, W.; Kong, G.; Chao, J.; Yin, T.; Tian, H.; Ya, H.; He, L.; Zhang, H. Genome-wide identification of the rubber tree superoxide dismutase (SOD) gene family and analysis of its expression under abiotic stress. PeerJ 2022, 10, e14251. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Waszczak, C.; Carmody, M.; Kangasjärvi, J. Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhang, R.; Yan, X.; Fan, K. Superoxide dismutase nanozymes: An emerging star for anti-oxidation. J. Mater. Chem. B 2021, 9, 6939–6957. [Google Scholar] [CrossRef]
- Tang, C.; Yang, M.; Fang, Y.; YingfengLuo Gao, S.; Xiao, X.; An, Z.; Zhou, B.; Zhang, B. The rubber tree genome reveals new insights into rubber production and species adaptation. Nat. Plants 2016, 2, 16073. [Google Scholar] [CrossRef]
- Liu, J.; Shi, C.; Shi, C.C.; Li, W.; Zhang, Q.J.; Zhang, Y.; Li, K.; Lu, H.F.; Shi, C.; Zhu, S.T.; et al. The chromosome-based rubber tree genome provides new insights into spurge genome evolution and rubber biosynthesis. Mol. Plant 2020, 13, 336–350. [Google Scholar] [CrossRef]
- Iqbal Qureshi, A.M.; Sofi, M.U.; Dar, N.A.; Khan, M.H.; Mahdi, S.S.; Dar, Z.A.; Bangroo, S.; El-Serehy, H.A.; Hefft, D.I.; Popescu, S.M. Insilco identification and characterization of superoxide dismutase gene family in Brassica rapa. Saudi J. Biol. Sci. 2021, 28, 5526–5537. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Li, W.; Zhou, Y.; Pei, L.; Liu, J.; Xia, X.; Che, R.; Li, H. Molecular characterization, expression and functional analysis of acyl-CoA-binding protein gene family in maize (Zea mays). BMC Plant Biol. 2021, 21, 94. [Google Scholar] [CrossRef]
- Huo, C.; He, L.; Yu, T.; Ji, X.; Li, R.; Zhu, S.; Zhang, F.; Xie, H.; Liu, W. The superoxide dismutase gene family in Nicotiana tabacum: Genome-wide identification, characterization, expression profiling and functional analysis in response to heavy metal stress. Front. Plant Sci. 2022, 13, 904105. [Google Scholar] [CrossRef]
- Rudić, J.; Dragićević, M.B.; Momčilović, I.; Simonović, A.D.; Pantelić, D. In silico study of superoxide dismutase gene family in potato and effects of elevated temperature and salicylic acid on gene expression. Antioxidants 2022, 11, 488. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Raza, A.; Gao, A.; Jia, Z.; Zhang, Y.; Hussain, M.A.; Mehmood, S.S.; Cheng, Y.; Lv, Y.; Zou, X. Genome-wide analysis and expression profile of superoxide dismutase (SOD) gene family in rapeseed (Brassica napus L.) under different hormones and abiotic stress conditions. Antioxidants 2021, 10, 1182. [Google Scholar] [CrossRef] [PubMed]
- Huseynova, I.M.; Aliyeva, D.R.; Aliyev, J.A. Subcellular localization and responses of superoxide dismutase isoforms in local wheat varieties subjected to continuous soil drought. Plant Physiol. Biochem. 2014, 81, 54–60. [Google Scholar] [CrossRef]
- Zhou, C.; Zhu, C.; Fu, H.; Li, X.; Chen, L.; Lin, Y.; Lai, Z.; Guo, Y. Genome-wide investigation of superoxide dismutase (SOD) gene family and their regulatory miRNAs reveal the involvement in abiotic stress and hormone response in tea plant (Camellia sinensis). PLoS ONE 2019, 14, e0223609. [Google Scholar] [CrossRef]
- Han, L.; Hua, W.; Cao, X.; Yan, J.; Chen, C.; Wang, Z. Genome-wide identification and expression analysis of the superoxide dismutase (SOD) gene family in Salvia miltiorrhiza. Gene 2020, 742, 144603. [Google Scholar] [CrossRef]
- Zhang, G.; Ding, Q.; Wei, B. Genome-wide identification of superoxide dismutase gene families and their expression patterns under low-temperature, salt and osmotic stresses in watermelon and melon. 3 Biotech 2021, 11, 194. [Google Scholar] [CrossRef]
- Huang, H.; Wang, H.; Tong, Y.; Wang, Y. Insights into the superoxide dismutase gene family and its roles in Dendrobium catenatum under abiotic stresses. Plants 2020, 9, 1452. [Google Scholar] [CrossRef]
- Xu, J.; Duan, X.; Yang, J.; Beeching, J.R.; Zhang, P. Coupled expression of Cu/Zn-superoxide dismutase and catalase in cassava improves tolerance against cold and drought stresses. Plant Signal. Behav. 2014, 8, e24525. [Google Scholar] [CrossRef]
- Li, Y.; Yu, W.; Chen, Y.; Yang, S.; Wu, S.; Chao, J.; Wang, X.; Tian, W.-M. Genome-wide identification and characterization of heat-shock transcription factors in rubber tree. Forests 2019, 10, 1157. [Google Scholar] [CrossRef]
- Yu, W.; Kong, G.; Ya, H.; He, L.; Wu, Y.; Zhang, H. Comprehensive analysis of the catalase (CAT) gene family and expression patterns in rubber tree (Hevea brasiliensis) under various abiotic stresses and multiple hormone treatments. Int. J. Mol. Sci. 2023, 25, 70. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef] [PubMed]
- Michael, B.; Christof, D.; Katharina, N.; Sierralta, W.D.; Jutta, P. Intracellular localization of Arabidopsis sulfurtransferases. Plant Physiol. 2004, 135, 916–926. [Google Scholar] [CrossRef]
- Yin, H.; Sun, Q.; Lu, X.; Zhang, L.; Yuan, Y.; Gong, C.; He, X.; Ma, W.; Mu, P. Identification of the glutamine synthetase (GS) gene family in four wheat species and functional analysis of Ta4D.GSe in Arabidopsis thaliana. Plant Mol. Biol. 2022, 110, 93–106. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, W.; Kong, G.; Ya, H.; Zhang, H. Overexpression of Hevea brasiliensis HbCDS2 Gene Enhances Cold Tolerance in Transgenic Arabidopsis. Plants 2025, 14, 3591. https://doi.org/10.3390/plants14233591
Yu W, Kong G, Ya H, Zhang H. Overexpression of Hevea brasiliensis HbCDS2 Gene Enhances Cold Tolerance in Transgenic Arabidopsis. Plants. 2025; 14(23):3591. https://doi.org/10.3390/plants14233591
Chicago/Turabian StyleYu, Wencai, Guanghong Kong, Huajin Ya, and Hanyao Zhang. 2025. "Overexpression of Hevea brasiliensis HbCDS2 Gene Enhances Cold Tolerance in Transgenic Arabidopsis" Plants 14, no. 23: 3591. https://doi.org/10.3390/plants14233591
APA StyleYu, W., Kong, G., Ya, H., & Zhang, H. (2025). Overexpression of Hevea brasiliensis HbCDS2 Gene Enhances Cold Tolerance in Transgenic Arabidopsis. Plants, 14(23), 3591. https://doi.org/10.3390/plants14233591






