Correlative Transcriptome and Metabolome Analysis of the Maize Shoot Response to Salt Stress
Abstract
1. Introduction
2. Results
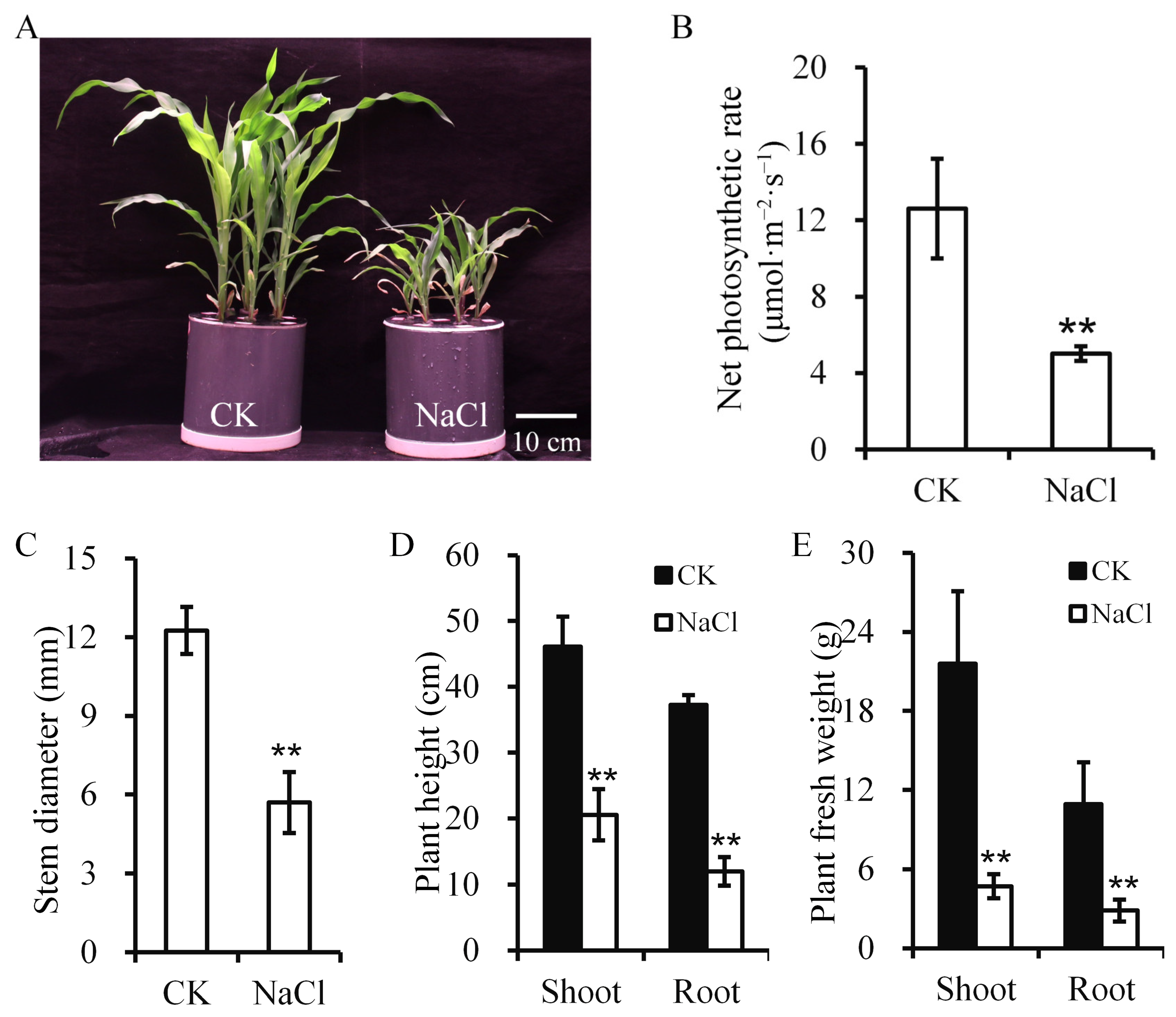
2.1. Salt-Stress-Induced Changes in Maize Growth and Physiology
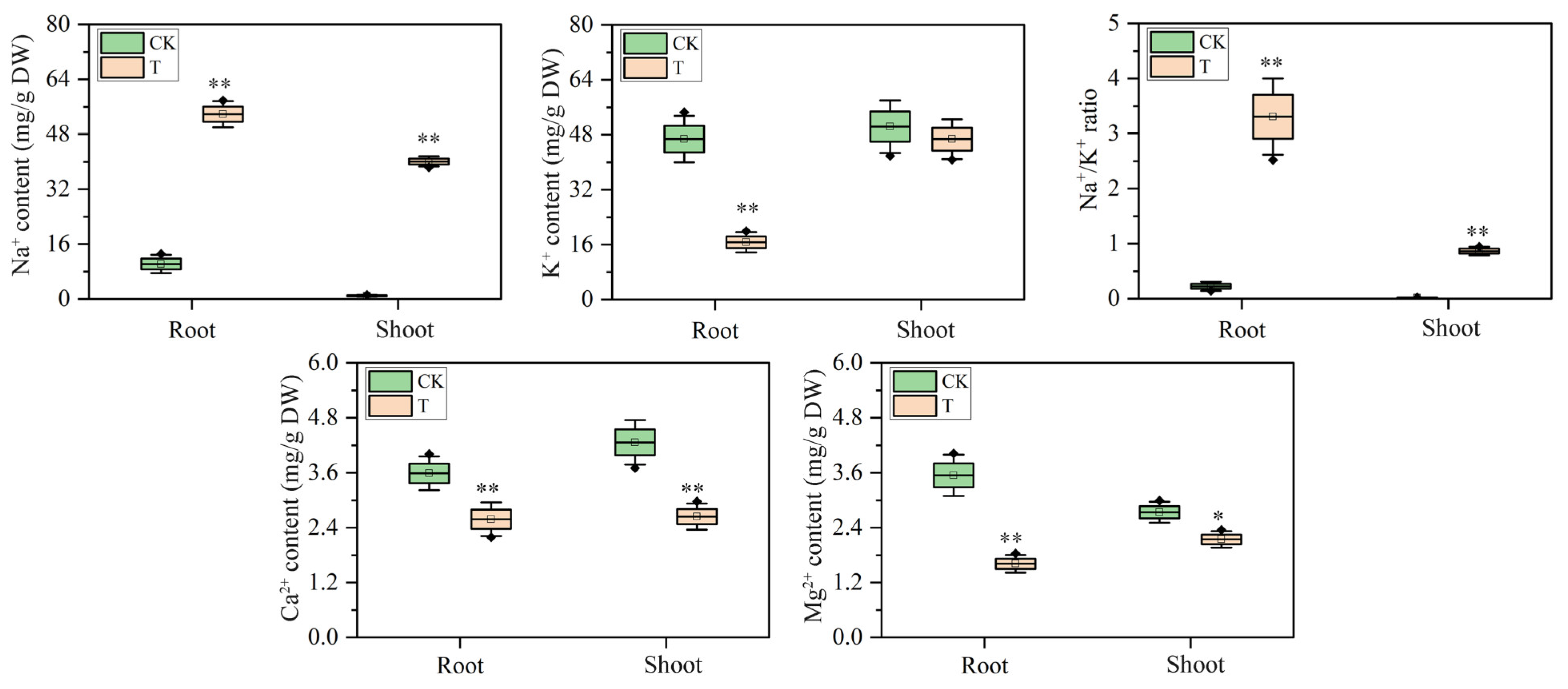
2.2. Mineral Elements Content Responses to Salt Stress
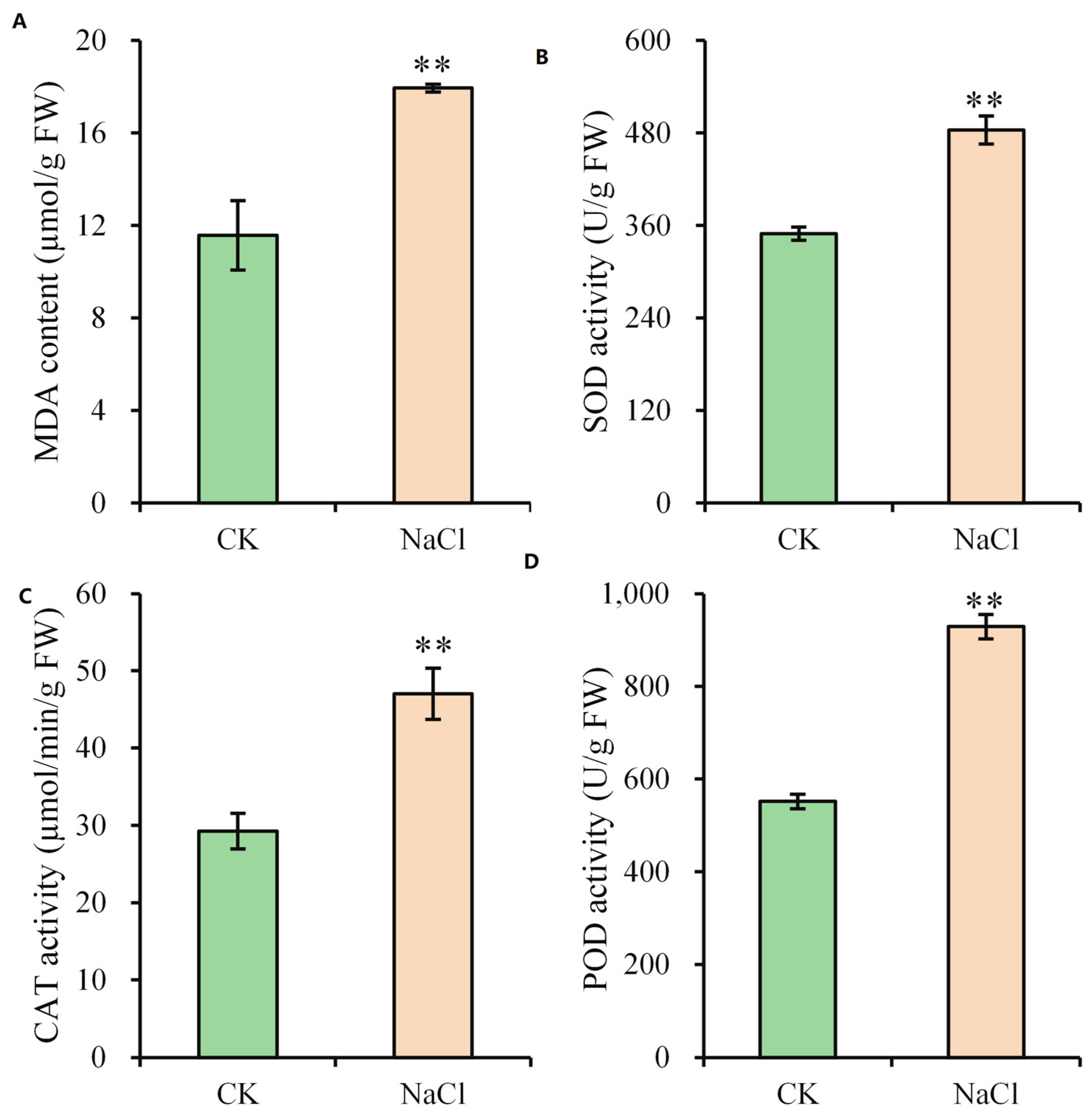
2.3. Antioxidative Enzyme Responses to Salt Stress
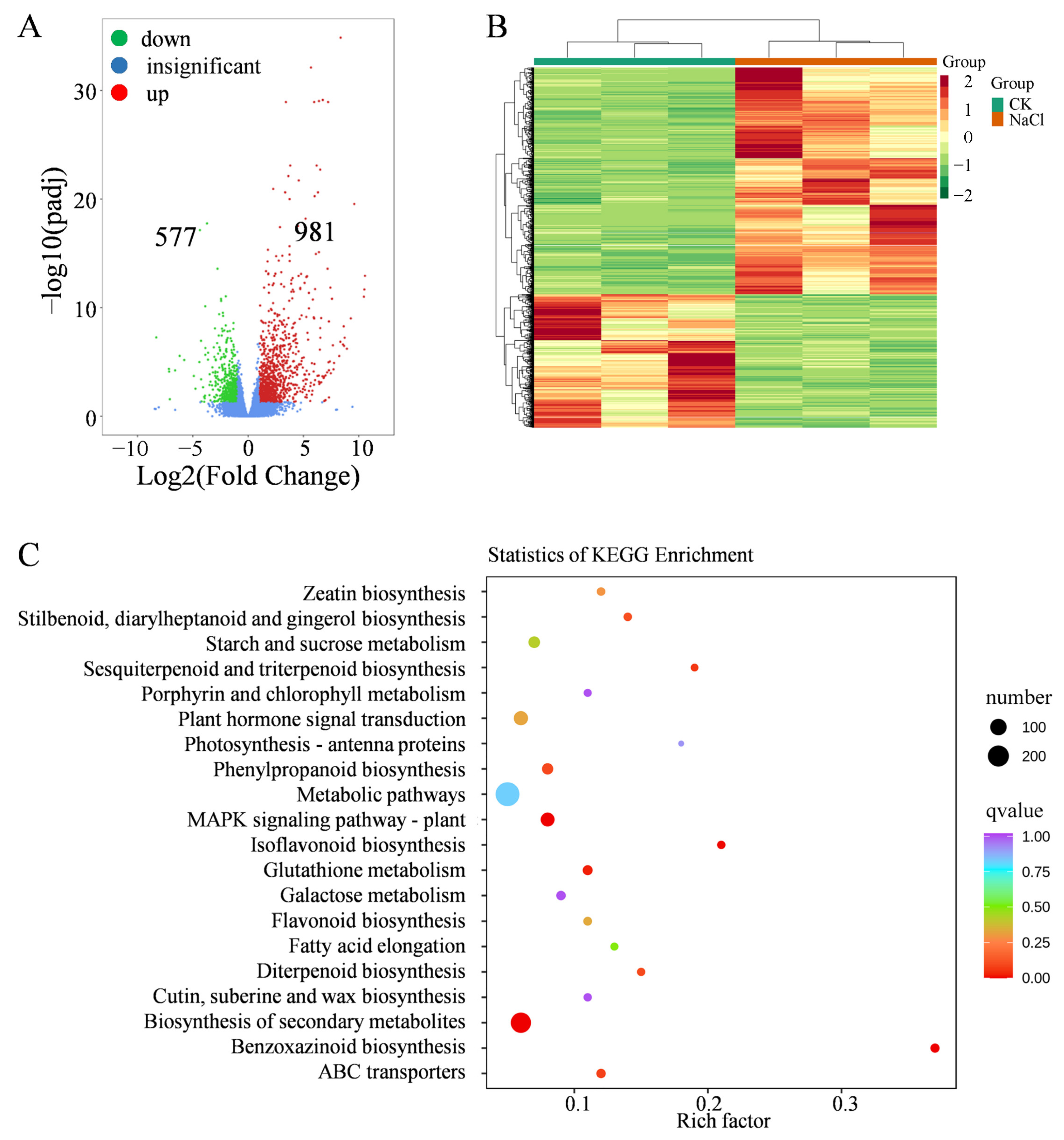
2.4. Transcriptomic Profiling in Leaves Under Salt Stress
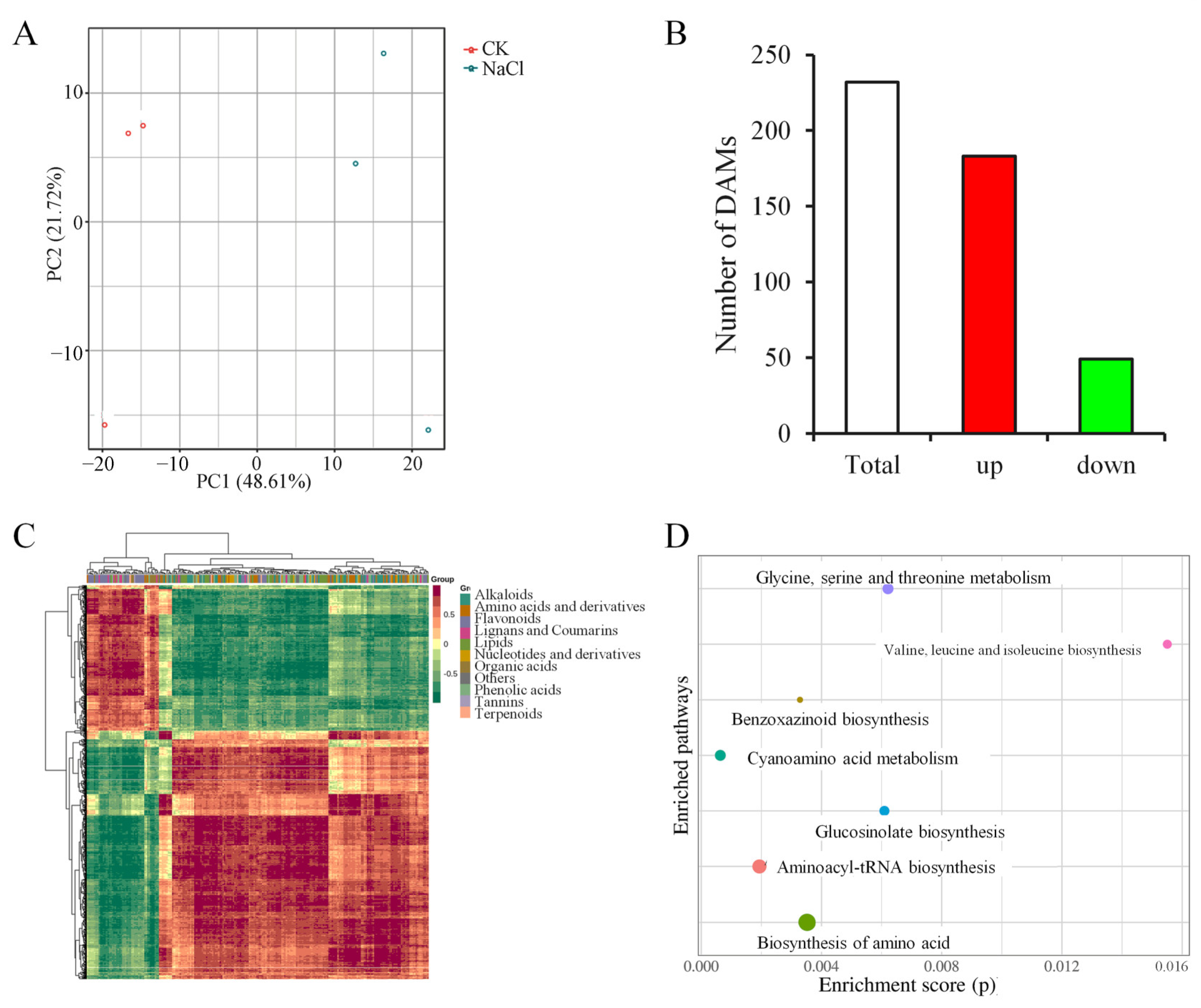
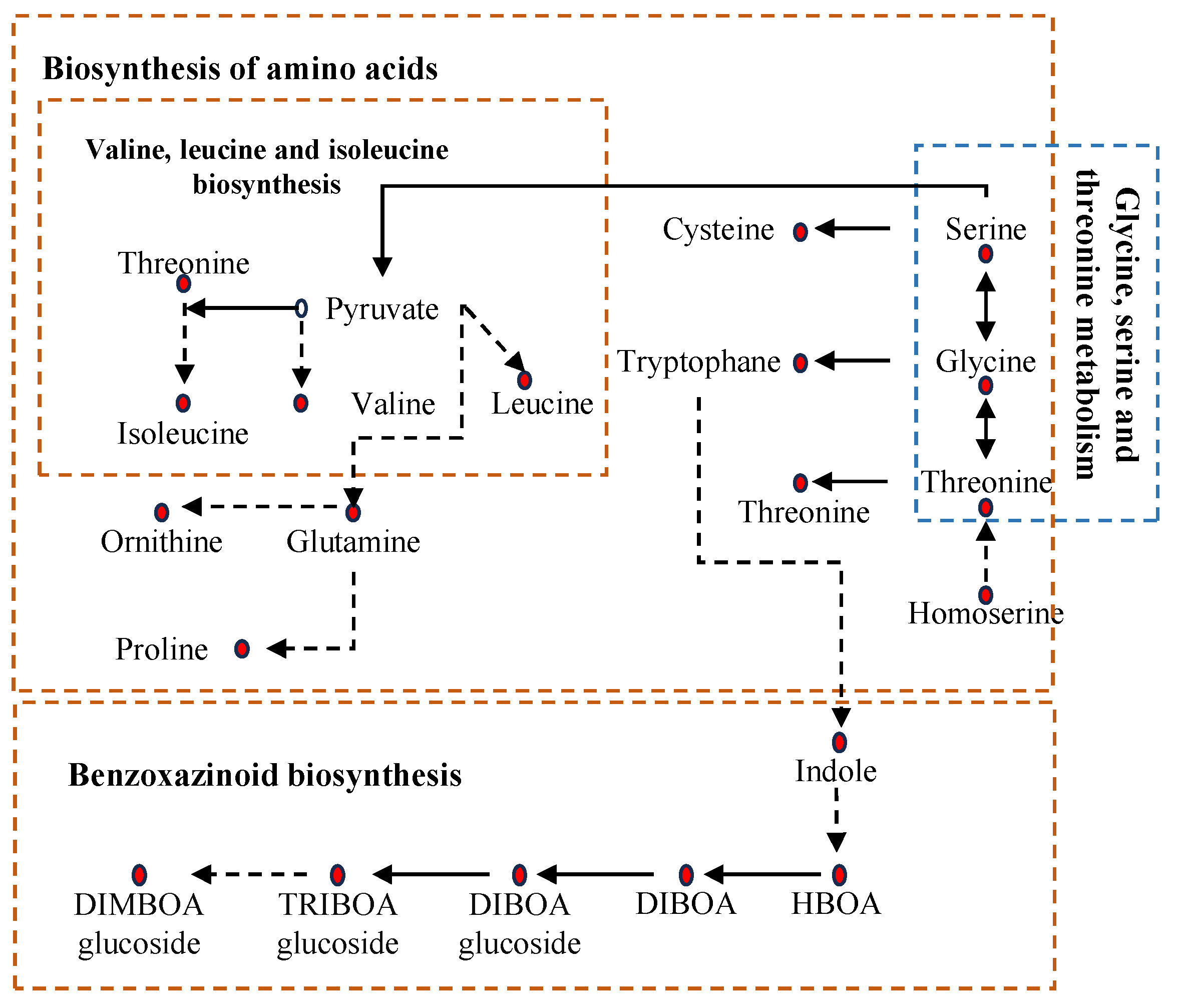
2.5. Metabolomic Analysis in Leaves Under Salt Stress
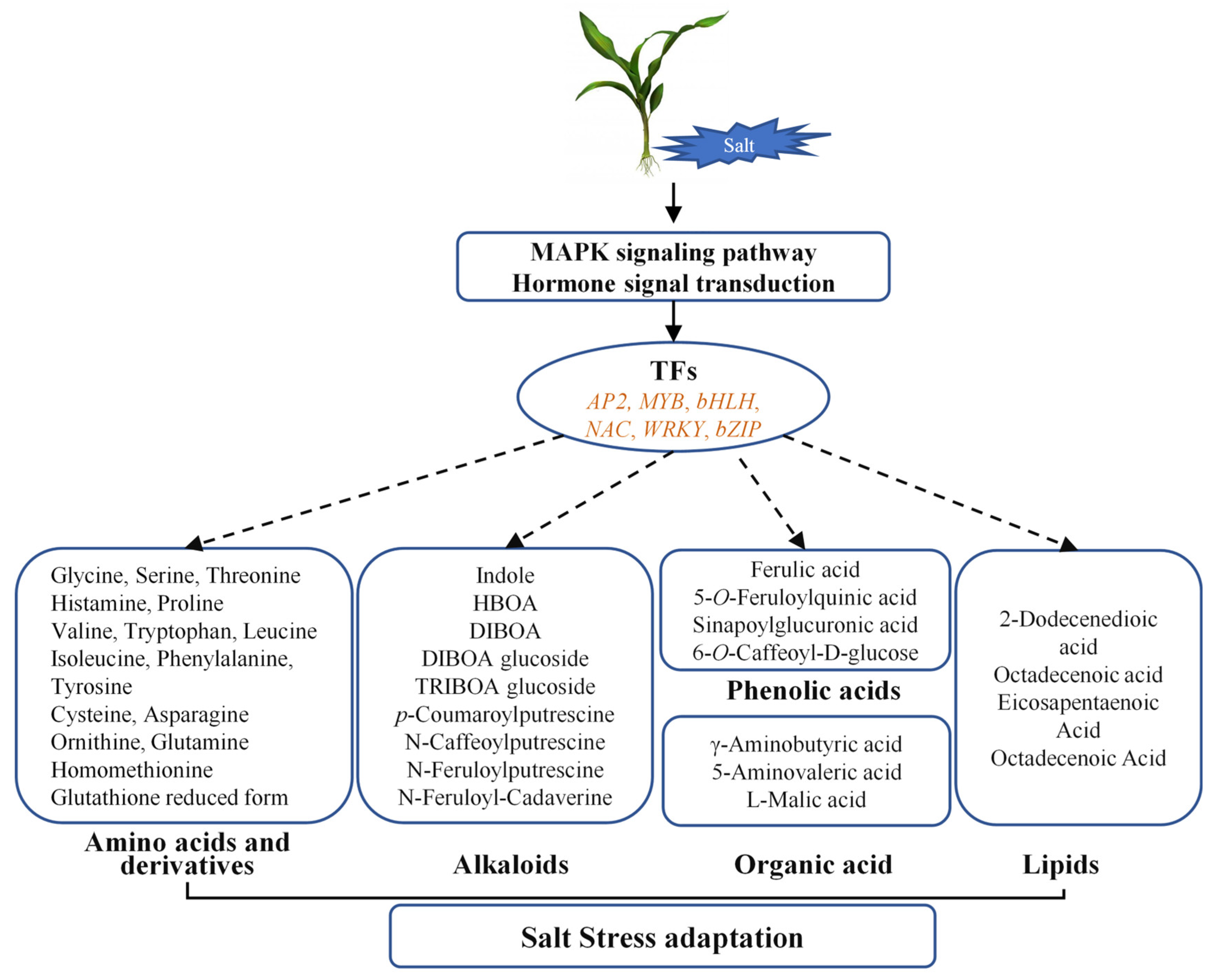
3. Discussion
3.1. Integrated Phenotypic–Physiological–Omics Responses to Salt Stress
3.2. Transcriptional Reprogramming to Salt Stress
3.3. Metabolic Reprogramming and Its Coordination with Transcriptional Changes
4. Materials and Methods
4.1. Plant Materials and Salt Stress Treatment
4.2. Measurement of Leaf Photosynthetic Rate
4.3. Measurement of Ion Concentration
4.4. Quantification of Antioxidant Enzyme Activities and MDA Content
4.5. RNA Isolation, Transcriptome Analysis and qRT-PCR Analysis
4.6. Metabolite Measurements
4.7. Correlation Analysis of Transcript and Metabolite Pairs
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ABA | Abscisic Acid |
| CAT | Catalase |
| DAM | Differentially Accumulated Metabolite |
| DEG | Differentially Expressed Gene |
| GO | Gene Ontology |
| ICP-MS | Inductively Coupled Plasma Mass Spectrometry |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MAPK | Mitogen-Activated Protein Kinase |
| MDA | Malondialdehyde |
| NBT | Nitroblue Tetrazolium |
| PCA | Principal Component Analysis |
| POD | Peroxidase |
| ROS | Reactive Oxygen Species |
| SOD | Superoxide Dismutase |
| TF | Transcription Factor |
| TBA | Thiobarbituric Acid |
| UPLC-MS/MS | Ultra-Performance Liquid Chromatography–Tandem Mass Spectrometry |
References
- Landi, S.; Hausman, J.F.; Guerriero, G.; Esposito, S. Poaceae vs. Abiotic stress: Focus on drought and salt stress, recent insights and perspectives. Front. Plant Sci. 2017, 8, 1214. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liang, X.; Wang, L.; Cao, Y.; Song, W.; Shi, J.; Lai, J.; Jiang, C. A HAK family Na+ transporter confers natural variation of salt tolerance in maize. Nat. Plants 2019, 5, 1297–1308. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; James, R.A.; Xu, B.; Athman, A.; Conn, S.J.; Jordans, C.; Byrt, C.S.; Hare, R.A.; Tyerman, S.D.; Tester, M.; et al. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat. Biotechnol. 2012, 30, 360–364. [Google Scholar] [CrossRef]
- Farooq, M.; Hussain, M.; Wakeel, A.; Siddique, K.H.M. Salt stress in maize: Effects, resistance mechanisms, and management. A review. Agron. Sustain. Dev. 2015, 35, 461–481. [Google Scholar] [CrossRef]
- Zamani, E.; Bakhtari, B.; Razi, H.; Hildebrand, D.; Moghadam, A.; Alemzadeh, A. Comparative morphological, physiological, and biochemical traits in sensitive and tolerant maize genotypes in response to salinity and Pb stress. Sci. Rep. 2024, 14, 31036. [Google Scholar] [CrossRef]
- Zhao, K.; Song, J.; Fan, H.; Zhou, S.; Zhao, M. Growth response to ionic and osmotic stress of NaCl in salt-tolerant and salt-sensitive maize. J. Integr. Plant Biol. 2010, 52, 468–475. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Fujita, M. Plant responses and tolerance to salt stress: Physiological and molecular interventions. Int. J. Mol. Sci. 2022, 23, 4810. [Google Scholar] [CrossRef]
- Farooq, M.; Zahra, N.; Ullah, A.; Nadeem, F.; Rehman, A.; Kapoor, R.; Al-Hinani, M.S.; Siddique, K.H.M. Salt stress in wheat: Effects, tolerance mechanisms, and management. J. Soil. Sci. Plant Nutr. 2024, 24, 8151–8173. [Google Scholar] [CrossRef]
- Wei, G.; Chen, Y.; Wang, J.; Feng, L. Molecular cloning and characterization of farnesyl diphosphate synthase from Rosa rugosa Thunb associated with salinity stress. PeerJ 2024, 12, e16929. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Choudhary, M.; Halder, T.; Prakash, N.R.; Singh, V.; Vineeth, T.V.; Sheoran, S.; Ravikiran, K.T.; Longmei, N.; Rakshit, S.; et al. Salinity stress tolerance and omics approaches: Revisiting the progress and achievements in major cereal crops. Heredity 2022, 128, 497–518. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.A.; Sarkhosh, A.; Khan, N.; Balal, R.M.; Ali, S.; Rossi, L.; Gómez, C.; Mattson, N.; Nasim, W.; Garcia-Sanchez, F. Insights into the physiological and biochemical impacts of salt stress on plant growth and development. Agronomy 2020, 10, 938. [Google Scholar] [CrossRef]
- Rizk, M.S.; Assaha, D.V.M.; Mekawy, A.M.M.; Shalaby, N.E.; Ramadan, E.A.; El-Tahan, A.M.; Ibrahim, O.M.; Metwelly, H.I.F.; Okla, M.K.; Maridueña-Zavala, M.G.; et al. Comparative analysis of salinity tolerance mechanisms in two maize genotypes: Growth performance, ion regulation, and antioxidant responses. BMC Plant Biol. 2024, 24, 818. [Google Scholar] [CrossRef]
- Maimaiti, A.; Gu, W.; Yu, D.; Guan, Y.; Qu, J.; Qin, T.; Wang, H.; Ren, J.; Zheng, H.; Wu, P. Dynamic molecular regulation of salt stress responses in maize (Zea mays L.) seedlings. Front. Plant. Sci. 2025, 16, 1535943. [Google Scholar] [CrossRef]
- Wang, C.; Wang, Y.; Cao, X.; Wu, C.; Wei, X.; Jiao, P.; Liu, S.; Ma, Y.; Guan, S. Unraveling saline-alkali stress tolerance: Contrasting morpho-physiological, biochemical, and ionic responses in maize (Zea mays L.) genotypes. Plant Physiol. Biochem. 2025, 229, 110349. [Google Scholar] [CrossRef]
- Fang, X.; Mo, J.J.; Zhou, H.; Shen, X.; Xie, Y.; Xu, J.; Yang, S. Comparative transcriptome analysis of gene responses of salt-tolerant and salt-sensitive rice cultivars to salt stress. Sci. Rep. 2023, 13, 19065. [Google Scholar] [CrossRef]
- Han, Y.; Wu, C.; Ji, X.; Yang, M.; Zhu, H.; Pei, Z.; Qu, M.; Qu, L.; Li, Z.; Yan, S. Molecular mechanisms between salt-tolerant and salt-sensitive rice (Oryza sativa L.) varieties under salt stress. Curr. Issues Mol. Biol. 2025, 47, 832. [Google Scholar] [CrossRef]
- Yu, W.; Wu, W.; Zhang, N.; Wang, L.; Wang, Y.; Wang, B.; Lan, Q.; Wang, Y. Research advances on molecular mechanism of salt tolerance in Suaeda. Biology 2022, 11, 1273. [Google Scholar] [CrossRef]
- Quan, C.; Huang, S.; Yu, Q.; Chen, Z.; Kashif, M.; Xu, M.; Wei, F.; Tang, D. Physiological, biochemical, and transcriptomic analyses reveal potential candidate genes of Platostoma palustre in response to salt stress. BMC Plant Biol. 2025, 25, 848. [Google Scholar] [CrossRef]
- Nakagami, H.; Pitzschke, A.; Hirt, H. Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci. 2005, 10, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Lv, W.; Zhang, D.; Jiang, S.; Zhang, S.; Li, D. Genome-wide identification and analysis of expression profiles of maize mitogen-activated protein kinase kinase kinase. PLoS ONE 2013, 8, e57714. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Pan, J.; Zhang, M.; Xing, X.; Zhou, Y.; Liu, Y.; Li, D.; Li, D. ZmMKK4, a novel group C mitogen-activated protein kinase kinase in maize (Zea mays), confers salt and cold tolerance in transgenic Arabidopsis. Plant Cell Environ. 2011, 34, 1291–1303. [Google Scholar] [CrossRef] [PubMed]
- Cai, G.; Wang, G.; Wang, L.; Liu, Y.; Pan, J.; Li, D. A maize mitogen-activated protein kinase kinase, ZmMKK1, positively regulated the salt and drought tolerance in transgenic. J. Plant Physiol. 2014, 171, 1003–1016. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, B.; Zhang, P.; Han, Q.; Zhao, G.; Zhao, F. Comparative transcriptome analysis reveals the underlying response mechanism to salt stress in maize seedling roots. Metabolites 2023, 13, 1155. [Google Scholar] [CrossRef]
- Reshi, Z.A.; Ahmad, W.; Lukatkin, A.S.; Bin Javed, S. From nature to lab: A review of secondary metabolite biosynthetic pathways, environmental influences, and in vitro approaches. Metabolites 2023, 13, 895. [Google Scholar] [CrossRef]
- Wang, C.; Wei, X.; Wang, Y.; Wu, C.; Jiao, P.; Jiang, Z.; Liu, S.; Ma, Y.; Guan, S. Metabolomics and transcriptomic analysis revealed the response mechanism of maize to saline-alkali stress. Plant Biotechnol. J. 2025. early view. [Google Scholar] [CrossRef]
- Liu, Q.; Kang, J.; Du, L.; Liu, Z.; Liang, H.; Wang, K.; He, H.; Zhang, X.; Wang, Q.; Hong, Y.; et al. Single-cell multiome reveals root hair-specific responses to salt stress. New Phytol. 2025, 246, 2634–2651. [Google Scholar] [CrossRef]
- Ren, S.; Bai, T.; Ma, Y.; Zhao, Y.; Ci, J.; Ren, X.; Zang, Z.; Ma, C.; Xiong, R.; Song, X.; et al. Molecular mechanisms underlying salt tolerance in maize: A combined transcriptome and metabolome analysis. Plants 2025, 14, 2031. [Google Scholar] [CrossRef]
- Ullah, M.S.; Mahmood, A.; Alawadi, H.F.N.; Seleiman, M.F.; Khan, B.A.; Javaid, M.M.; Wahid, A.; Abdullah, F.; Wasonga, D.O. Silicon-mediated modulation of maize growth, metabolic responses, and antioxidant mechanisms under saline conditions. BMC Plant Biol. 2025, 25, 3. [Google Scholar] [CrossRef]
- Chérel, I.; Gaillard, I. The complex fine-tuning of K+ fluxes in plants in relation to osmotic and ionic abiotic stresses. Int. J. Mol. Sci. 2019, 20, 715. [Google Scholar] [CrossRef] [PubMed]
- Danquah, A.; de Zelicourt, A.; Colcombet, J.; Hirt, H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 2014, 32, 40–52. [Google Scholar] [CrossRef]
- Sun, Y.; Zhao, N.; Sun, H.; Xu, S.; Lu, Y.; Xi, H.; Guo, Z.; Shi, H. Transcriptome profiling reveals molecular responses to salt stress in common vetch (Vicia sativa L.). Plants 2024, 13, 714. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Osuna, A.; Calatrava, V.; Galvan, A.; Fernandez, E.; Llamas, A. Identification of the MAPK cascade and its relationship with nitrogen metabolism in the green alga Chlamydomonas reinhardtii. Int. J. Mol. Sci. 2020, 21, 3417. [Google Scholar] [CrossRef]
- Yan, Z.; Li, K.; Li, Y.; Wang, W.; Leng, B.; Yao, G.; Zhang, F.; Mu, C.; Liu, X. The ZmbHLH32-ZmIAA9-ZmARF1 module regulates salt tolerance in maize. Int. J. Biol. Macromol. 2023, 253, 126978. [Google Scholar] [CrossRef]
- Bao, L.; Sun, W.; Wang, J.; Zhou, Y.; Wang, J.; Wang, Q.; Sun, D.Q.; Lin, H.; Fan, J.; Zhou, Y.; et al. The transcription factor ZmMYBR24 gene is involved in a variety of abiotic stresses in maize (Zea mays L.). Plants 2025, 14, 2054. [Google Scholar] [CrossRef]
- Bo, C.; Cai, R.; Fang, X.; Wu, H.; Ma, Z.; Yuan, H.; Cheng, B.; Fan, J.; Ma, Q. Transcription factor ZmWRKY20 interacts with ZmWRKY115 to repress expression of ZmbZIP111 for salt tolerance in maize. Plant J. 2022, 111, 1660–1675. [Google Scholar] [CrossRef]
- Tian, T.; Wang, J.; Wang, H.; Cui, J.; Shi, X.; Song, J.; Li, W.; Zhong, M.; Qiu, Y.; Xu, T. Nitrogen application alleviates salt stress by enhancing osmotic balance, ROS scavenging, and photosynthesis of rapeseed seedlings (Brassica napus). Plant Signal. Behav. 2022, 17, 2081419. [Google Scholar] [CrossRef]
- Zhang, W.; Tang, S.; Li, X.; Chen, Y.; Li, J.; Wang, Y.; Bian, R.; Jin, Y.; Zhu, X.; Zhang, K. Arabidopsis WRKY1 promotes monocarpic senescence by integrative regulation of flowering, leaf senescence, and nitrogen remobilization. Mol. Plant. 2024, 17, 1289–1306. [Google Scholar] [CrossRef]
- Vignesh, P.; Mahadevaiah, C.; Parimalan, R.; Valarmathi, R.; Dharshini, S.; Nisha, S.; Suresha, G.S.; Swathi, S.; Swamy, H.K.M.; Sreenivasa, V.; et al. Comparative de novo transcriptome analysis identifies salinity stress responsive genes and metabolic pathways in sugarcane and its wild relative Erianthus arundinaceus [Retzius] Jeswiet. Sci. Rep. 2021, 11, 24514. [Google Scholar] [CrossRef] [PubMed]
- Hoque, M.A.; Banu, M.N.; Okuma, E.; Amako, K.; Nakamura, Y.; Shimoishi, Y.; Murata, Y. Exogenous proline and glycinebetaine increase NaCl-induced ascorbate-glutathione cycle enzyme activities, and proline improves salt tolerance more than glycinebetaine in tobacco Bright Yellow-2 suspension-cultured cells. J. Plant Physiol. 2007, 164, 1457–1468. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Shi, L.; Yan, C.; Zhong, X.; Gu, F.; Liu, Q.; Xia, X.; Li, H. Ionomic and metabolic responses to neutral salt or alkaline salt stresses in maize (Zea mays L.) seedlings. BMC Plant Biol. 2017, 17, 41. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Hu, X.; Liao, L.; Xu, T.; Sun, Q.; Tang, M.; Chen, Z.; Wang, Z. Lipid metabolism and antioxidant system contribute to salinity tolerance in halophytic grass seashore paspalum in a tissue-specific manner. BMC Plant Biol. 2023, 23, 337. [Google Scholar] [CrossRef]
- Ullah, A.; Ali, I.; Noor, J.; Zeng, F.J.; Bawazeer, S.; Eldin, S.M.; Asghar, M.A.; Javed, H.H.; Saleem, K.; Ullah, S.; et al. Exogenous γ-aminobutyric acid (GABA) mitigated salinity-induced impairments in mungbean plants by regulating their nitrogen metabolism and antioxidant potential. Front. Plant Sci. 2023, 13, 1081188. [Google Scholar] [CrossRef]
- Zhao, M.; Tai, H.; Sun, S.; Zhang, F.; Xu, Y.; Li, W. Cloning and characterization of maize miRNAs involved in responses to nitrogen deficiency. PLoS ONE 2012, 7, e29669. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Wei, G.; Chen, Y.; Wang, M.; Xi, Y.; Xu, Y.; Hussain, H.; Zhu, K.; Xu, Y.; Bai, M.; Wang, J.; et al. Integrative application of metabolomics and transcriptomics provides new insights into carotenoid biosynthesis during Rosa rugosa hips ripening. Food Biosci. 2024, 60, 104422. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Xiong, W.; Wang, Y.; Guo, Y.; Tang, W.; Zhao, Y.; Yang, G.; Pei, Y.; Chen, J.; Song, X.; Sun, J. Transcriptional and metabolic responses of maize shoots to long-term potassium deficiency. Front. Plant Sci. 2022, 13, 922581. [Google Scholar] [CrossRef]
- Yan, H.; Nie, Y.; Cui, K.; Sun, J. Integrative transcriptome and metabolome profiles reveal common and unique pathways involved in seed initial imbibition under artificial and natural salt stresses during germination of halophyte quinoa. Front. Plant Sci. 2022, 13, 853326. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, W.; Zhang, L.; Wang, Y.; Wei, G.; Zhu, K.; Zhao, K.; Wu, Z. Correlative Transcriptome and Metabolome Analysis of the Maize Shoot Response to Salt Stress. Plants 2025, 14, 3554. https://doi.org/10.3390/plants14233554
Xiong W, Zhang L, Wang Y, Wei G, Zhu K, Zhao K, Wu Z. Correlative Transcriptome and Metabolome Analysis of the Maize Shoot Response to Salt Stress. Plants. 2025; 14(23):3554. https://doi.org/10.3390/plants14233554
Chicago/Turabian StyleXiong, Wangdan, Lingxin Zhang, Yujian Wang, Guo Wei, Kaikai Zhu, Kai Zhao, and Zhenying Wu. 2025. "Correlative Transcriptome and Metabolome Analysis of the Maize Shoot Response to Salt Stress" Plants 14, no. 23: 3554. https://doi.org/10.3390/plants14233554
APA StyleXiong, W., Zhang, L., Wang, Y., Wei, G., Zhu, K., Zhao, K., & Wu, Z. (2025). Correlative Transcriptome and Metabolome Analysis of the Maize Shoot Response to Salt Stress. Plants, 14(23), 3554. https://doi.org/10.3390/plants14233554






