Grape Leaf Cultivar Identification in Complex Backgrounds with an Improved MobileNetV3-Small Model
Abstract
1. Introduction
Research Contributions
2. Related Work
3. Materials and Experimental Approaches
3.1. Dataset
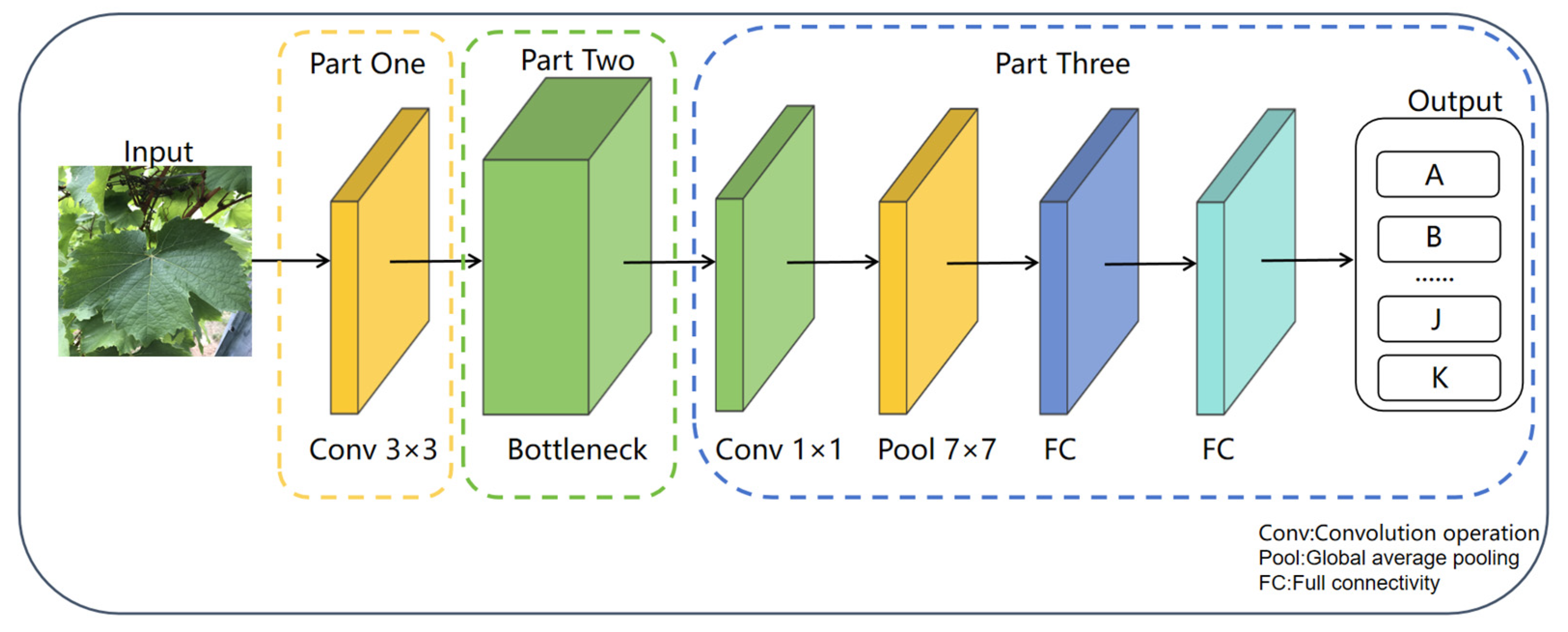
3.2. MobileNetV3-Small Network
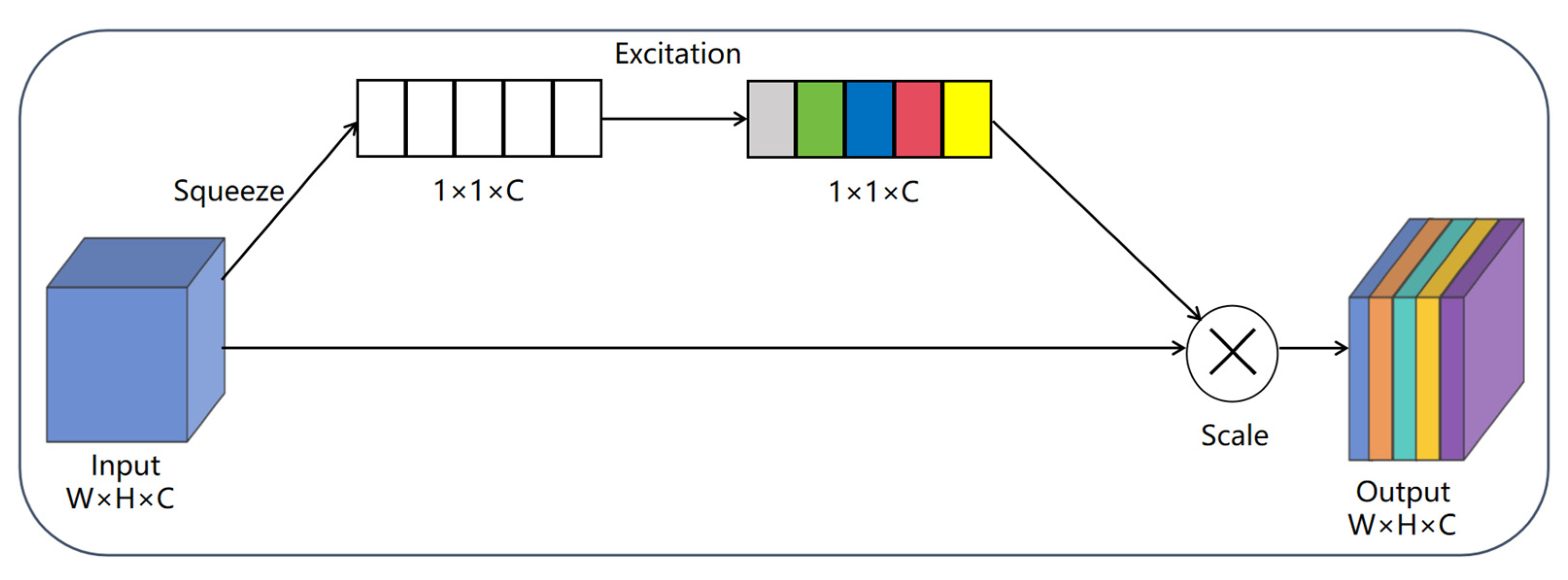
3.2.1. Squeeze-And-Excitation (SE) Attention Mechanism
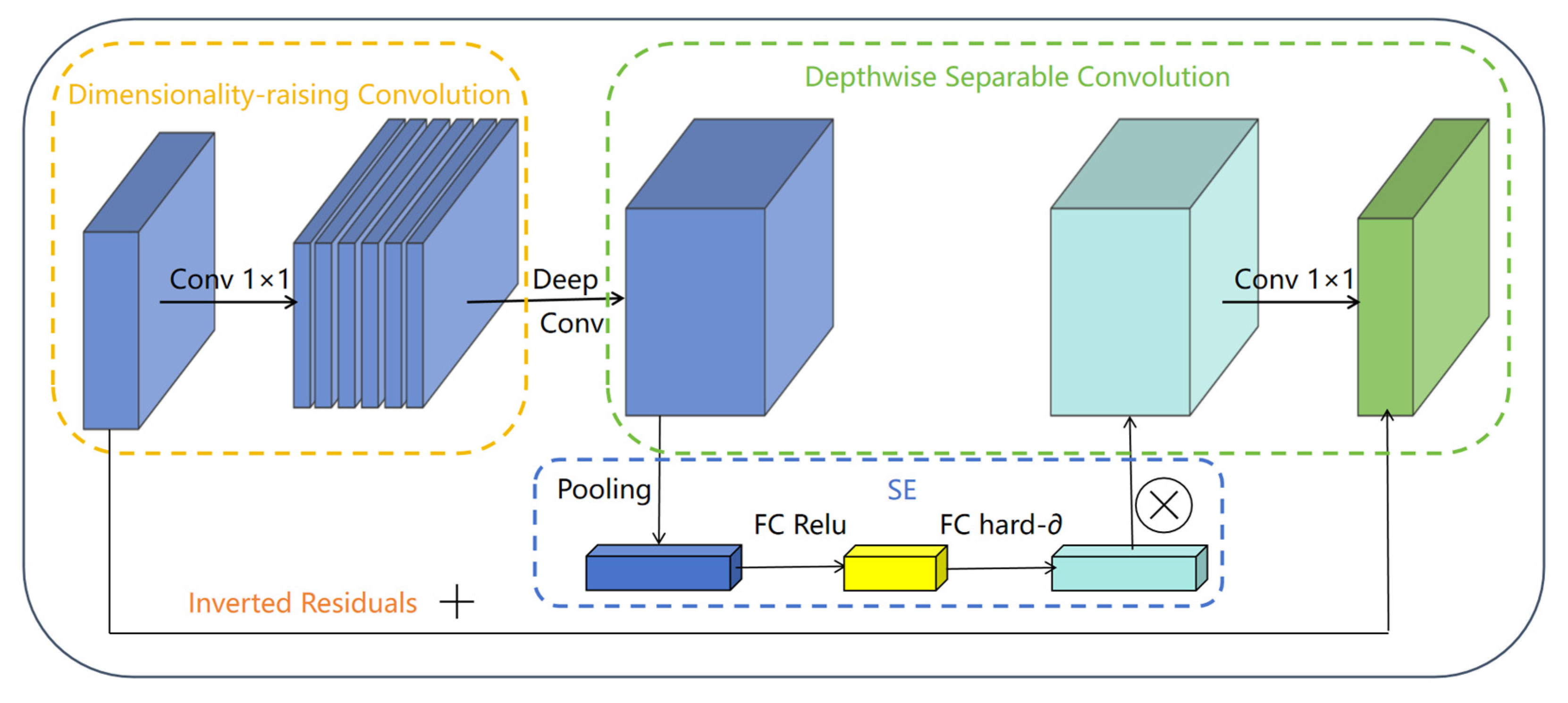
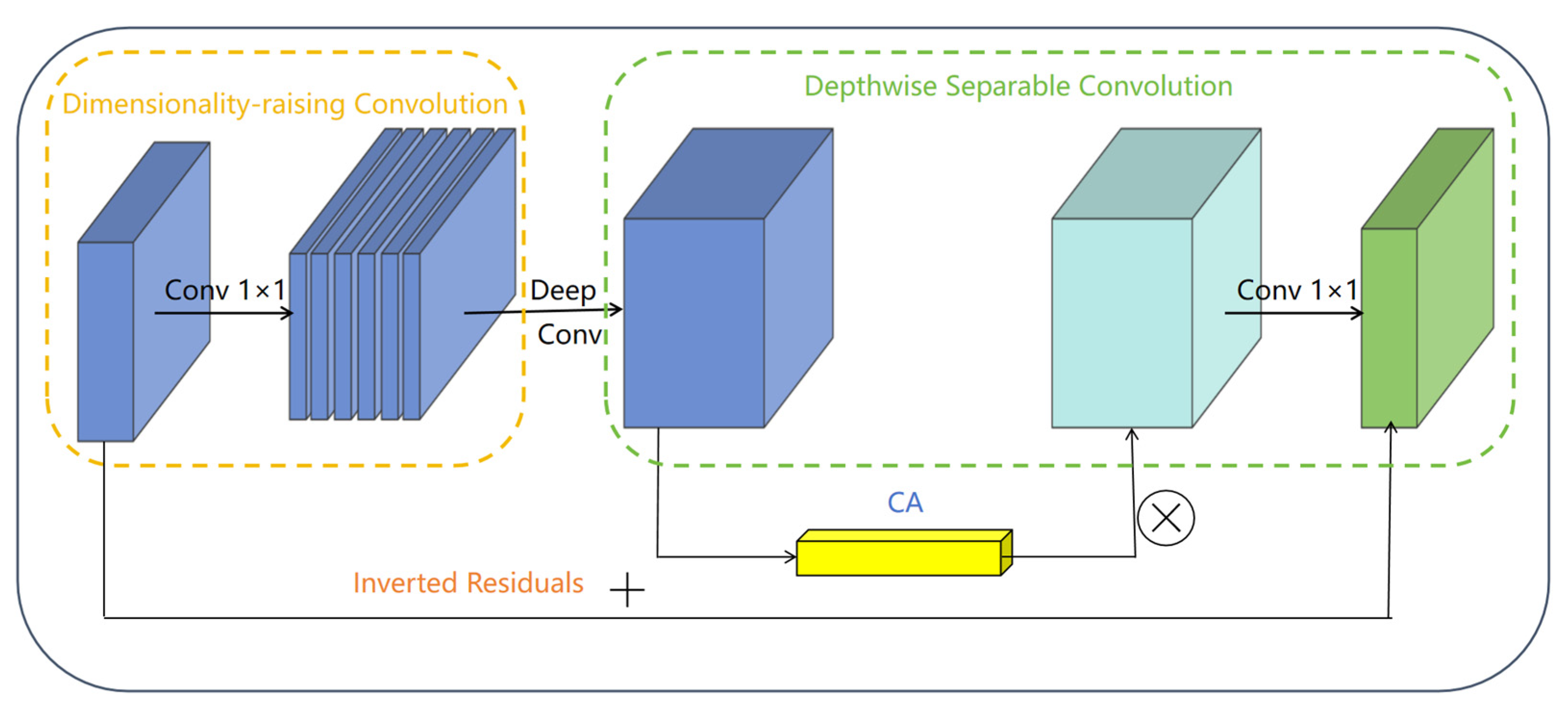
3.2.2. Bottleneck Module
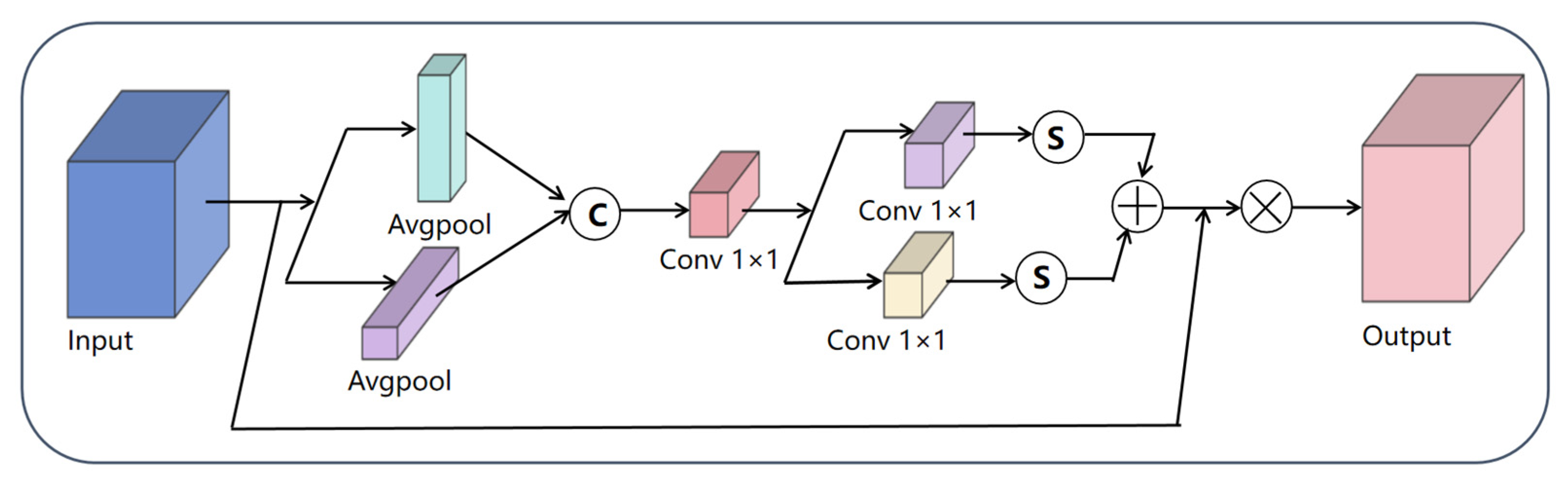
3.3. Coordinate Attention Mechanism
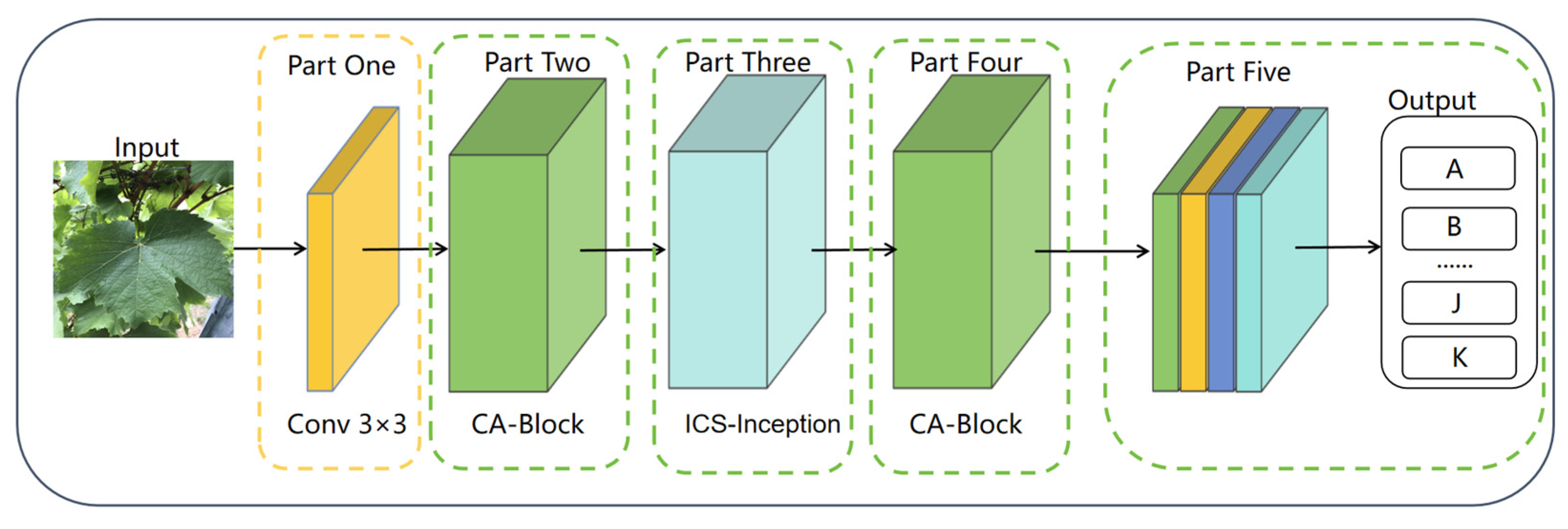
3.4. Key Innovations of the Proposed ICS-MS Model
- CA Mechanism Integration: The incorporation of CA mechanism enhances spatial perception of leaf serrations and vein patterns through coordinate encoding. This modification significantly improves morphological discrimination while reducing parameters, achieving better lightweight performance;
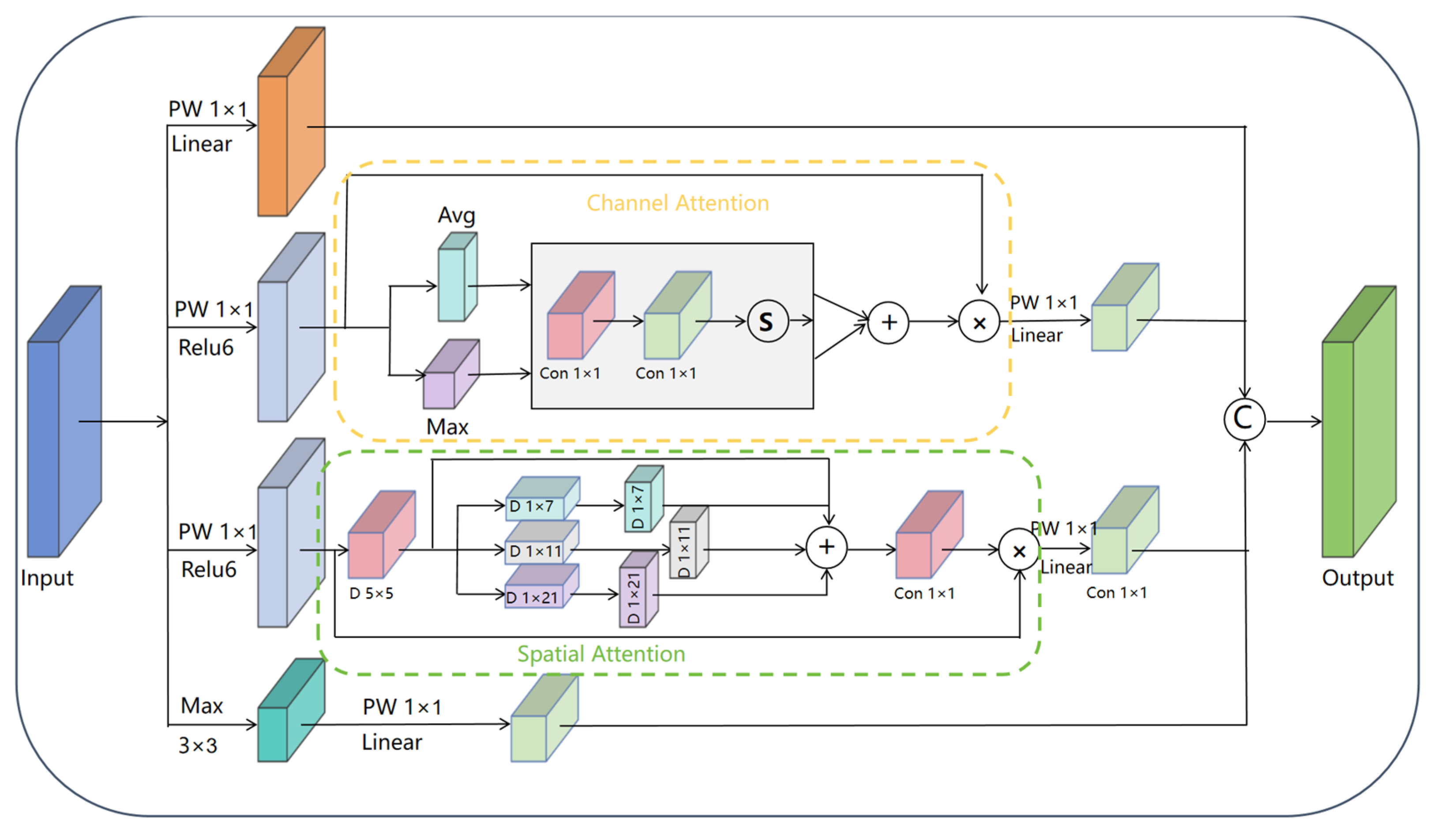
- ICS-Inception Architecture: A novel ICS-Inception structure employs parallel 1 × 1 convolutions to simultaneously capture and fuse both macro-morphological and micro-textural features. This design effectively addresses the complex variations in grape leaves and enhances fine-grained feature discrimination;
- Joint Supervision Strategy: The model employs a combination of cross-entropy and center-based loss terms to refine the distribution of learned features. Such a composite optimization scheme enhances both the robustness and generalization of the network, making it well suited for deployment in resource-limited agricultural environments.
3.4.1. CA-Block Module
3.4.2. The ICS-Inception Structure
3.4.3. Joint Supervised Loss Function
3.5. Metrics for Evaluation
4. Results and Analysis
4.1. Experimental Setup
4.2. Baseline Model Structure Improvement Experiments
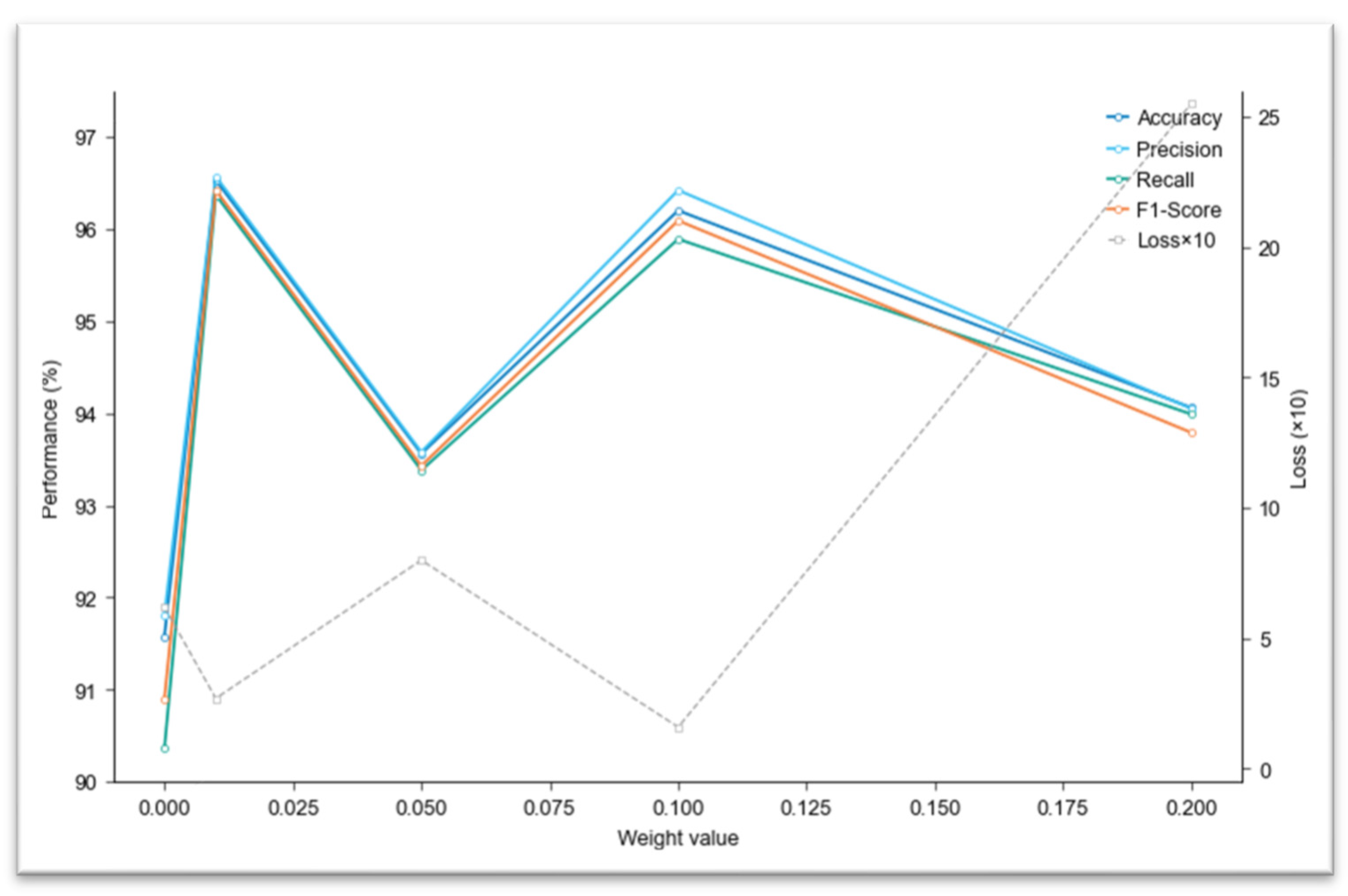
4.3. Loss Function Optimization Experiments
4.4. Module Contribution Analysis
4.5. Comparative Evaluation of Various Models’ Performance
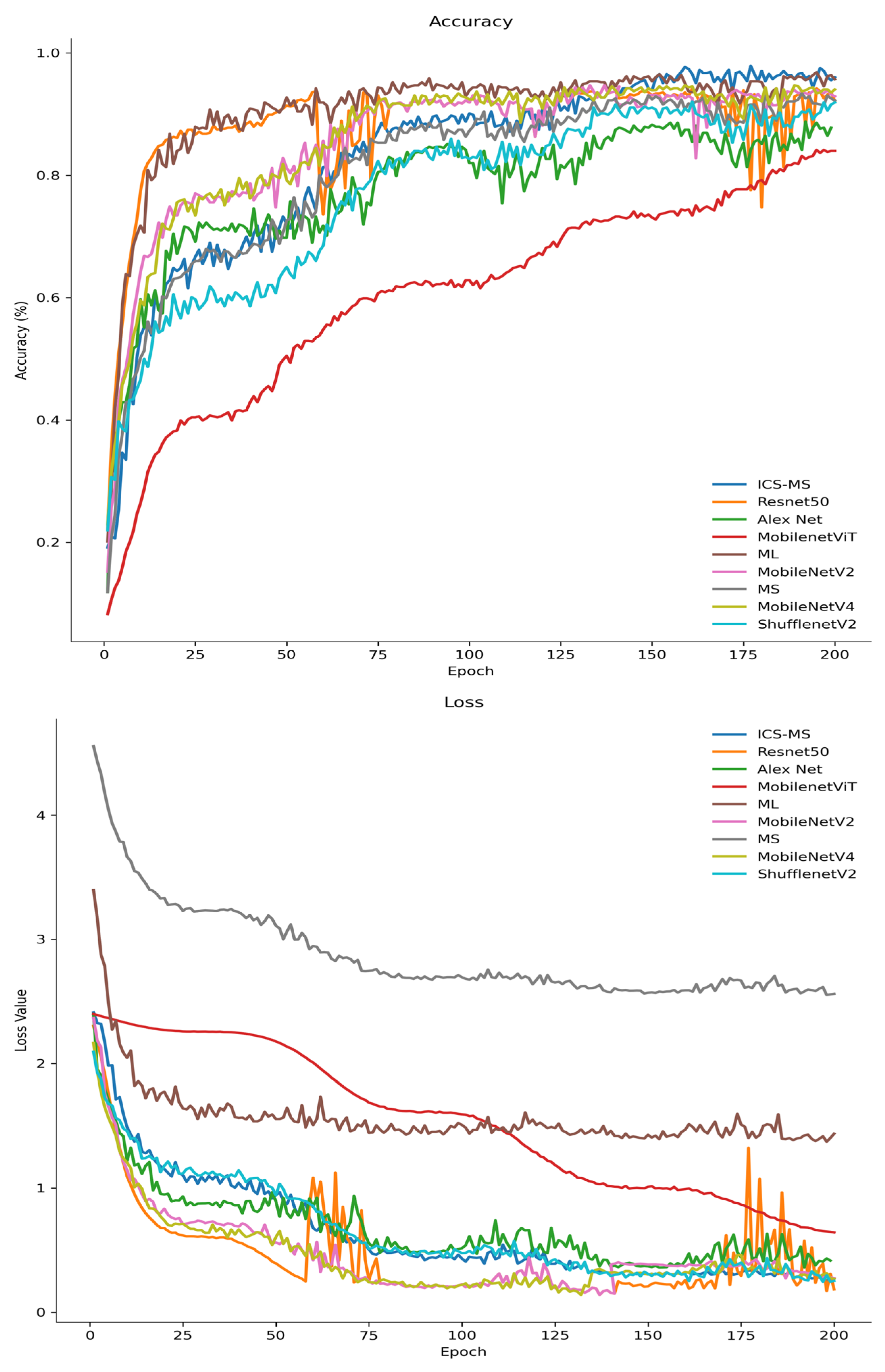
4.5.1. Comparison with State-of-the-Art Lightweight Models
4.5.2. Comparison with Related Works in the Literature
4.6. Computational Efficiency and Model Compactness
4.7. Summary of Experimental Findings
- Enhanced Accuracy: The ICS-MS model outperformed both lightweight models such as MobileNetV3 and more complex architectures like ResNet50 in terms of test-set accuracy, highlighting its exceptional ability to discern subtle morphological features essential for accurate classification.
- Efficient Model Design: By incorporating the CA-Block for precise feature selection and the ICS-Inception module for multi-scale feature aggregation, the ICS-MS model significantly reduced the number of parameters by over 95% compared to ResNet50 and by more than 20% compared to baseline models. This optimization resulted in a lightweight design without compromising performance.
- Improved Robustness: The joint supervision loss function used by the ICS-MS model strengthened its performance in dealing with class-imbalanced scenarios, thereby enhancing the model’s robustness and generalization capabilities.
5. Discussion
5.1. Model Adaptability Analysis
5.2. Model Constraints
5.3. Practical Implications and Positive Impacts
6. Conclusions
- (1)
- A CA mechanism to enhance spatial feature capture, realizing lightweight and efficient feature recalibration;
- (2)
- An ICS-Inception structure for multi-dimensional feature extraction with parameter reduction, effectively fusing macro and micro features of grape leaves;
- (3)
- A joint loss function combining cross-entropy and center loss to optimize feature space distribution, improving the discriminability of similar varieties.
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, W.; Garmali, M.; Li, H.P.; Wang, M.; Han, S.A.; Xie, H.; Turup, M.; Zhou, X.W.; Caik Aaasimu, A.; Pan, M.Q. Packing and Edibility Characteristics Evaluation of 80 Grape Cultivars’ Leaves. Chin. Agric. Sci. Bull. 2022, 38, 130–137. [Google Scholar] [CrossRef]
- Alston, J.; Sambucci, O. Grapes in the World Economy. In Handbook of Agricultural Economics, 2nd ed.; Springer: Cham, Switzerland, 2019; Volume 5. [Google Scholar] [CrossRef]
- Eli-Chukwu, N.C. Applications of Artificial Intelligence in Agriculture: A Review. Eng. Technol. Appl. Sci. Res. 2019, 9, 4377–4383. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.; Wang, J.; Wang, L.; Li, B.; Liu, S.; Sun, X.; Ma, Z. Fungal Chromatin Remodeler Isw1 Modulates Translation via Regulating tRNA Transcription. Nucleic Acids Res. 2025, 53, gkaf225. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, S.; Gan, P.; Xie, H.T.; Li, C.; Tang, T.X.; Hu, Q.; Zhu, Z.H.; Zhang, Z.K.; Zhang, J.S.; Zhu, Y.S.; et al. Virulence Effectors Encoded in the Rice Yellow Dwarf Phytoplasma Genome Participate in Pathogenesis. Plant Physiol. 2025, 197, kiae601. [Google Scholar] [CrossRef]
- Chen, X.; Li, P.; Liu, H.; Chen, X.; Huang, J.; Luo, C.; Li, G.; Hsiang, T.; Collinge, D.B.; Zheng, L. A Novel Transcription Factor UvCGBP1 Regulates Development and Virulence of Rice False Smut Fungus Ustilaginoidea virens. Virulence 2021, 12, 1563–1579. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lv, K.; Xie, Y.; Yu, Q.; Zhang, N.; Zheng, Q.; Wu, J.; Zhang, J.; Li, J.; Zhao, H.; Xu, W. Amur Grape VaMYB4a-VaERF054-Like Module Regulates Cold Tolerance Through a Regulatory Feedback Loop. Plant Cell Environ. 2025, 48, 1130–1148. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Bing, S.Y.; Ji, Y.H.; Yan, B.B.; Xu, J.P. Grading Method of Fresh Cut Rose Flowers Based on Improved YOLOv8s. Smart Agric. 2024, 6, 118–127. [Google Scholar]
- Gill, H.S.; Murugesan, G.; Mehbodniya, A.; Sajja, G.S.; Gupta, G.; Bhatt, A. Fruit Type Classification Using Deep Learning and Feature Fusion. Comput. Electron. Agric. 2023, 211, 107990. [Google Scholar] [CrossRef]
- Kiran, K.; Kaur, A. Deep Learning-Based Plant Species Identification Using VGG16 and Transfer Learning: Enhancing Accuracy in Biodiversity and Agricultural Applications. In Proceedings of the 3rd International Conference on Intelligent Data Communication Technologies and Internet of Things (IDCIoT), Bengaluru, India, 5–7 February 2025; pp. 1538–1543. [Google Scholar] [CrossRef]
- Devi, V.; Govu, P.; Sande, R.; Nakka, P. How Deep Learning Identifies and Learns Aspects of Plant for Classification. ITM Web Conf. 2025, 74, 01013. [Google Scholar] [CrossRef]
- Aissaoui, A.; Aloui, I.; Boudibi, S.; Khomri, Z.E.; Foughalia, A. Deep Learning Approaches for Plant Diseases Identification and Classification: A Comprehensive Review. In Proceedings of the 8th International Conference on Image and Signal Processing and their Applications (ISPA), Biskra, Algeria, 21–22 April 2024; pp. 1–11. [Google Scholar] [CrossRef]
- Rajab, M.; Abdullatif, F. Classification of Grapevine Leaves Images Using VGG-16 and VGG-19 Deep Learning Net-s. TELKOMNIKA (Telecommun. Comput. Electron. Control) 2024, 22, 445–453. [Google Scholar] [CrossRef]
- Nijhum, S.M.R.H.; Rahman, M.M. Enhancing Bangladeshi Plants Classification Using Transfer Learning and CNN-Based Architecture. In Proceedings of the 6th International Conference on Electrical Engineering and Information & Communication Technology (ICEEICT), Dhaka, Bangladesh, 2–4 May 2024; pp. 746–751. [Google Scholar] [CrossRef]
- Elnemr, H. Convolutional Neural Network Architecture for Plant Seedling Classification. Int. J. Adv. Comput. Sci. Appl. 2019, 10, 319–325. [Google Scholar] [CrossRef]
- Xu, T.B.; Yang, P.P.; Zhang, X.Y.; Liu, C.L. Lightweight Net: Toward Fast and Lightweight Convolutional Neural Networks via Architecture Distillation. Pattern Recognit. 2019, 88, 272–284. [Google Scholar] [CrossRef]
- Wang, A.; Chen, H.; Lin, Z.; Han, J.; Ding, G. Rep ViT: Revisiting Mobile CNN From ViT Perspective. In Proceedings of the 2024 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 16–22 June 2024; pp. 15909–15920. [Google Scholar] [CrossRef]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar] [CrossRef]
- Koklu, M.; Unlersen, M.F.; Ozkan, I.A.; Aslan, M.F.; Sabanci, K. A CNN-SVM Study Based on Selected Deep Features for Grapevine Leaves Classification. Measurement 2022, 188, 110425. [Google Scholar] [CrossRef]
- Elfatimi, E.; Eryigit, R.; Elfatimi, L. Beans Leaf Diseases Classification Using MobileNet Models. IEEE Access 2022, 10, 9471–9482. [Google Scholar] [CrossRef]
- Elkassar, A. Deep Learning Based Grapevine Leaf Classification Using Augmented Images and Multi-Classifier Fusion for Improved Accuracy and Precision. In Proceedings of the 14th International Conference on Electrical Engineering (ICEENG), Cairo, Egypt, 21–23 May 2024; pp. 190–192. [Google Scholar] [CrossRef]
- Pereira, C.S.; Morais, R.; Reis, M.J.C.S. Deep Learning Techniques for Grape Plant Species Identification in Natural Images. Sensors 2019, 19, 4850. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.; Hama, H.; Jalal, S.; Ahmed, M. Deep Learning in Grapevine Leaves Varieties Classification Based on Dense Convolutional Network. J. Image Graph. 2023, 11, 98–103. [Google Scholar] [CrossRef]
- Dogan, G.; Imak, A.; Ergen, B.; Sengur, A. A New Hybrid Approach for Grapevine Leaves Recognition Based on ESRGAN Data Augmentation and GASVM Feature Selection. Neural Comput. Appl. 2024, 36, 7669–7683. [Google Scholar] [CrossRef]
- Tian, Y.; Zhao, C.; Zhang, T.; Wu, H.; Zhao, Y. Recognition Method of Cabbage Heads at Harvest Stage under Complex Background Based on Improved YOLOv8n. Agriculture 2024, 14, 1125. [Google Scholar] [CrossRef]
- Wang, C.; Wang, Z.; Chen, L.; Liu, W.; Wang, X.; Cao, Z.; Zhao, J.; Zou, M.; Li, H.; Yuan, W.; et al. Intelligent Identification of Tea Plant Seedlings Under High-Temperature Conditions via YOLOv11-MEIP Model Based on Chlorophyll Fluorescence Imaging. Plants 2025, 14, 1965. [Google Scholar] [CrossRef]
- Nishankar, S.; Mithuran, T.; Thuseethan, S.; Sebastian, Y.; Yeo, K.C.; Shanmugam, B. TOM-SSL: Tomato Disease Recognition Using Pseudo-Labeling-Based Semi-Supervised Learning. AgriEngineering 2025, 7, 248. [Google Scholar] [CrossRef]
- Vlah, M. Grapevine Leaves [Data Set]; Kaggle: San Francisco, CA, USA, 2021. [Google Scholar] [CrossRef]
- Abd Elaziz, E.; Al-qaness, M.; Dahou, A.; Alsamhi, S.; Abualigah, L.; Ibrahim, R.; Ewees, A. Evolution toward Intelligent Communications: Impact of Deep Learning Applications on the Future of 6G Technology. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2023, 14, e1521. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-Excitation Networks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar] [CrossRef]
- Liang, X.R.; Liang, J.F.; Yin, T.; Tang, X.Y. A Lightweight Method for Face Expression Recognition Based on Improved MobileNetV3. IET Image Process. 2023, 17, 2375–2384. [Google Scholar] [CrossRef]
- Hou, Q.; Zhou, D.; Feng, J. Coordinate Attention for Efficient Mobile Network Design. In Proceedings of the 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA, 20–25 June 2021; pp. 13708–13717. [Google Scholar] [CrossRef]

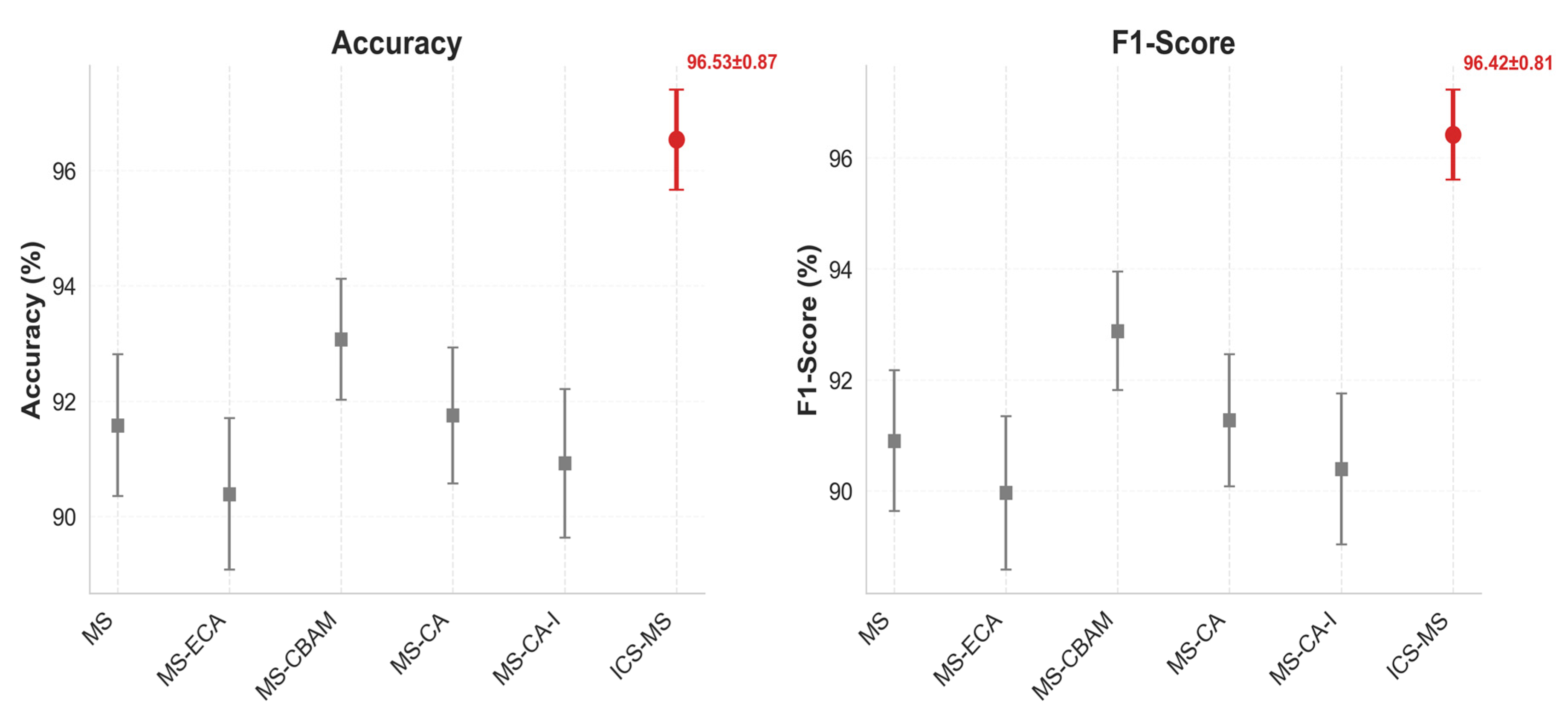
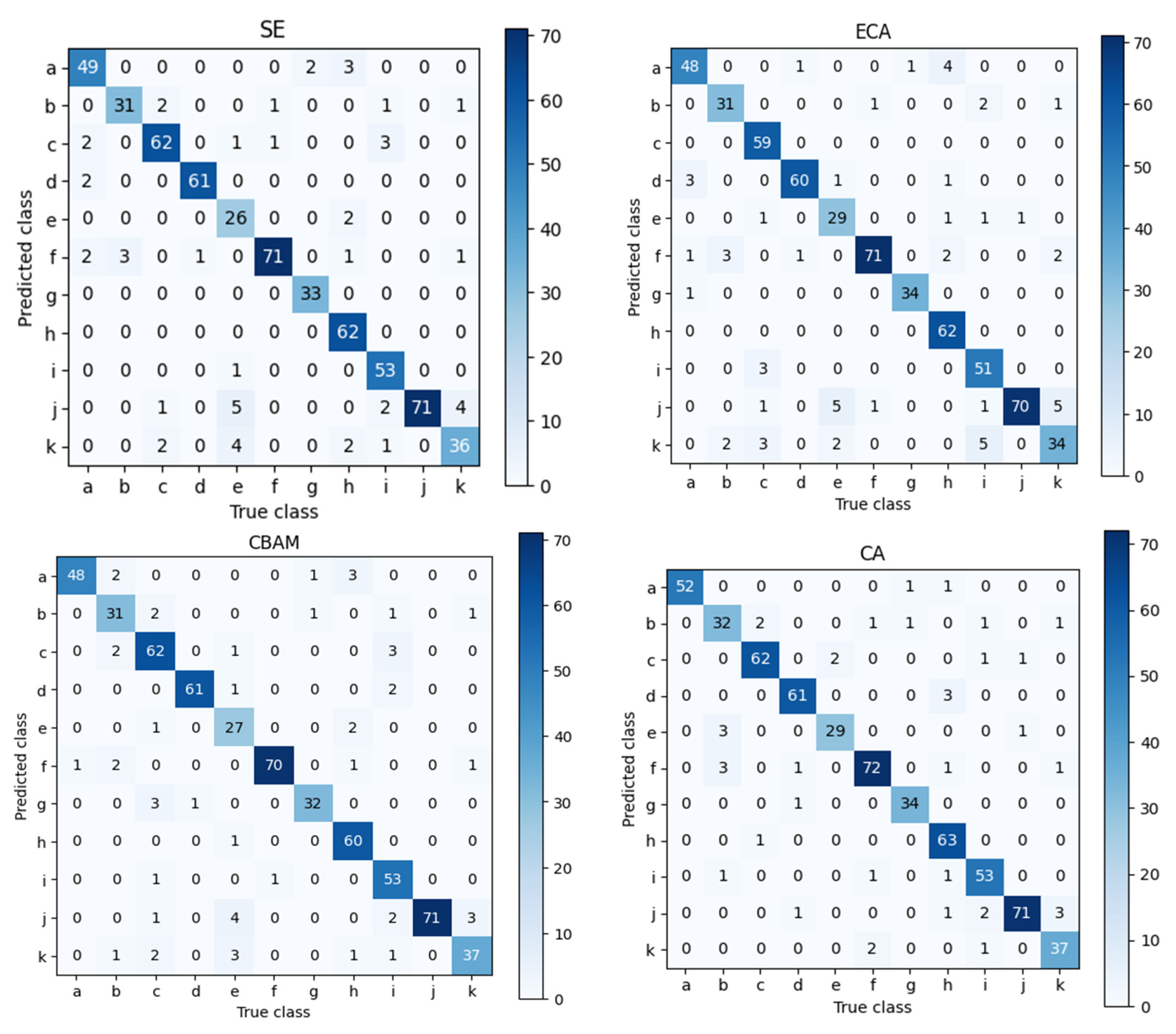
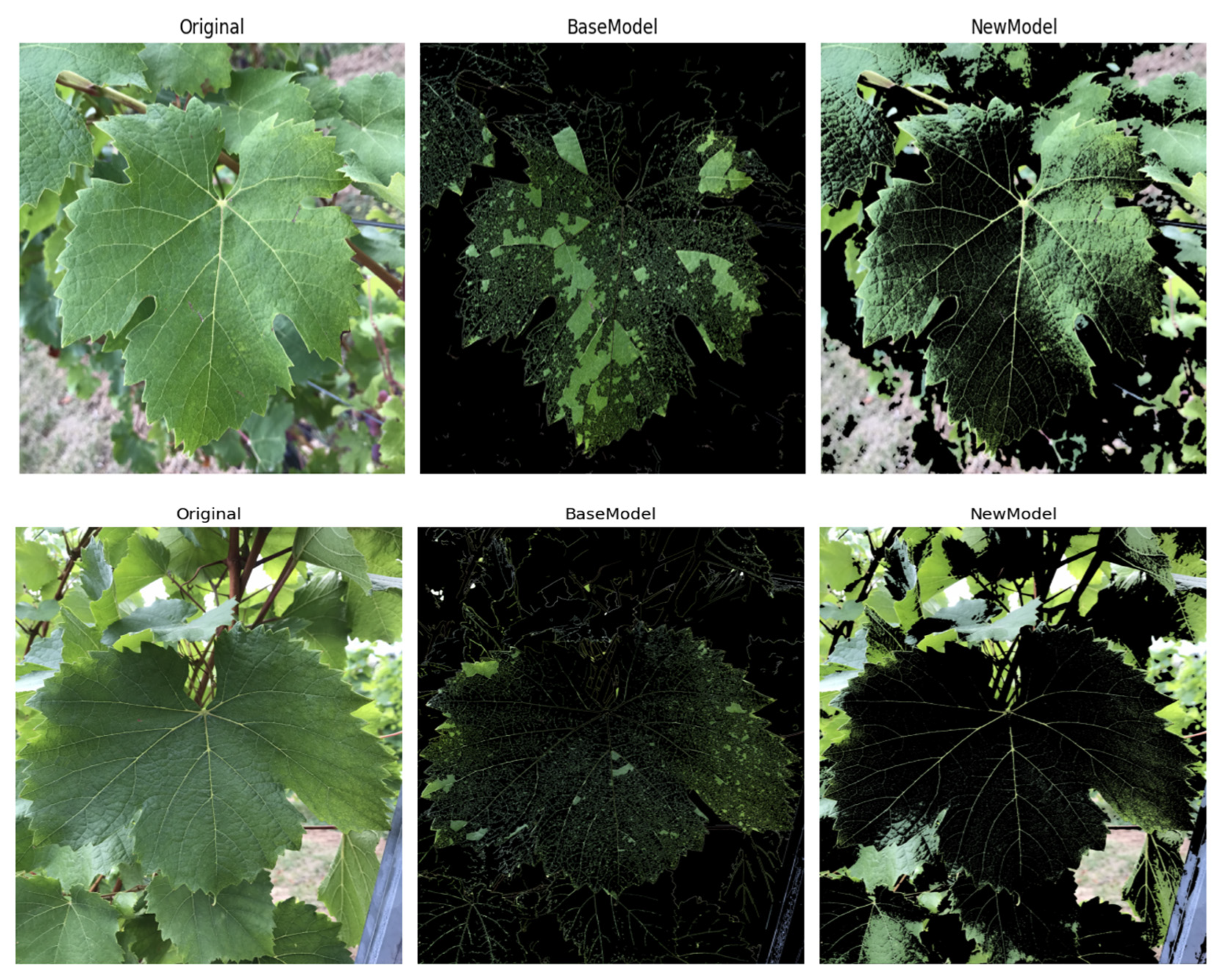
| Model | Parameters (M) | Accuracy (%) | F1-Score (%) | Precision (%) | Recall (%) |
|---|---|---|---|---|---|
| MS | 1.53 | 91.58 ± 1.23 | 90.90 ± 1.27 | 91.81 ± 1.15 | 90.36 ± 1.32 |
| MS-ECA | 1.52 ↓ | 90.39 ± 1.31 | 89.96 ± 1.38 | 90.57 ± 1.28 | 89.76 ± 1.45 |
| MS-CBAM | 1.52 ↓ | 93.07 ± 1.05 | 92.88 ± 1.07 | 92.96 ± 0.98 | 92.46 ± 1.12 |
| MS-CA | 1.37 ↓ | 91.75 ± 1.18 | 91.27 ± 1.19 | 91.93 ± 1.09 | 90.99 ± 1.25 |
| MS-CA-I | 1.07 ↓ | 90.92 ± 1.29 | 90.39 ± 1.36 | 90.38 ± 1.34 | 90.59 ± 1.41 |
| ICS-MS | 1.17 ↓ | 96.53 ± 0.87 | 96.42 ± 0.81 | 96.57 ± 0.79 | 96.37 ± 0.85 |
| Weight | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) | Loss |
|---|---|---|---|---|---|
| 0 | 91.93 | 91.42 | 91.61 | 91.35 | 0.62 |
| 0.01 | 96.53 | 96.57 | 96.37 | 96.42 | 0.27 |
| 0.05 | 93.56 | 93.58 | 93.38 | 93.43 | 0.80 |
| 0.1 | 96.20 | 96.42 | 95.89 | 96.09 | 0.16 |
| 0.2 | 94.06 | 94.05 | 93.99 | 93.79 | 2.55 |
| Model | Parameters (M) | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) |
|---|---|---|---|---|---|
| MS (CE-only) | 1.53 | 91.58 | 91.81 | 90.36 | 90.90 |
| MS-CA (CE-only) | 1.37 ↓ | 91.75 ↑ | 91.93 ↑ | 90.99 ↑ | 91.27 ↑ |
| ICS-MS (CE-only) | 1.12 ↓ | 91.93 ↑ | 91.42 ↑ | 91.61 ↑ | 91.35 ↑ |
| ICS-MS (Joint Loss) | 1.17 ↓ | 96.53 ↑ | 96.57 ↑ | 96.37 ↑ | 96.42 ↑ |
| Model | Parameters (M) | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) |
|---|---|---|---|---|---|
| Resnet50 | 23.53 | 93.78 ± 1.02 | 93.06 | 93.28 | 93.49 |
| Alex Net | 14.6 | 88.78 ± 1.35 | 88.30 | 88.51 | 88.28 |
| MobileNetVit | 4.95 | 84.16 ± 1.42 | 86.36 | 80.38 | 80.09 |
| ML | 4.2 | 96.20 ± 0.93 | 96.42 | 95.89 | 96.09 |
| MobileNetV4 | 2.5 | 94.51 ± 1.08 | 94.38 | 94.33 | 94.46 |
| EfficientNet-Lite1 | 2.4 | 93.87 ± 1.05 | 93.92 | 93.56 | 93.74 |
| MobileNetV2 | 2.24 | 93.89 ± 1.12 | 93.92 | 93.28 | 93.76 |
| MS | 1.53 | 91.58 ± 1.23 | 91.81 | 90.36 | 90.81 |
| EfficientNet-Lite0 | 1.3 | 92.45 ± 1.17 | 92.68 | 92.13 | 92.35 |
| ShufflenetV2 | 1.26 | 92.08 ± 1.09 | 92.22 | 91.79 | 91.88 |
| ICS-MS | 1.17 | 96.53 ± 0.87 | 96.57 | 96.37 | 96.42 |
| Model | Task and Dataset Details | Parameters | Metric |
|---|---|---|---|
| MobileNetV2 [21] | Grape leaf classification (5 cultivars) | - | Accuracy: 96.00% |
| AlexNet [22] | Grape leaf classification (2 vineyards) | 60 | Accuracy: 77.30% |
| DenseNet-30 [23] | Grape leaf classification (5 cultivars, 500 images) | 12 | Accuracy: 98.00% |
| ESRGAN + GASVM [24] | Grape leaf classification (small-sample) | - | Accuracy: ~94.00% |
| YOLOv8n-Cabbage [25] | Cabbage detection (complex background) | 4.8 | mAP50: 94.5% |
| YOLOv11-MEIP [26] | Tea seedling recognition (high temperature) | mAP50: 99.46% | |
| TOM-SSL [27] | Tomato disease classification (10% labeled data) | Accuracy: 72.51% | |
| ICS-MS (Our Study) | Grape leaf classification (11 cultivars) | 1.17 | Accuracy: 96.53 ± 0.87% |
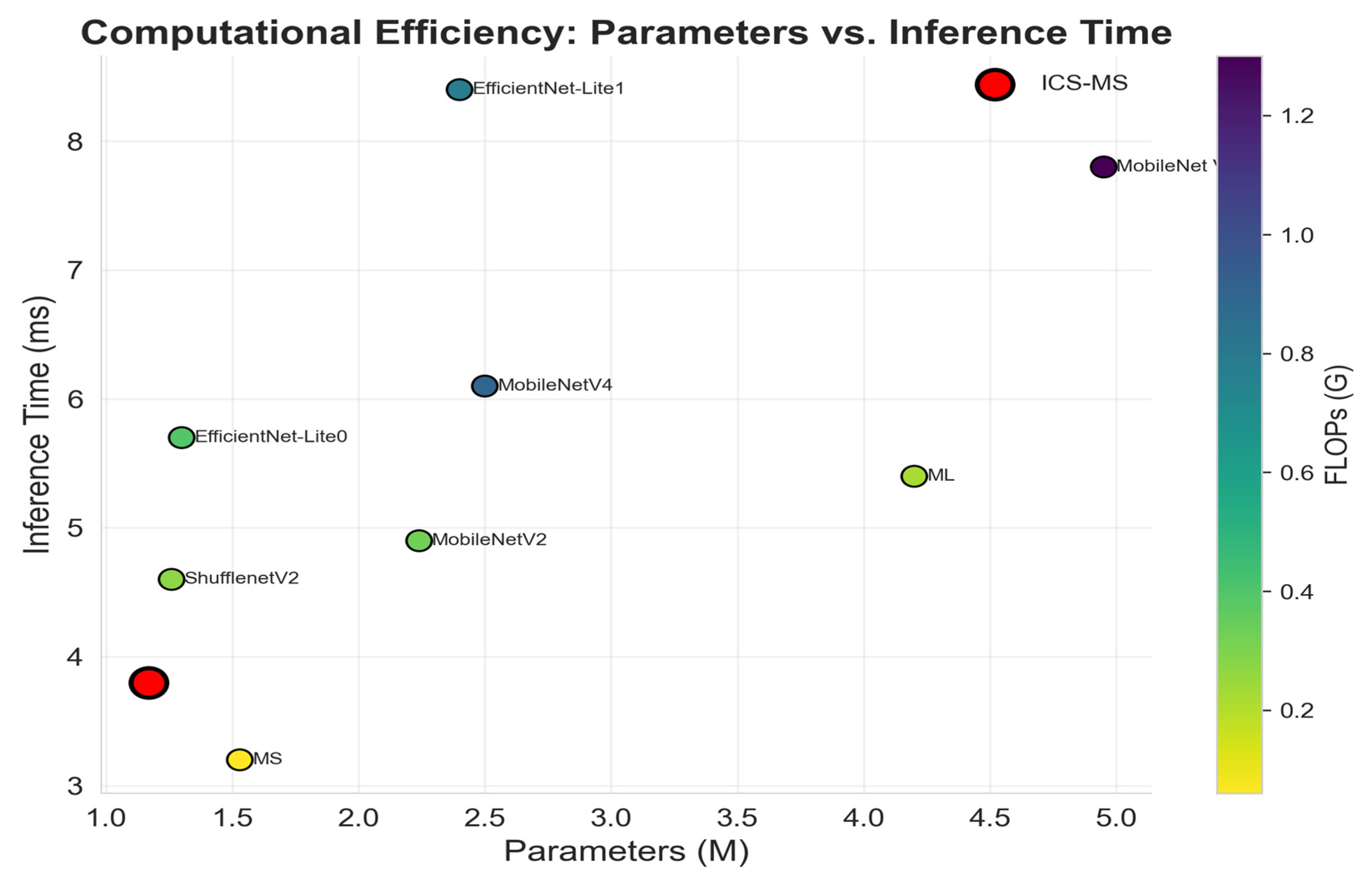
| Model | Parameters (M) | FLOPs(G) | Inference Time (ms/Image) |
|---|---|---|---|
| MobileNet Vit | 4.95 | 1.30 | 7.8 |
| ML | 4.2 | 0.22 | 5.4 |
| MobileNetV4 | 2.5 | 0.90 | 6.1 |
| EfficientNet-Lite1 | 2.4 | 0.78 | 8.4 |
| MobileNetV2 | 2.24 | 0.33 | 4.9 |
| MS | 1.53 | 0.06 | 3.2 |
| EfficientNet-Lite0 | 1.3 | 0.39 | 5.7 |
| ShufflenetV2 | 1.26 | 0.27 | 4.6 |
| ICS-MS | 1.17 | 0.21 | 3.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, L.; Du, Z.; Liu, X.; Wu, Z.; Lin, X.; Wen, B. Grape Leaf Cultivar Identification in Complex Backgrounds with an Improved MobileNetV3-Small Model. Plants 2025, 14, 3581. https://doi.org/10.3390/plants14233581
Deng L, Du Z, Liu X, Wu Z, Lin X, Wen B. Grape Leaf Cultivar Identification in Complex Backgrounds with an Improved MobileNetV3-Small Model. Plants. 2025; 14(23):3581. https://doi.org/10.3390/plants14233581
Chicago/Turabian StyleDeng, Liuyun, Zhiguo Du, Xiaoyong Liu, Zhihui Wu, Xudong Lin, and Bin Wen. 2025. "Grape Leaf Cultivar Identification in Complex Backgrounds with an Improved MobileNetV3-Small Model" Plants 14, no. 23: 3581. https://doi.org/10.3390/plants14233581
APA StyleDeng, L., Du, Z., Liu, X., Wu, Z., Lin, X., & Wen, B. (2025). Grape Leaf Cultivar Identification in Complex Backgrounds with an Improved MobileNetV3-Small Model. Plants, 14(23), 3581. https://doi.org/10.3390/plants14233581





