Characterization of the Soybean (Glycine max) Heavy-Metal-Associated Isoprenylated Plant Protein (HIPP) Gene Family in Response to Aluminum
Abstract
1. Introduction
2. Results
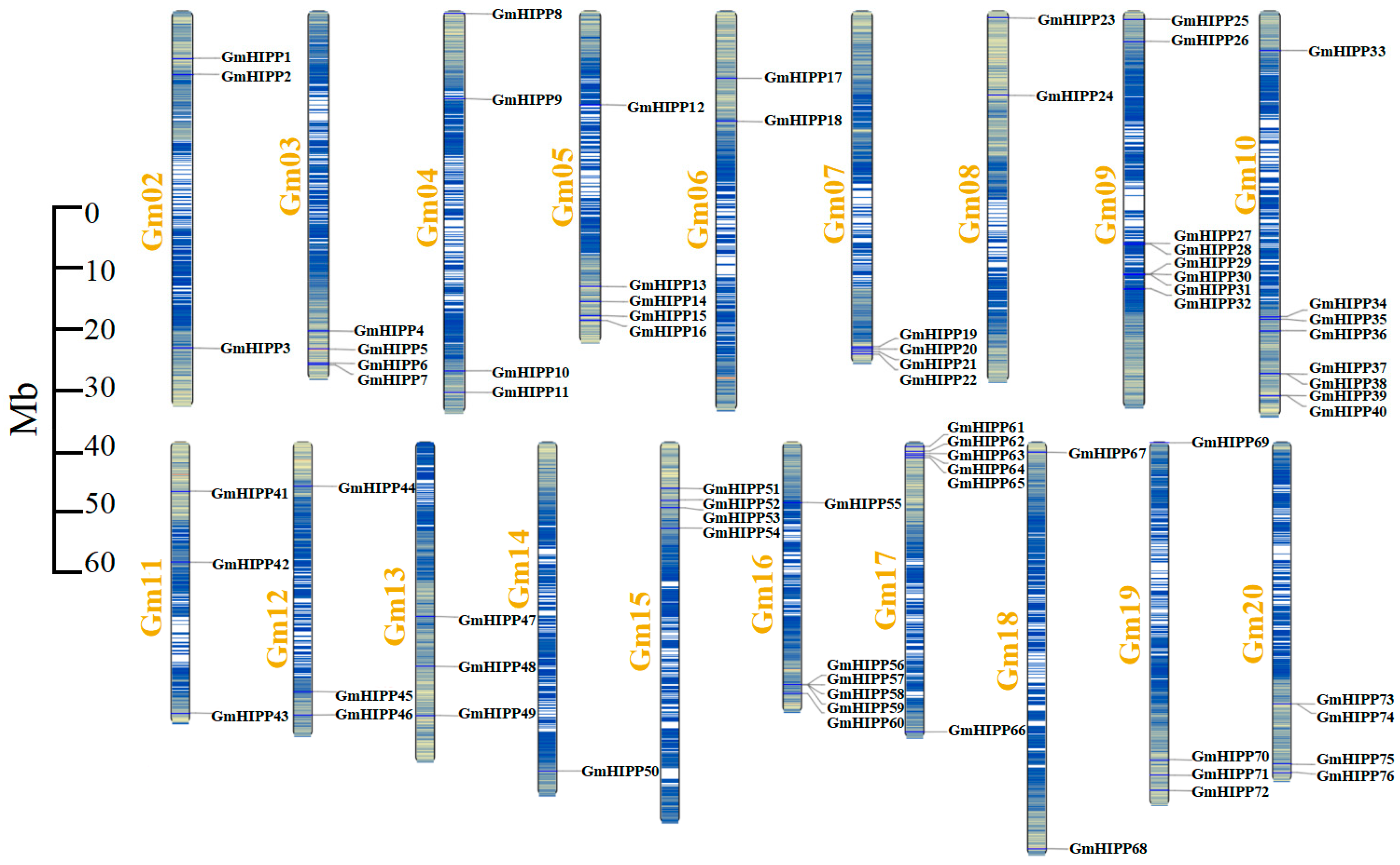
2.1. Identification and Chromosomal Distribution of GmHIPP Genes in the Soybean Genome
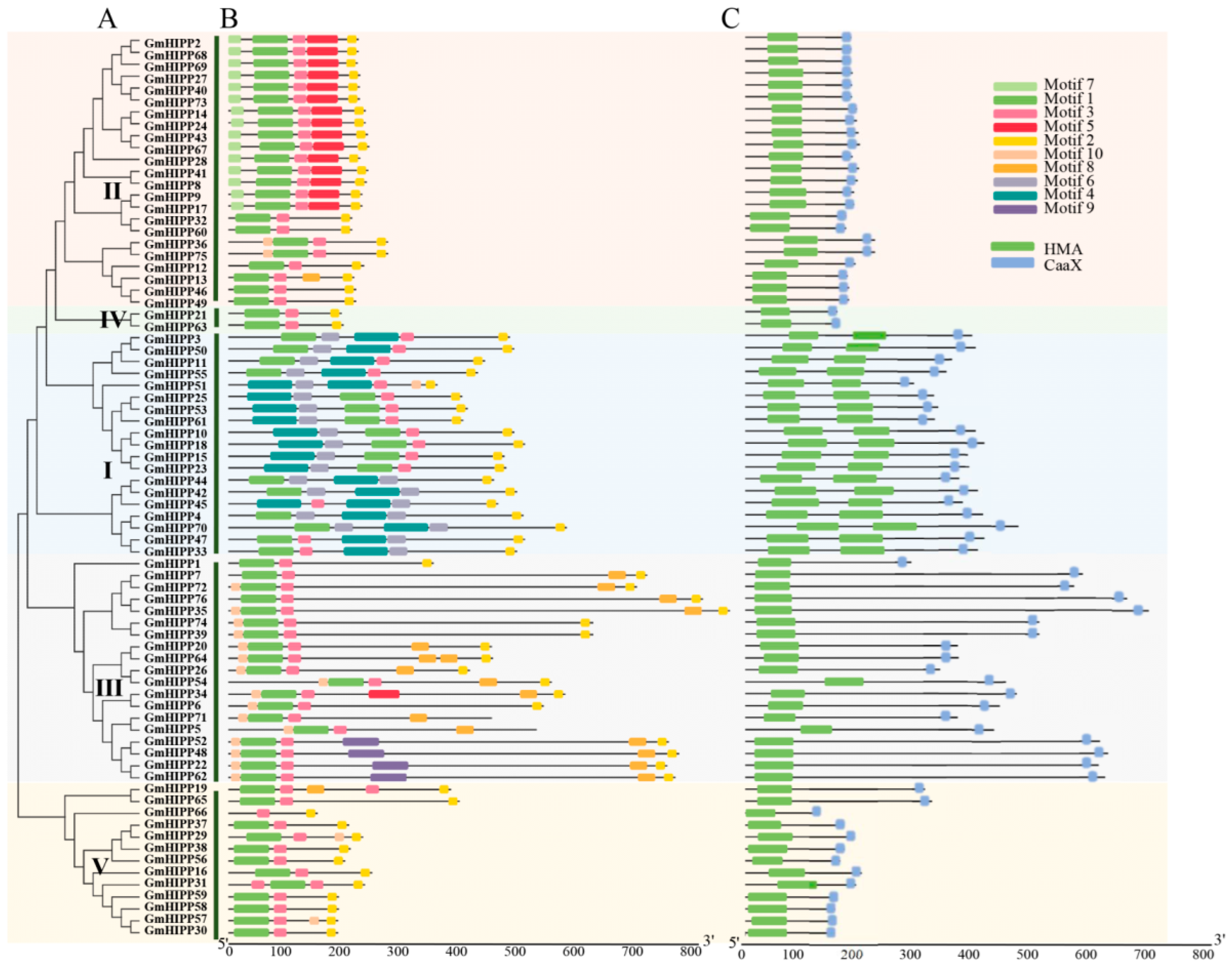
2.2. Phylogenetic Tree, Domains, Conserved Motifs, and Gene Structure of GmHIPP Family
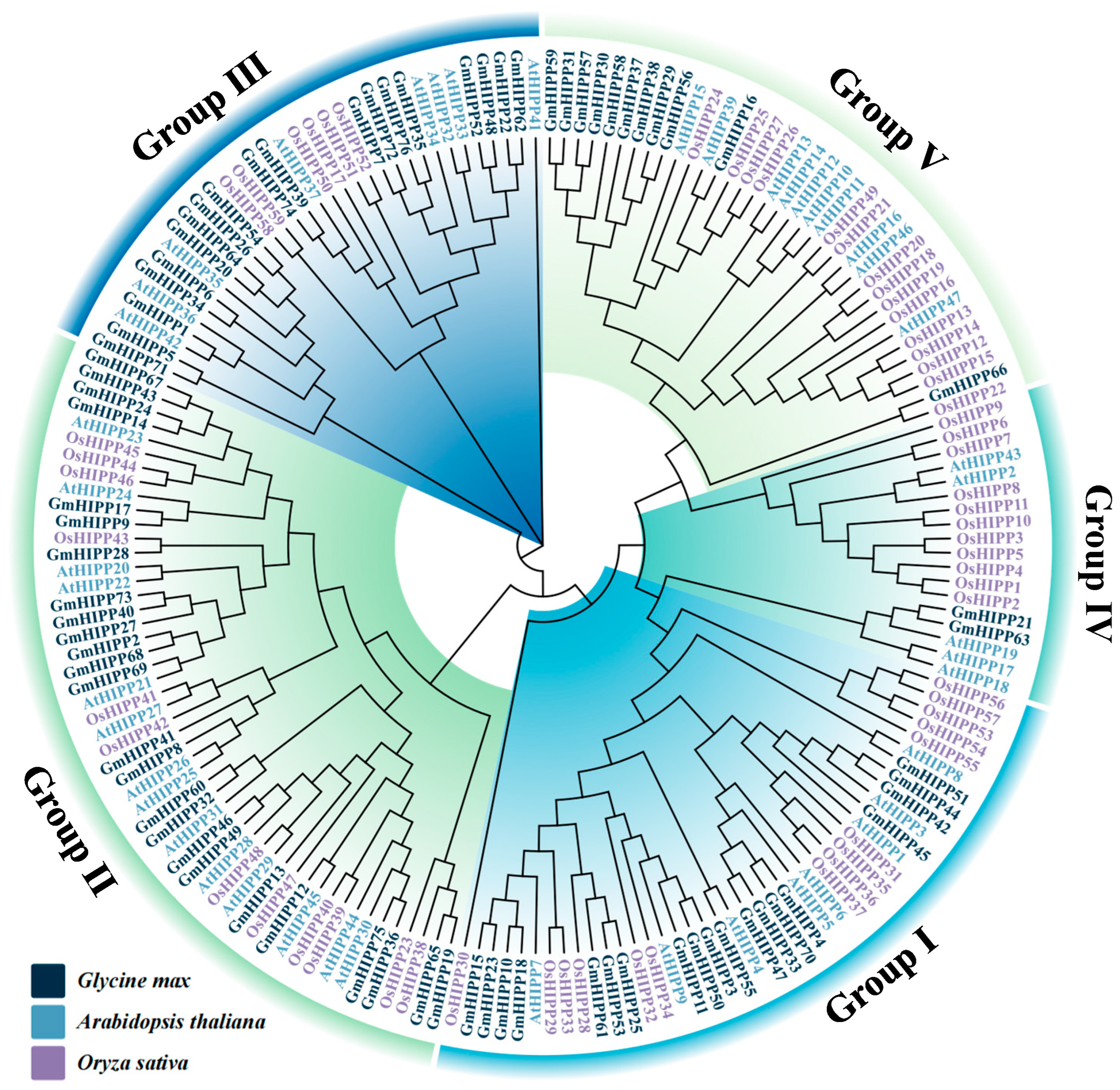
2.3. Phylogenetic Analysis of HIPP Proteins from Three Species
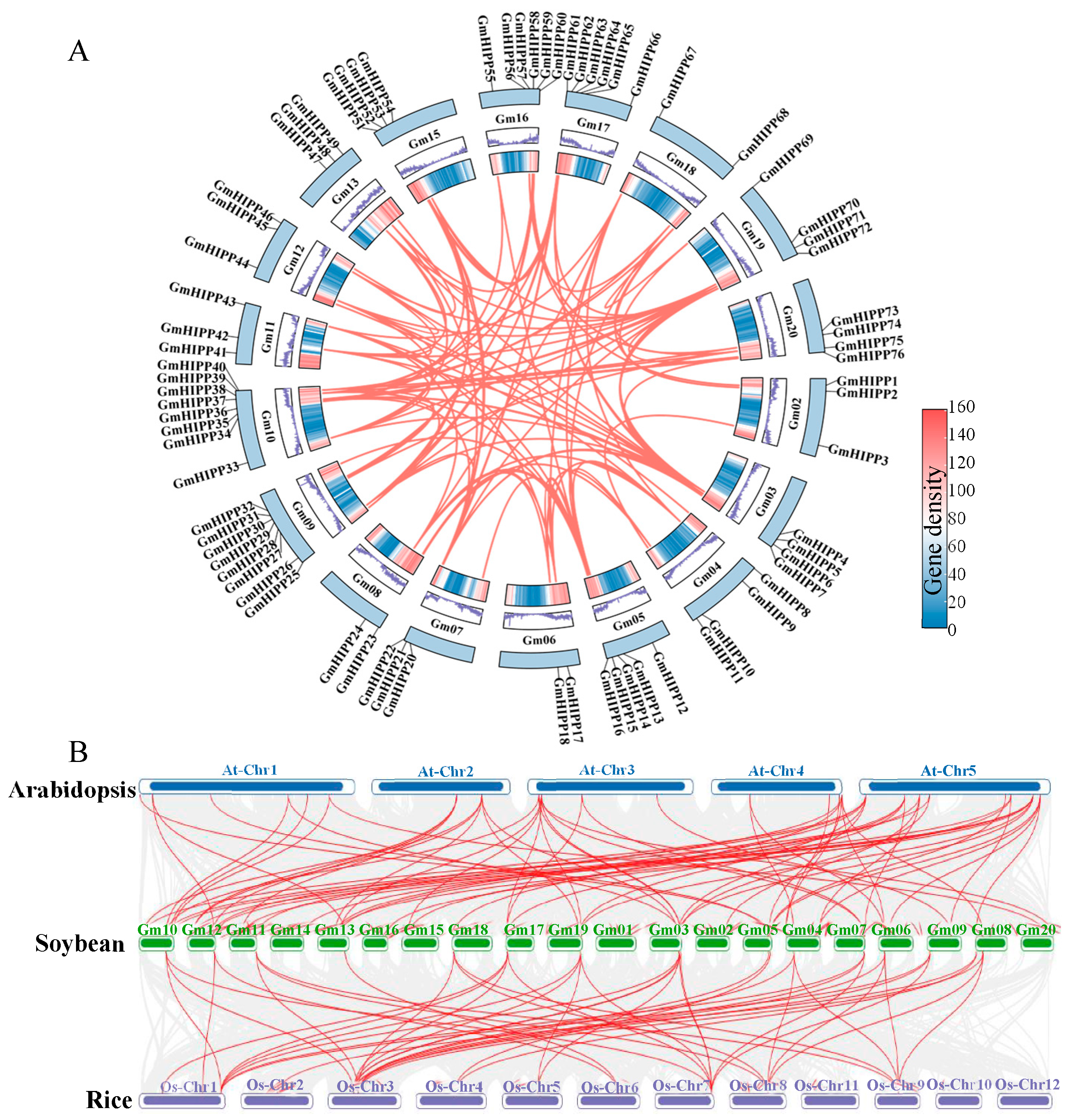
2.4. Evolutionary and Synteny Analysis of HIPP Gene in Soybean
2.5. Analysis of the Promoter Region of the GmHIPP Gene Family
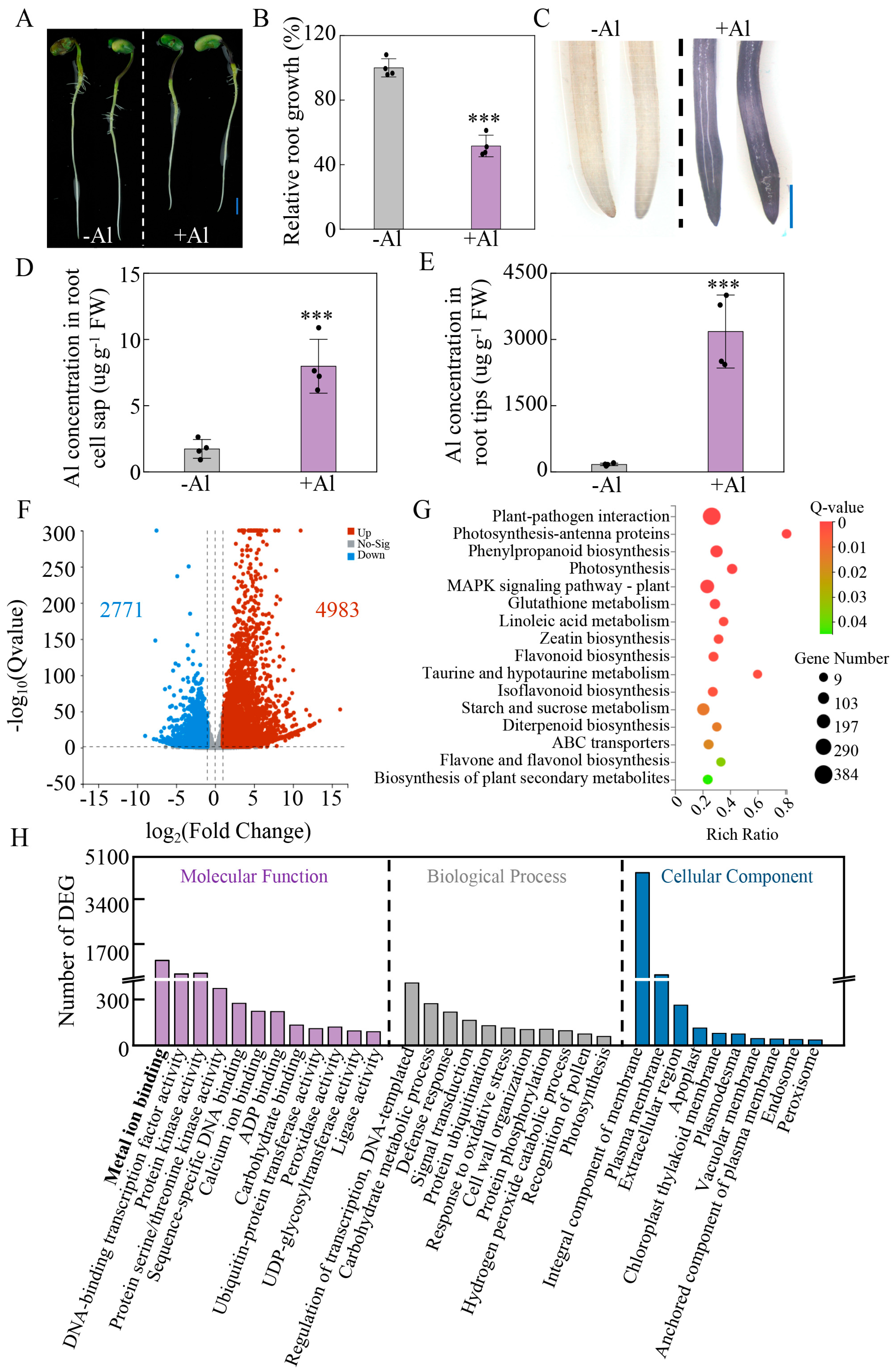
2.6. Transcriptomic Changes in Soybean Roots Exposed to Al Toxicity
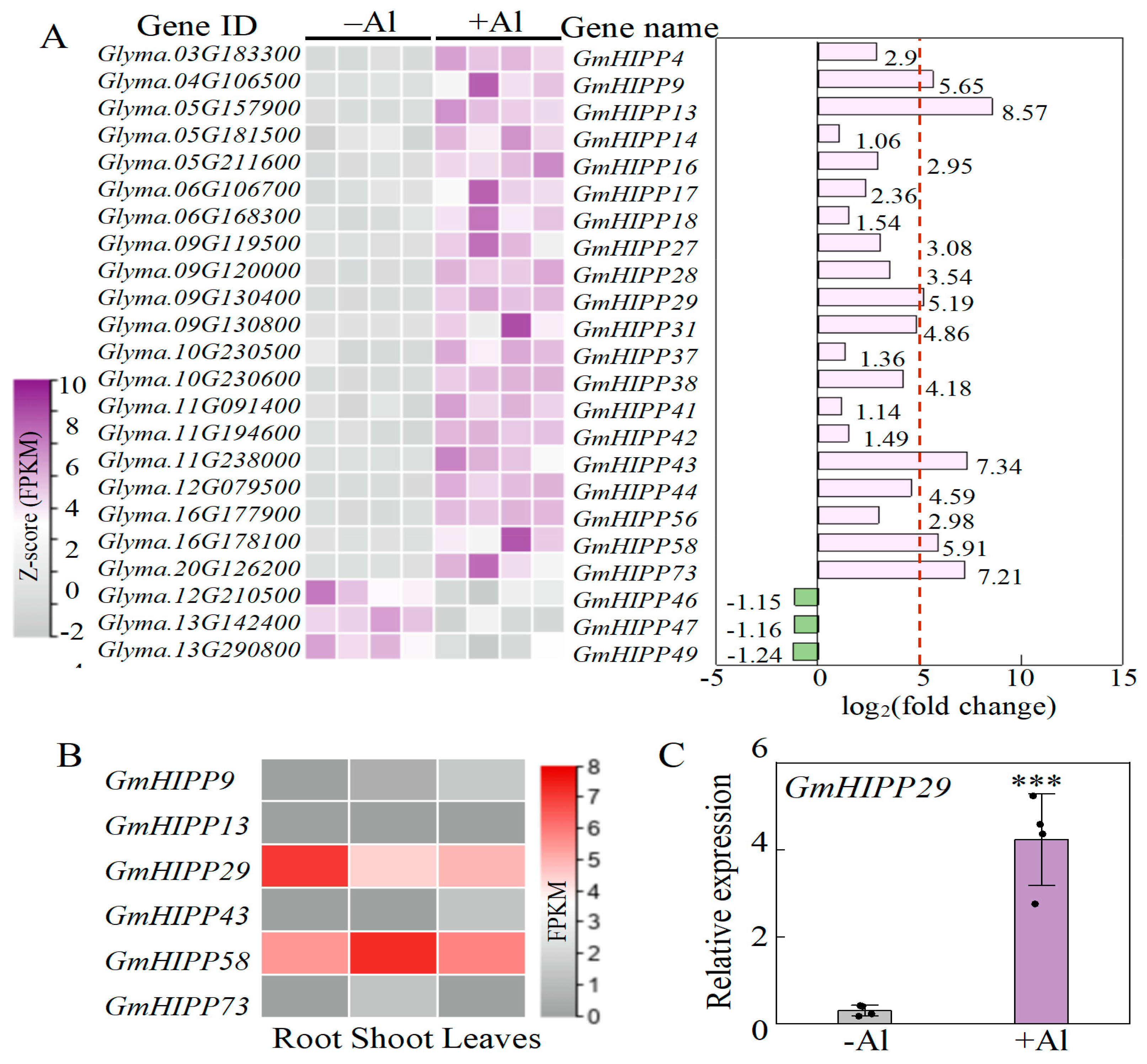
2.7. Identification of Al-Responsive Genes Related to Heavy Metal-Associated Isoprenylated Plant Protein (HIPP)
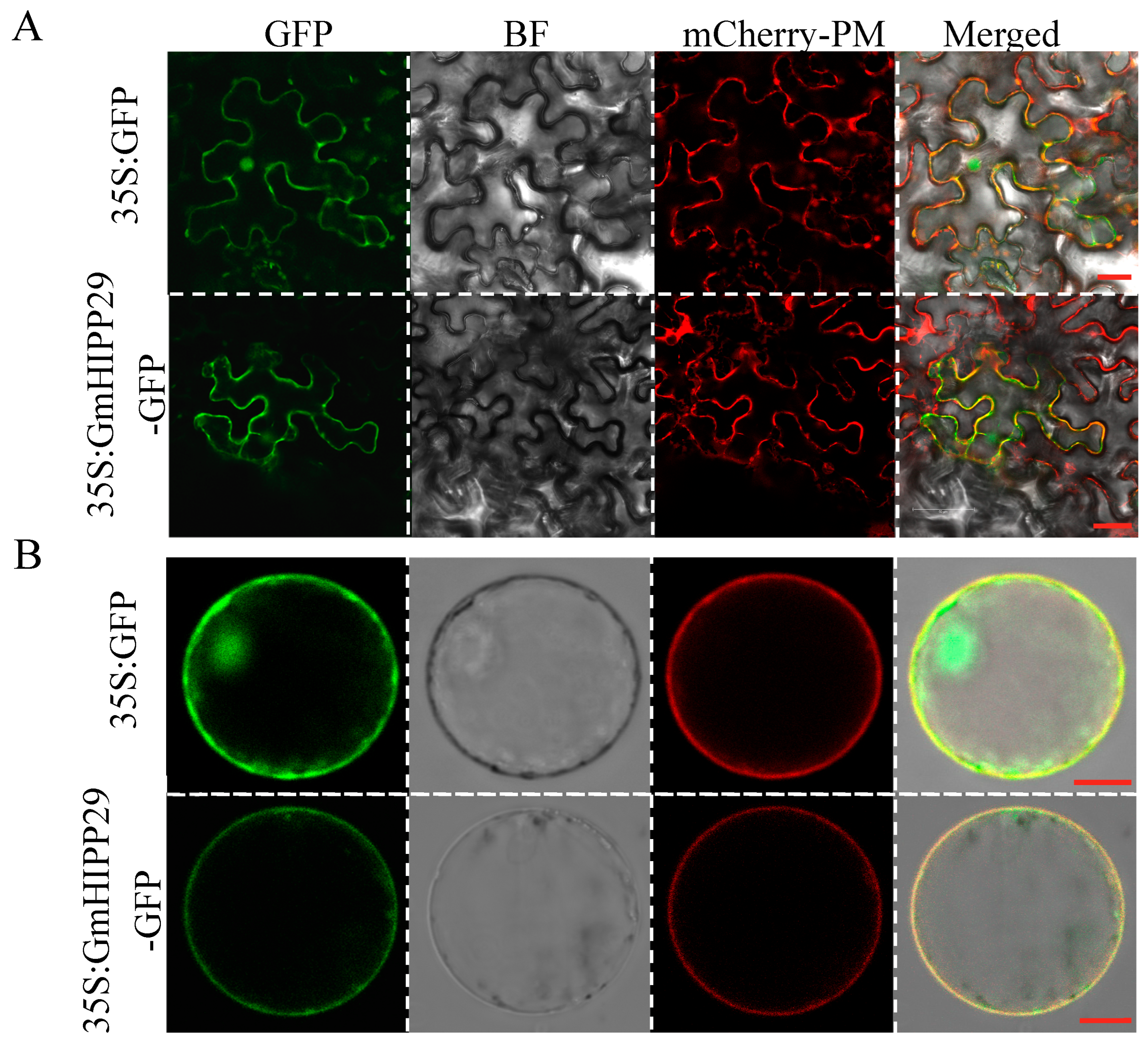
2.8. Subcellular Localization of GmHIPP29
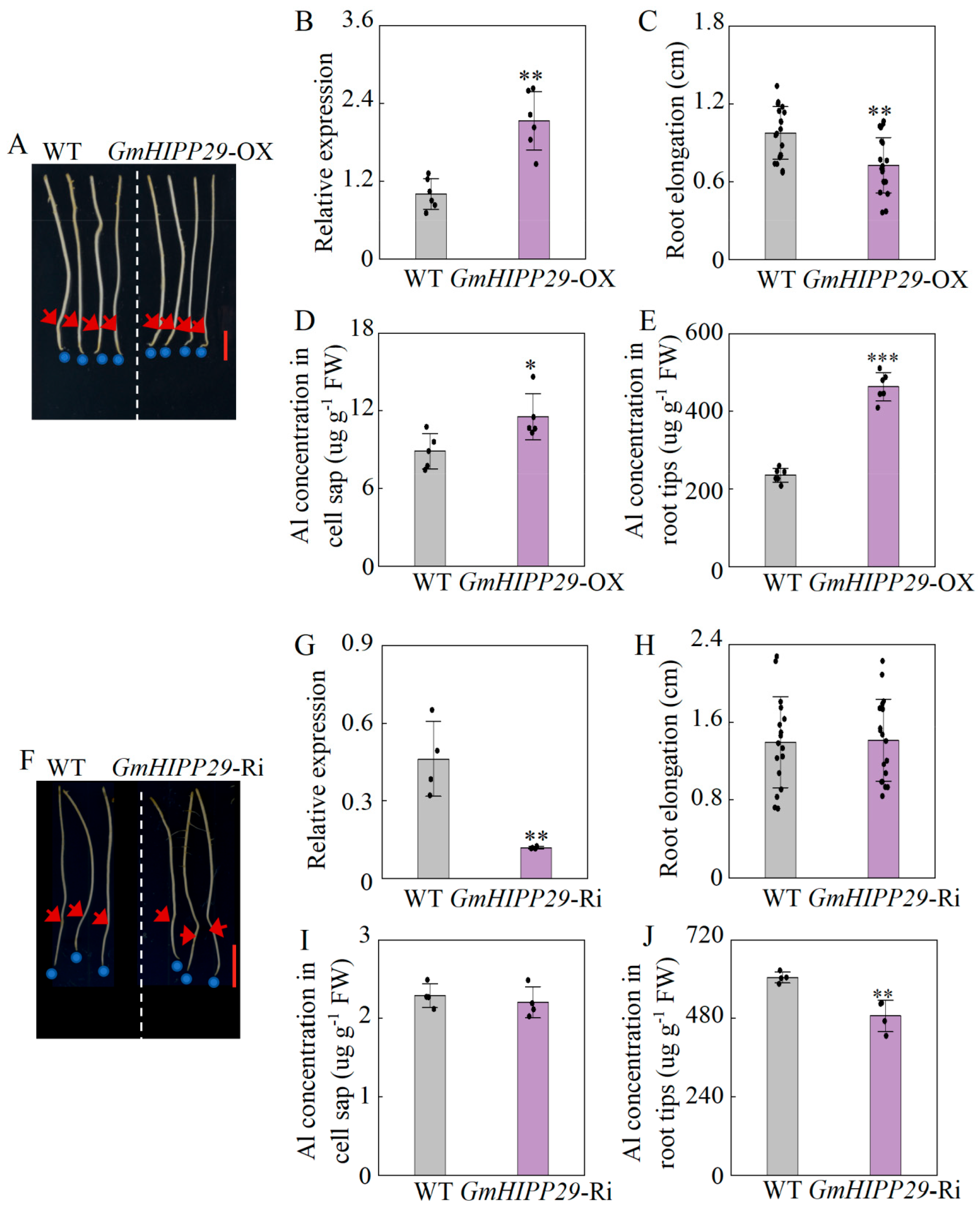
2.9. Alternative Expression of GmHIPP29 Affects Soybean Hairy Roots Growth in Response to Al Toxicity
3. Discussion
3.1. Structure Characteristics of GmHIPP
3.2. GmHIPP Genes in Response to Al Toxicity
3.3. The Potential Mechanism of GmHIPP29 in Regulating Al Tolerance in Soybean
4. Materials and Methods
4.1. Identification of GmHIPP Family Genes in Soybean
4.2. Chromosome Distribution and Protein Structure Analysis of the HIPP Gene Family
4.3. Phylogenetic Analysis of the HIPP Gene Family
4.4. Collinearity Analysis
4.5. Cis-Regulatory Element Analysis in the Promoter Regions of HIPP Genes in Soybean
4.6. Plant Growth Conditions and Al Treatment
4.7. Root Length Measurement
4.8. Spatial Localization and Quantification of Al Accumulation in Root Tips of Al Accumulation in Root Tips
4.9. RNA Isolation and Transcriptomic Analysis
4.10. Quantitative Real-Time PCR (qRT-PCR) Analysis
4.11. Subcellular Localization of the Soybean HIPP Family Member GmHIPP29
4.12. Functional Analysis of GmHIPP29 in Soybean Hairy Roots
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kochian, L.; Pineros, M.; Liu, P.; Magalhaes, J. Plant adaptation to acid soils: The molecular basis for crop aluminum resistance. Annu. Rev. Plant Biol. 2015, 66, 571–598. [Google Scholar] [CrossRef] [PubMed]
- Che, J.; Zhao, X.; Shen, R. Molecular mechanisms of plant adaptation to acid soils: A review. Pedosphere 2023, 33, 12–24. [Google Scholar] [CrossRef]
- Li, X.; Zhang, X.; Zhao, Q.; Liao, H. Genetic improvement of legume roots for adaption to acid soils. Crop J. 2023, 11, 1022–1033. [Google Scholar] [CrossRef]
- Ofoe, R.; Thomas, R.; Asiedu, S.; Wang-Pruski, G.; Fofana, B.; Abbey, L. Aluminum in plant: Benefits, toxicity and tolerance mechanisms. Front. Plant Sci. 2023, 13, 1085998. [Google Scholar] [CrossRef]
- de Castro, L.M.R.; Vinson, C.C.; da Gordo, S.M.C.; Williams, T.C.R.; Cury, N.F.; de Souza, M.C.; Pereira, L.A.R. Molecular and physiological aspects of plant responses to aluminum: What do we know about Cerrado plants? Braz. J. Bot. 2022, 45, 545–562. [Google Scholar] [CrossRef]
- Chandra, J.; Keshavkant, S. Mechanisms underlying the phytotoxicity and genotoxicity of aluminum and their alleviation strategies: A review. Chemosphere 2021, 278, 130384. [Google Scholar] [CrossRef]
- Chauhan, D.; Yadav, V.; Vaculík, M.; Gassmann, W.; Pike, S.; Arif, N.; Singh, V.; Deshmukh, R.; Sahi, S.; Tripathi, D. Aluminum toxicity and aluminum stress-induced physiological tolerance responses in higher plants. Crit. Rev. Biotechnol. 2021, 41, 715–730. [Google Scholar] [CrossRef]
- Zhang, X.; Long, Y.; Huang, J.; Xia, J. Molecular mechanisms for coping with Al toxicity in plants. Int. J. Mol. Sci. 2019, 20, 1551. [Google Scholar] [CrossRef]
- Huang, C.; Yamaji, N.; Chen, Z.; Ma, J. A tonoplast-localized half-size ABC transporter is required for internal detoxification of aluminum in rice. Plant J. 2012, 69, 857–867. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Yamaji, N.; Kasai, T.; Ma, J.F. Plasma membrane-localized transporter for aluminum in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 18381–18385. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, N.; Hu, W.; Yan, W.; Jin, X.; Yu, Y.; Du, C.; Liu, C.; He, W.; Zhang, S. ZmNRAMP4 Enhances the Tolerance to Aluminum Stress in Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 8162. [Google Scholar] [CrossRef]
- Ahmad, M.Z.; Chen, S.; Qi, X.; Feng, J.; Chen, H.; Liu, X.; Sun, M.; Deng, Y. Genome wide analysis of HMA gene family in Hydrangea macrophylla and characterization of HmHMA2 in response to aluminum stress. Plant Physiol. Bioch. 2024, 216, 109182. [Google Scholar] [CrossRef] [PubMed]
- Barr, Z.K.; Werner, T.; Tilsner, J. Heavy metal-associated isoprenylated plant proteins (HIPPs) at plasmodesmata: Exploring the link between localization and function. Plants 2023, 12, 3015. [Google Scholar] [CrossRef] [PubMed]
- Zschiesche, W.; Barth, O.; Daniel, K.; Böhme, S.; Rausche, J.; Humbeck, K. The zinc-binding nuclear protein HIPP3 acts as an upstream regulator of the salicylate-dependent plant immunity pathway and of flowering time in Arabidopsis thaliana. New Phytol. 2015, 207, 1084–1096. [Google Scholar] [CrossRef]
- Khan, I.U.; Rono, J.K.; Zhang, B.Q.; Liu, X.S.; Wang, M.Q.; Wang, L.L.; Wu, X.C.; Chen, X.; Cao, H.W.; Yang, Z.M. Identification of novel rice (Oryza sativa) HPP and HIPP genes tolerant to heavy metal toxicity. Ecotox. Environ. Safe. 2019, 175, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Peng, X.; Wang, X.; Wang, C. The heavy metal-associated isoprenylated plant protein (HIPP) gene family plays a crucial role in cadmium resistance and accumulation in the tea plant (Camellia sinensis L.). Ecotox. Environ. Safe 2023, 260, 115077. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, X.; Liu, J.; Niu, Y.; Chen, Y.; Hao, Y.; Zhao, J.; Sun, L.; Wang, H.; Xiao, J.; et al. Characterization of the heavy-metal-associated isoprenylated plant protein (HIPP) Gene Family from Triticeae Species. Int. J. Mol. Sci. 2020, 21, 6191. [Google Scholar] [CrossRef]
- Zheng, Q.; Yu, Q.; Wu, N.; Yao, W.; Li, J.; Lv, K.; Xu, W. A grape VvHOS1-interacting HIPP protein (VvHIPP21) negatively regulates cold and drought stress. Environ. Exp. Bot. 2023, 207, 105203. [Google Scholar] [CrossRef]
- Xu, J.; Cui, J.; He, Q.; Liu, Y.; Lu, X.; Qi, J.; Xiong, J.; Yu, W.; Li, C. Genome-wide identification of HIPP and mechanism of SlHIPP4/7/9/21/26/32 mediated phytohormones response to Cd, osmotic, and salt stresses in tomato. Plant Physiol. Bioch. 2024, 217, 109220. [Google Scholar] [CrossRef]
- Zhang, H.; Zhai, G.; Ni, X.; Liu, Z.; Song, T.; Han, Y.; Wang, Y.; Shao, Y.; Wang, F.; Zou, G.; et al. Genome-wide identification of HIPP genes family in sorghum reveals the novel role of SbHIPP40 in accumulation of cadmium. J. Hazard. Mater. 2025, 494, 138478. [Google Scholar] [CrossRef]
- Xie, Z.; Yang, L.; Fan, M.; Xuan, S.; Jia, X.; Zhang, Z.; Li, N.; Liu, M.; Zhao, J.; Li, J. Genome-wide identification, characterization and expression analysis of the chalcone synthase gene family in Chinese cabbage. BMC Genom. 2025, 26, 168. [Google Scholar] [CrossRef]
- Guo, T.; Weber, H.; Niemann, M.C.E.; Theisl, L.; Leonte, G.; Novák, O.; Werner, T. Arabidopsis HIPP proteins regulate endoplasmic reticulum-associated degradation of CKX proteins and cytokinin responses. Mol. Plant 2021, 14, 1918–1934. [Google Scholar] [CrossRef] [PubMed]
- de Abreu-Neto, J.B.; Turchetto-Zolet, A.C.; de Oliveira, L.F.; Zanettini, M.H.; Margis-Pinheiro, M. Heavy metal-associated isoprenylated plant protein (HIPP): Characterization of a family of proteins exclusive to plants. Febs J. 2013, 280, 1604–1616. [Google Scholar] [CrossRef]
- Tehseen, M.; Cairns, N.; Sherson, S.; Cobbett, C.S. Metallochaperone-like genes in Arabidopsis thaliana. Metallomics 2010, 2, 556–564. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.Q.; Liu, X.S.; Feng, S.J.; Zhao, Y.N.; Wang, L.L.; Rono, J.K.; Li, H.; Yang, Z.M. Developing a cadmium resistant rice genotype with OsHIPP29 locus for limiting cadmium accumulation in the paddy crop. Chemosphere 2020, 247, 125958. [Google Scholar] [CrossRef]
- Chen, G.; Xiong, S. OsHIPP24 is a copper metallochaperone which affects rice Growth. J. Plant Biol. 2021, 64, 145–153. [Google Scholar] [CrossRef]
- Niu, M.; Bao, C.; Zhan, J.; Yue, X.; Zou, J.; Su, N.; Cui, J. Plasma membrane-localized protein BcHIPP16 promotes the uptake of copper and cadmium in planta. Ecotoxicol. Environ. Saf. 2021, 227, 112920. [Google Scholar] [CrossRef]
- Graham, P.; Vance, C. Legumes: Importance and constraints to greater use. Plant Physiol. 2003, 131, 872–877. [Google Scholar] [CrossRef]
- Herridge, D.; Peoples, M.; Boddey, R. Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil. 2008, 311, 1–18. [Google Scholar] [CrossRef]
- Wilson, R. Soybean: Market driven research needs. In G. Stacey, Genetics and genomics of soybean. Plant Genet. Genom. Crops Models 2008, 2, 3–15. [Google Scholar]
- Zhang, H.; Jiang, H.; Hu, Z.; Song, Q.; An, Y.C. Development of a versatile resource for post-genomic research through consolidating and characterizing 1500 diverse wild and cultivated soybean genomes. BMC Genom. 2022, 23, 250. [Google Scholar] [CrossRef]
- Li, J.; Tian, J.; Zhou, M.; Tian, J.; Liang, C. Research progress on the physiological and molecular mechanisms underlying soybean aluminum resistance. New Crops 2025, 2, 100034. [Google Scholar] [CrossRef]
- Liang, C.; Piñeros, M.; Tian, J.; Yao, Z.; Sun, L.; Liu, J.; Shaff, J.; Coluccio, A.; Kochian, L.; Liao, H. Low pH, aluminum, and phosphorus coordinately regulate malate exudation through GmALMT1 to improve soybean adaptation to acid soils. Plant Physiol. 2013, 161, 1347–1361. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, Z.; Gong, L.; Chen, A.; Liu, N.; Li, S.; Sun, H.; Yang, Z.; You, J. Functional characterization of three MATE genes in relation to aluminum-induced citrate efflux from soybean root. Plant Soil. 2019, 443, 121–138. [Google Scholar] [CrossRef]
- Wang, Z.; Li, H.; Wei, Z.; Sun, H.; He, Y.; Gao, J.; Yang, Z.; You, J. Overexpression of UDP-glycosyltransferase genes enhanced aluminum tolerance through disrupting cell wall polysaccharide components in soybean. Plant Soil. 2021, 469, 135–147. [Google Scholar] [CrossRef]
- Chen, Q.; Li, J.; Liu, G.; Lu, X.; Chen, K.; Tian, J.; Liang, C. A berberine bridge enzyme-Like protein, GmBBE-like43, confers soybean’s coordinated adaptation to aluminum toxicity and phosphorus deficiency. Front. Plant Sci. 2022, 13, 947986. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Wang, N.; Xing, C.; Wang, F.; Wu, K.; Du, X. Immobilization of aluminum with mucilage secreted by root cap and root border cells is related to aluminum resistance in Glycine max L. Environ. Sci. Pollut. Res. 2013, 20, 8924–8933. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Wang, F.; Li, R.; Zhang, S.; Wang, N.; Xu, G. Response and tolerance of root border cells to aluminum toxicity in soybean seedlings. J. Inorg. Biochem. 2011, 105, 966–971. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Cheng, Y.; Xian, P.; Lin, R.; Xia, Q.; He, X.; Liang, Q.; Lian, T.; Ma, Q.; Nian, H. Fine-mapping QTLs and the validation of candidate genes for aluminum tolerance using a high-density genetic map. Plant Soil. 2019, 444, 119–137. [Google Scholar] [CrossRef]
- Liu, G.; Li, D.; Mai, H.; Lin, X.; Lu, X.; Chen, K.; Wang, R.; Riaz, M.; Tian, J.; Liang, C. GmSTOP1-3 regulates flavonoid synthesis to reduce ROS accumulation and enhance aluminum tolerance in soybean. J. Hazard. Mater. 2024, 480, 136074. [Google Scholar] [CrossRef]
- Wu, W.; Lin, Y.; Chen, Q.; Peng, W.; Peng, J.; Tian, J.; Liang, C.; Liao, H. Functional conservation and divergence of soybean GmSTOP1 members in proton and aluminum tolerance. Front. Plant Sci. 2018, 9, 570. [Google Scholar] [CrossRef]
- Lin, Y.; Liu, G.; Xue, Y.; Guo, X.; Luo, J.; Pan, Y.; Chen, K.; Tian, J.; Liang, C. Liang, Functional characterization of aluminum (Al)-responsive membrane-bound NAC transcription factors in soybean roots. Int. J. Mol. Sci. 2021, 22, 12854. [Google Scholar] [CrossRef]
- Shu, W.; Zhou, Q.; Xian, P.; Cheng, Y.; Lian, T.; Ma, Q.; Zhou, Y.; Li, H.; Nian, H. GmWRKY81 encoding a WRKY transcription factor enhances aluminum tolerance in soybean. Int. J. Mol. Sci. 2022, 23, 6518. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Conery, S.S. The evolutionary fate and consequences of duplicate genes. Science 2020, 290, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Xiong, S.; Kong, X.; Chen, G.; Tian, L.; Qian, D.; Zhu, Z.; Qu, L.Q. Metallochaperone OsHIPP9 is involved in the retention of cadmium and copper in rice. Plant Cell Environ. 2023, 46, 1946–1961. [Google Scholar] [CrossRef]
- Che, J.; Tsutsui, T.; Yokosho, K.; Yamaji, N.; Ma, J.F. Functional characterization of an aluminum (Al)-inducible transcription factor, ART2, revealed a different pathway for Al tolerance in rice. New Phytol. 2018, 220, 209–218. [Google Scholar] [CrossRef]
- Guo, Q.; Zhu, S.; Lai, T.; Tian, C.; Hu, M.; Lu, X.; Xue, Y.; Liang, C.; Tian, J. A Phosphate-Starvation Enhanced Purple Acid Phosphatase, GmPAP23 Mediates Intracellular Phosphorus Recycling and Yield in Soybean. Plant Cell Environ. 2025, 22. [Google Scholar] [CrossRef]
- Liu, G.; Chen, Q.; Li, D.; Mai, H.; Zhou, Y.; Lin, M.; Feng, X.; Lin, X.; Lu, X.; Chen, K.; et al. GmSTOP1-3 Increases Soybean Manganese Accumulation Under Phosphorus Deficiency by Regulating GmMATE2/13 and GmZIP6/GmIREG3. Plant Cell Environ. 2025, 48, 1812–1828. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.L.; Zhang, X.H.; Zhong, L.J.; Wang, X.Y.; Jin, L.S.; Lyu, S.H. One-step generation of composite soybean plants with transgenic roots by Agrobacterium rhizogenes-mediated transformation. BMC Plant Biol. 2020, 12, 208. [Google Scholar] [CrossRef]
- Wang, L.; Wang, W.; Miao, Y.; Peters, M.; Schultze-Kraft, R.; Liu, G.; Chen, Z. Development of transgenic composite Stylosanthes plants to study root growth regulated by a β-expansin gene, SgEXPB1, under phosphorus deficiency. Plant Cell Rep. 2023, 42, 575–585. [Google Scholar] [CrossRef]
- Shi, Y.; Jiang, N.; Wang, M.; Du, Z.; Chen, J.; Huang, Y.; Li, M.; Jin, Y.; Li, J.; Wan, J.; et al. OsHIPP17 is involved in regulating the tolerance of rice to copper stress. Front. Plant Sci. 2023, 14, 1183445. [Google Scholar]
- Li, J.Y.; Liu, J.; Dong, D.; Jia, X.; McCouch, S.R.; Kochian, L.V. Natural variation underlies alterations in Nramp aluminum transporter (NRAT1) expression and function that play a key role in rice aluminum tolerance. Proc. Natl. Acad. Sci. USA 2014, 111, 6503–6508. [Google Scholar] [CrossRef]
- Larsen, P.B.; Cancel, J.; Rounds, M.; Ochoa, V. Arabidopsis ALS1 encodes a root tip and stele localized half type ABC transporter required for root growth in an aluminum toxic environment. Planta 2007, 225, 1447–1458. [Google Scholar] [CrossRef]
- Shi, Y.; Jiang, W.; Li, M.; Jiang, N.; Huang, Y.; Wang, M.; Du, Z.; Chen, J.; Li, J.; Wu, L.; et al. Metallochaperone protein OsHIPP17 regulates the absorption and translocation of cadmium in rice (Oryza sativa L.). Int. J. Biol. Macromol. 2023, 245, 125607. [Google Scholar] [CrossRef]
- Zhou, L.; Ye, L.; Pang, B.; Hou, Y.; Yu, J.; Du, X.; Gu, L.; Wang, H.; Zhu, B. Overexpression of ApHIPP26 from the Hyperaccumulator Arabis paniculata confers enhanced cadmium tolerance and accumulation to Arabidopsis thaliana. Int. J. Mol. Sci. 2023, 24, 15052. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Darzentas, N. Circoletto: Visualizing sequence similarity with circos. Bioinformatics 2010, 26, 2620–2621. [Google Scholar] [CrossRef]
- Zhao, L.; Cui, J.; Cai, Y.; Yang, S.; Liu, J.; Wang, W.; Gai, J.; Hu, Z.; Li, Y. Comparative transcriptome analysis of two contrasting soybean varieties in response to aluminum toxicity. Int. J. Mol. Sci. 2020, 21, 4316. [Google Scholar] [CrossRef]
- Lin, Y.; Liu, G.; Liu, P.; Chen, Q.; Guo, X.; Lu, X.; Cai, Z.; Sun, L.; Liu, J.; Chen, K.; et al. Border-like cell formation mediated by SgPG1 confers aluminum resistance in Stylosanthes guianensis. Plant J. 2024, 120, 1605–1624. [Google Scholar] [CrossRef] [PubMed]
- Li, C.X.; Yan, J.Y.; Ren, J.Y.; Sun, L.; Xu, C.; Li, G.X.; Ding, Z.J.; Zheng, S.J. A WRKY transcription factor confers aluminum tolerance via regulation of cell wall modifying genes. J. Integr. Plant Biol. 2020, 62, 1176–1192. [Google Scholar] [CrossRef]
- Li, J.; Liu, L.; Wang, L.; Rao, I.M.; Wang, Z.; Chen, Z. AcEXPA1, an alpha-expansin gene, participates in the aluminum tolerance of carpetgrass (Axonopus compressus) through root growth regulation. Plant Cell Rep. 2024, 43, 159. [Google Scholar] [CrossRef]
- Zhang, Z.; Mo, X.; Zhao, H.; Lu, X.; Fan, S.; Huang, X.; Mai, H.; Liao, H.; Zhang, Y.; Liang, C.; et al. Crystal structure and function of a phosphate starvation responsive protein phosphatase, GmHAD1-2 regulating soybean root development and flavonoid metabolism. New Phytol. 2024, 244, 2396–2412. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Liu, Q.; Yu, Z.; Wang, C.; Mai, H.; Liu, G.; Li, R.; Pang, G.; Chen, D.; Liu, H.; et al. eIF4E1 Regulates Arabidopsis embryo development and root growth by interacting with RopGEF7. Front. Plant Sci. 2022, 13, 938476. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Tian, J.; Liang, C.; Wang, T.; Lu, X. Characterization of the Soybean (Glycine max) Heavy-Metal-Associated Isoprenylated Plant Protein (HIPP) Gene Family in Response to Aluminum. Plants 2025, 14, 3582. https://doi.org/10.3390/plants14233582
Li J, Tian J, Liang C, Wang T, Lu X. Characterization of the Soybean (Glycine max) Heavy-Metal-Associated Isoprenylated Plant Protein (HIPP) Gene Family in Response to Aluminum. Plants. 2025; 14(23):3582. https://doi.org/10.3390/plants14233582
Chicago/Turabian StyleLi, Jifu, Jiang Tian, Cuiyue Liang, Tianqi Wang, and Xing Lu. 2025. "Characterization of the Soybean (Glycine max) Heavy-Metal-Associated Isoprenylated Plant Protein (HIPP) Gene Family in Response to Aluminum" Plants 14, no. 23: 3582. https://doi.org/10.3390/plants14233582
APA StyleLi, J., Tian, J., Liang, C., Wang, T., & Lu, X. (2025). Characterization of the Soybean (Glycine max) Heavy-Metal-Associated Isoprenylated Plant Protein (HIPP) Gene Family in Response to Aluminum. Plants, 14(23), 3582. https://doi.org/10.3390/plants14233582






