Fine Mapping of qSPJ_1 and Candidate Gene Identification for Soybean Seed Protein Content
Abstract
1. Introduction
1.1. Functions of Soybean Seed Protein
1.2. Influencing Factors of Soybean Seed Protein Content
1.3. Research Progress on QTLs Related to Soybean Seed Protein Content
1.4. Research Progress on Genes Related to Soybean Seed Protein Content
1.5. Significance of This Study
2. Results
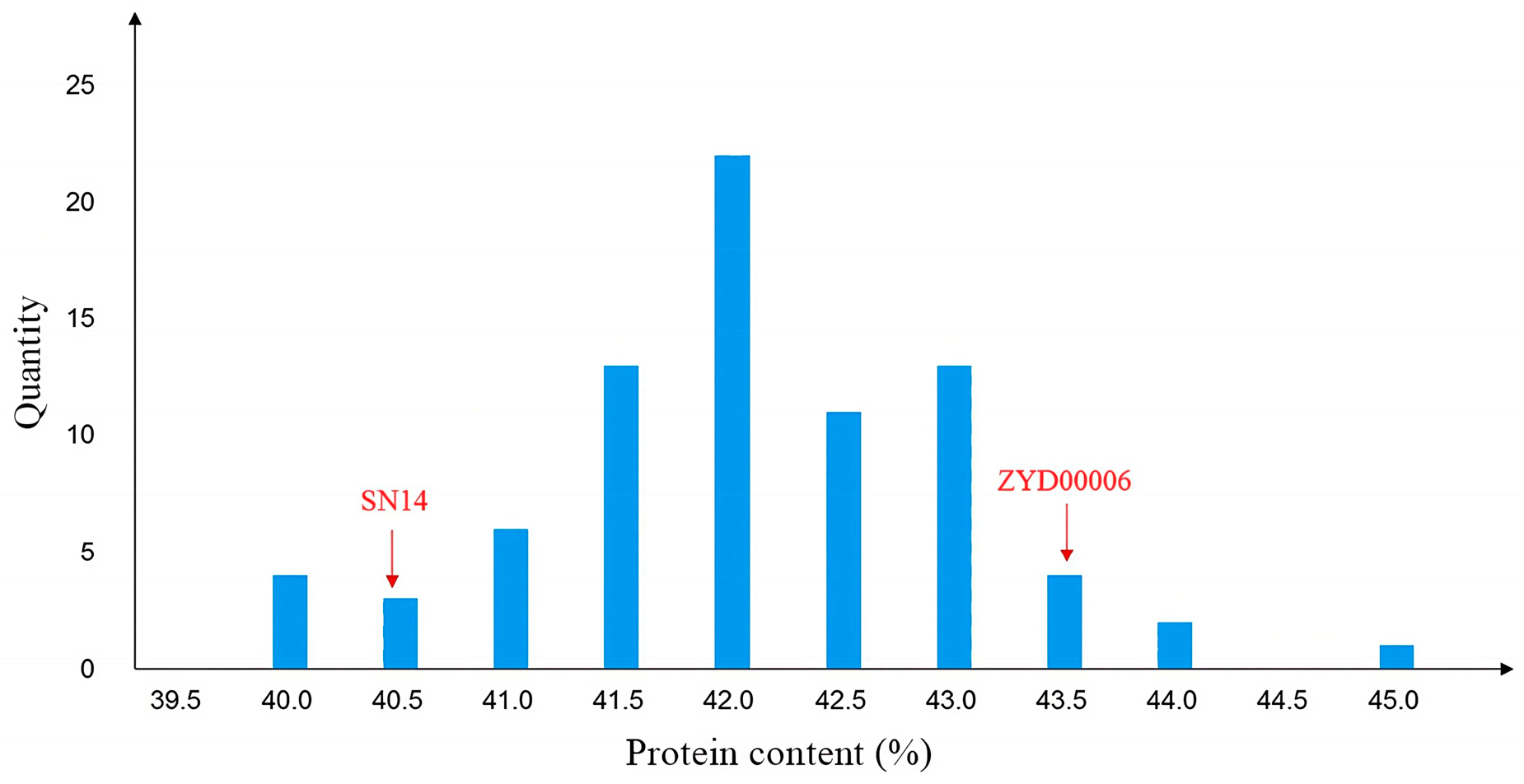
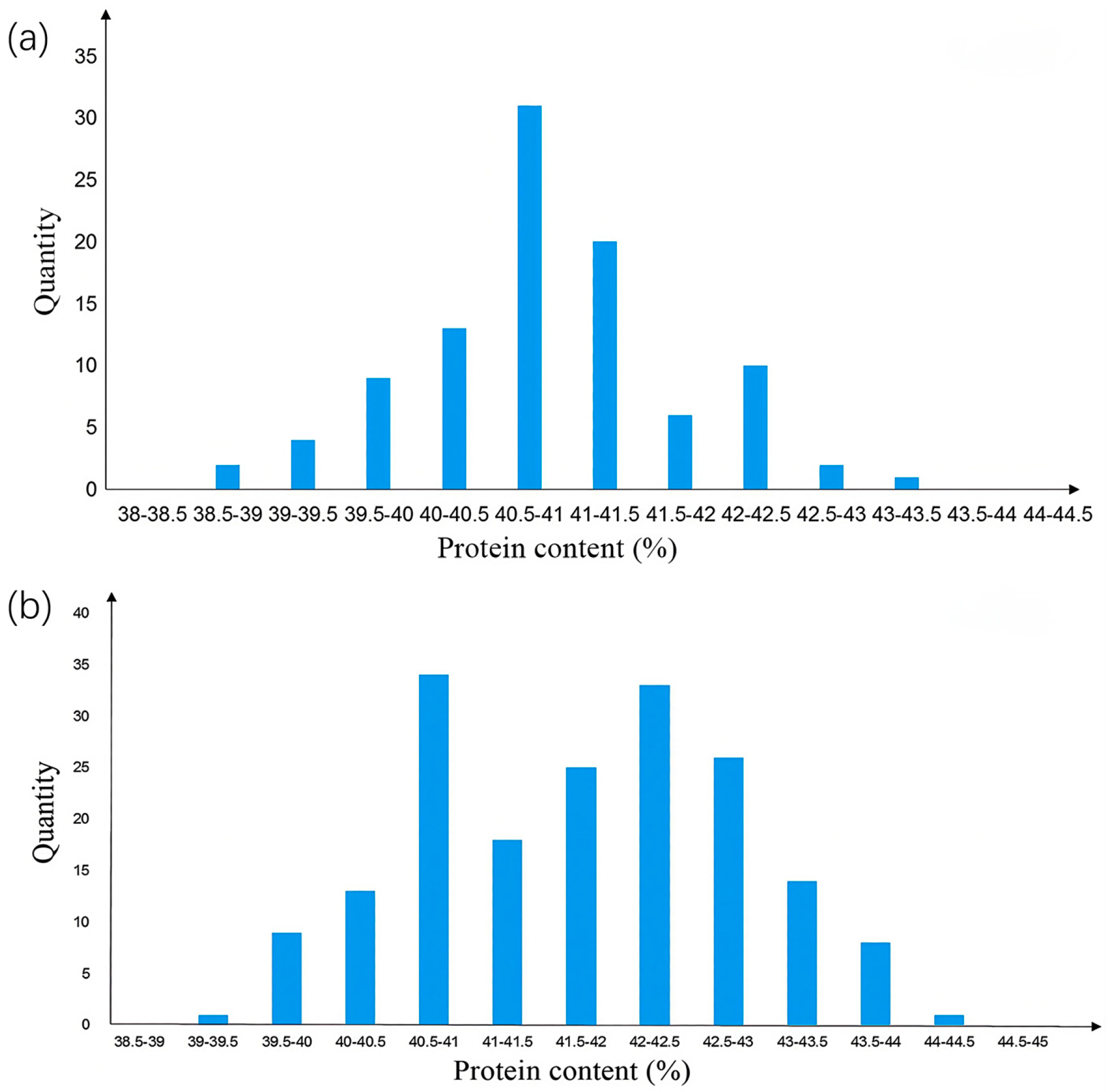
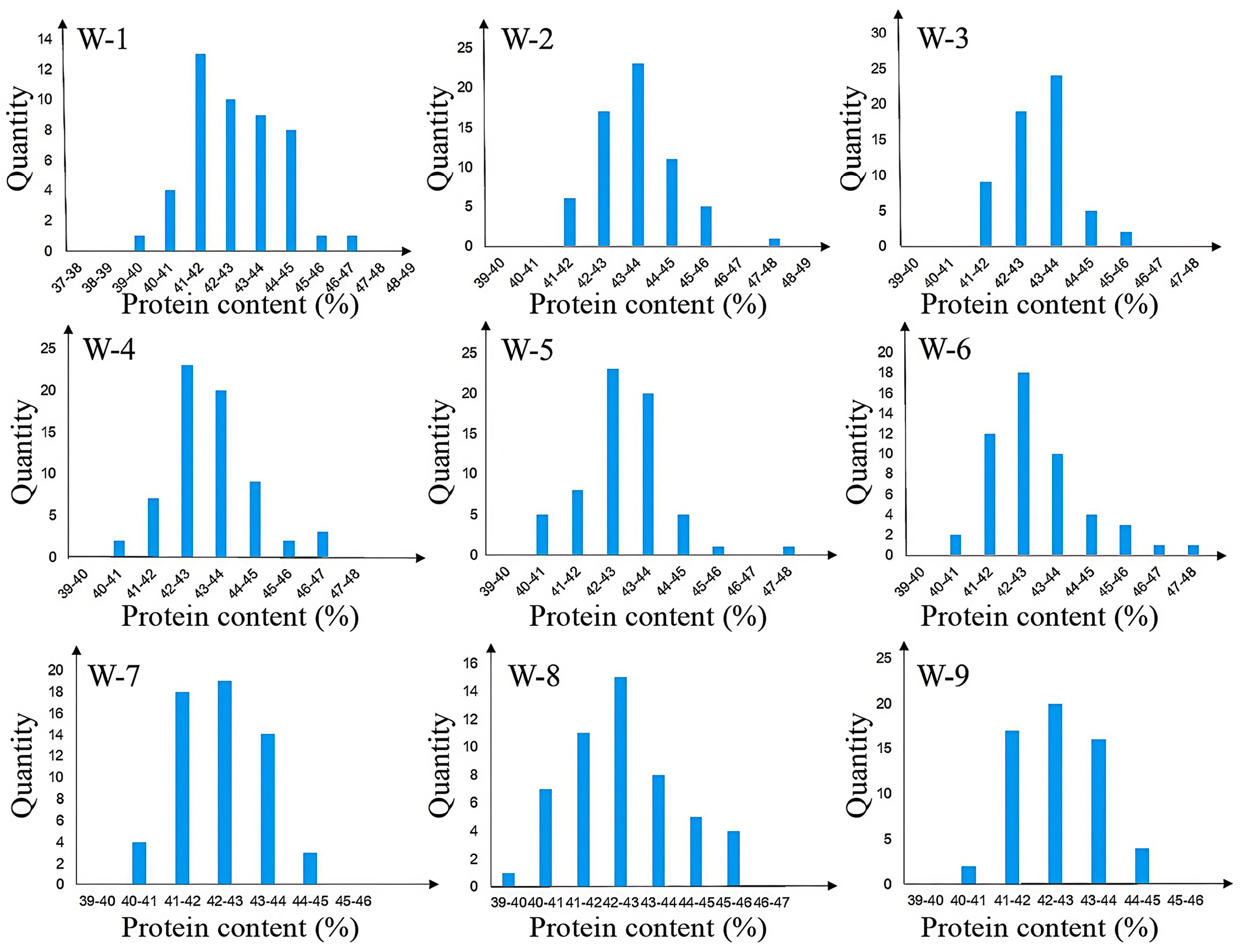
2.1. Initial Mapping of Protein Quantitative Trait Loci (Protein QTLs)
2.2. Fine Mapping of Protein Locus qSPJ_1
2.3. Gene Annotation and Parental Sequence Alignment of Candidate Genes
2.4. Quantitative Real-Time PCR Analysis (qRT-PCR Analysis)
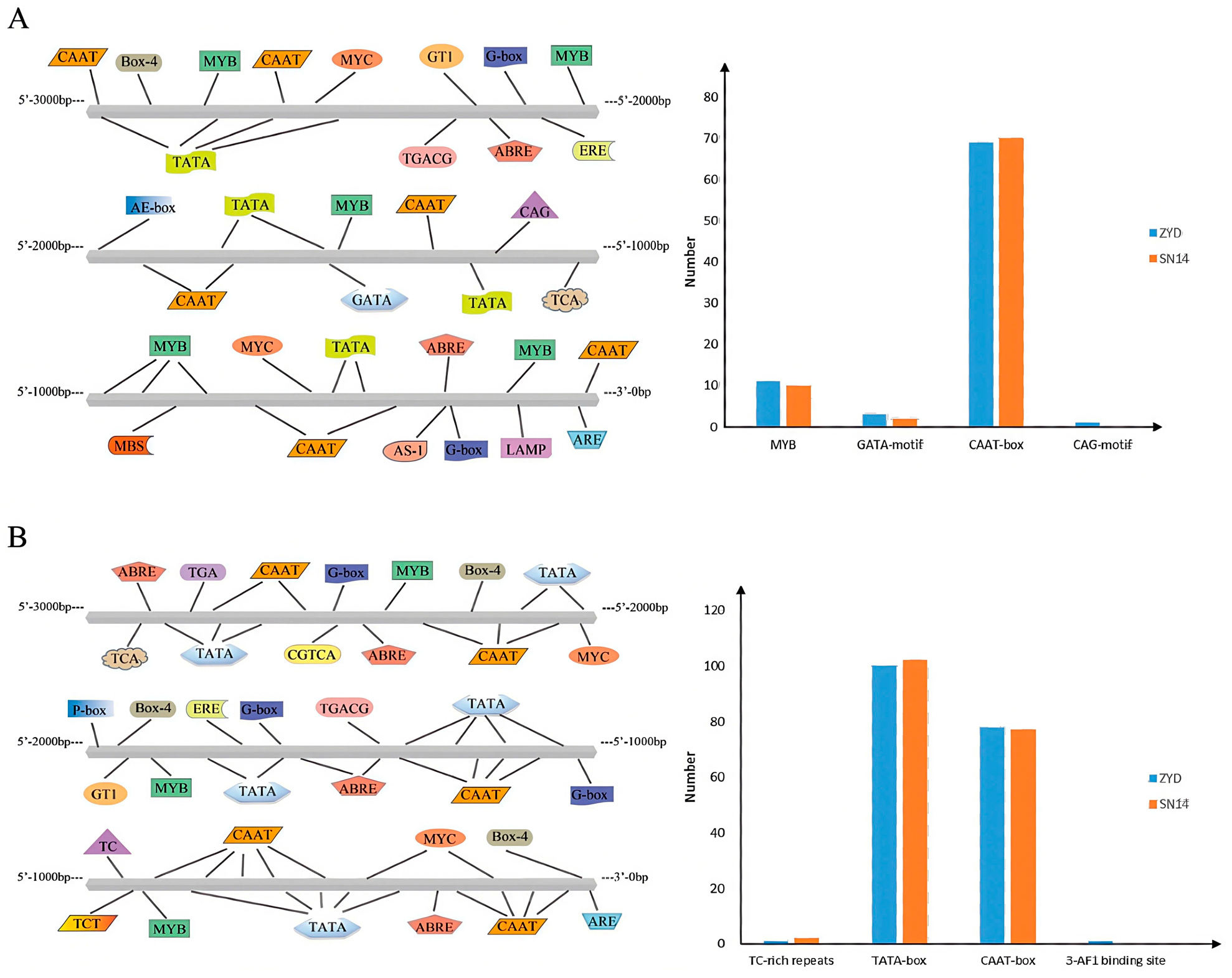
2.5. Analysis of Promoter Elements of Candidate Genes
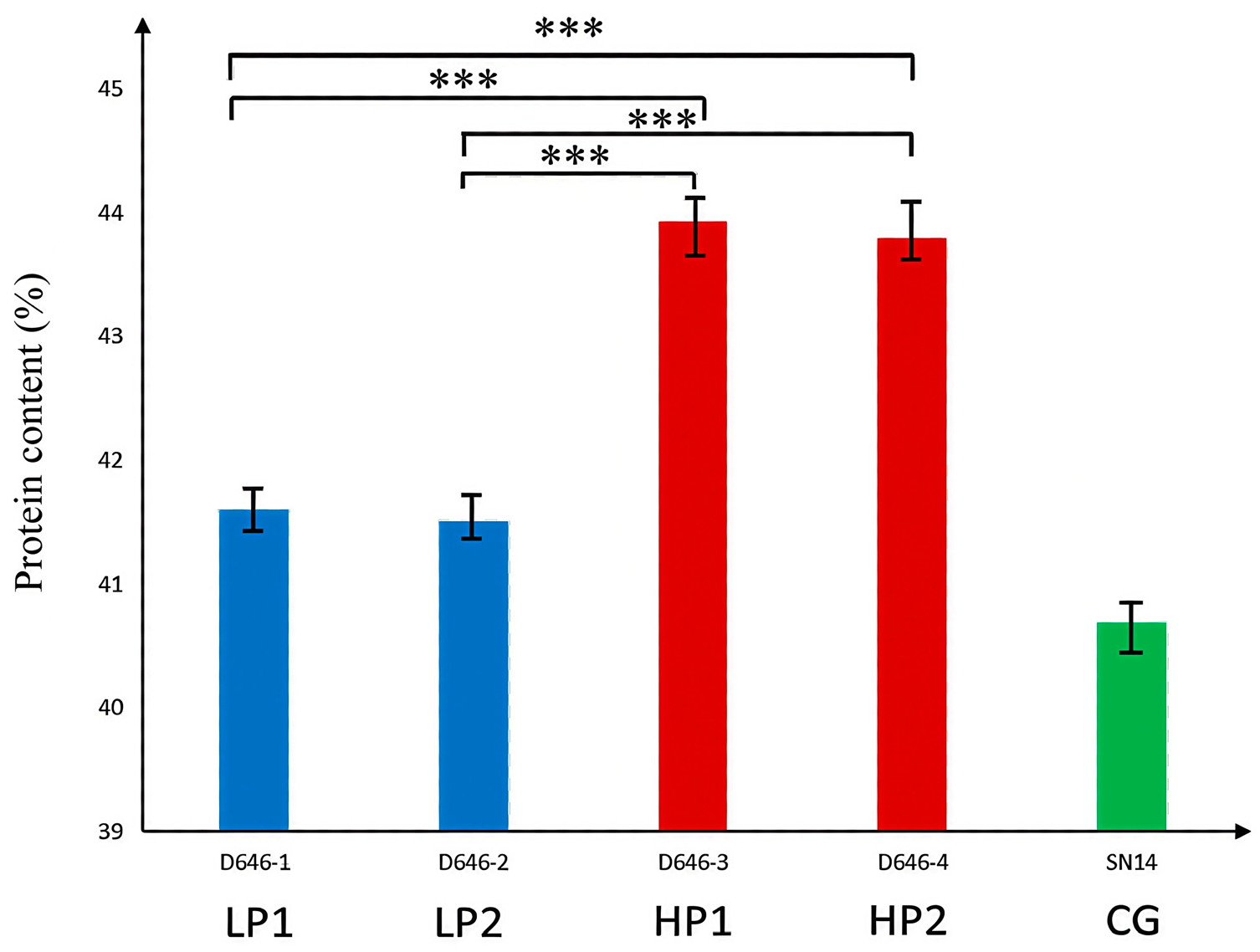
2.6. Haplotype Analysis of Candidate Genes
3. Discussion
4. Materials and Methods
4.1. Experimental Materials
4.1.1. Construction of the CSSL Population
4.1.2. Construction of Mapping Populations
4.1.3. Materials for Quantitative Real-Time PCR (qPCR)
4.1.4. Materials for Haplotype Analysis
4.2. Experimental Methods
4.2.1. Methods for Planting and Field Management of Experimental Materials
4.2.2. Determination of Soybean Seed Protein Content
4.2.3. DNA Extraction and Detection
4.2.4. Acquisition and Identification of SSR Markers
4.2.5. Segregation Analysis of Traits (SEA)
4.2.6. Single-Marker Analysis
4.2.7. Functional Annotation of Candidate Genes
4.2.8. Amino Acid Sequence Alignment of Candidate Genes
4.2.9. Quantitative Real-Time PCR (qPCR) Analysis
4.2.10. Promoter Element Analysis
4.2.11. Haplotype Analysis of Candidate Genes in the Germplasm Population
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| CSSL | Chromosome Segment Substitution Line |
| QTL | Quantitative Trait Locu |
| qRT-PCR | Quantitative Real-time PCR |
| SNP | Single Nucleotide Polymorphism |
| RIL | Recombinant Inbred Line |
| CDS | Coding Sequence |
| RHL | Residual Heterozygous Line |
| SSR | Simple sequence repeat |
| PAGE | Polyacrylamide Gel Electrophoresis |
| SEA | Segregation Analysis |
References
- Siddiqui, G.A.; Naeem, A. Connecting the dots: Macromolecular crowding and protein aggregation. J. Fluoresc. 2023, 33, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Mao, L.; Zeng, Z.; Yu, X.; Lian, J.; Feng, J.; Yang, W.; An, J.; Wu, H.; Zhang, M.; et al. Genetic mapping high protein content QTL from soybean ‘Nanxiadou 25’and candidate gene analysis. BMC Plant Biol. 2021, 21, 388. [Google Scholar] [CrossRef]
- Zhou, Z.; Jiang, Y.; Wang, Z.; Gou, Z.; Lyu, J.; Li, W.; Yu, Y.; Shu, L.; Zhao, Y.; Ma, Y. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 2015, 33, 408–414. [Google Scholar] [CrossRef]
- Krishnan, H.B.; Jez, J.M. Review: The promise and limits for enhancing sulfur-containing amino acid content of soybean seed. Plant Sci. 2018, 272, 14–21. [Google Scholar] [CrossRef]
- Mejia, S.B.; Messina, M.; Li, S.S.; Viguiliouk, E.; Chiavaroli, L.; Khan, T.A.; Srichaikul, K.; Mirrahimi, A.; Sievenpiper, J.L.; Kris-Etherton, P.; et al. A meta-analysis of 46 studies identified by the FDA demonstrates that soy protein decreases circulating LDL and total cholesterol concentrations in adults. J. Nutr. 2019, 149, 968–981. [Google Scholar] [CrossRef] [PubMed]
- Gao, F. Progress on Factors Influencing Protein Formation and Accumulation in Soybean. Soybean Sci. Technol. 2014, 14–19. (In Chinese) [Google Scholar]
- Piper, E.L.; Boote, K.I. Temperature and cultivar effects on soybean seed oil and protein concentrations. J. Am. Oil Chem. Soc. 1999, 76, 1233–1241. [Google Scholar] [CrossRef]
- Xu, J.; Fang, Y.; Cheng, Y.; Wang, Y.; Guo, C. Soybean Cultivation in Low-Latitude Regions: Adaptive Strategies for Sustainable Production. Plant Cell Environ. 2025. [Google Scholar] [CrossRef]
- Thomas, J.; Boote, K.; Allen, L.; Gallo-Meagher, M.; Davis, J. Elevated temperature and carbon dioxide effects on soybean seed composition and transcript abundance. Crop Sci. 2003, 43, 1548–1557. [Google Scholar] [CrossRef]
- Haq, M.U.; Mallarino, A.P. Response of soybean grain oil and protein concentrations to foliar and soil fertilization. Agron. J. 2005, 97, 910–918. [Google Scholar] [CrossRef]
- Fasoula, V.A.; Harris, D.K.; Boerma, H.R. Validation and designation of quantitative trait loci for seed protein, seed oil, and seed weight from two soybean populations. Crop Sci. 2004, 44, 1218–1225. [Google Scholar] [CrossRef]
- Patil, G.; Mian, R.; Vuong, T.; Pantalone, V.; Song, Q.; Chen, P.; Shannon, G.J.; Carter, T.C.; Nguyen, H.T. Molecular mapping and genomics of soybean seed protein: A review and perspective for the future. Theor. Appl. Genet. 2017, 130, 1975–1991. [Google Scholar] [CrossRef] [PubMed]
- Chiluwal, A.; Haramoto, E.R.; Hildebrand, D.; Naeve, S.; Poffenbarger, H.; Purcell, L.C.; Salmeron, M. Late-season nitrogen applications increase soybean yield and seed protein concentration. Front. Plant Sci. 2021, 12, 715940. [Google Scholar] [CrossRef]
- Diers, B.W.; Keim, P.; Fehr, W.; Shoemaker, R. RFLP analysis of soybean seed protein and oil content. Theor. Appl. Genet. 1992, 83, 608–612. [Google Scholar] [CrossRef]
- Warrington, C.; Abdel-Haleem, H.; Hyten, D.; Cregan, P.; Orf, J.; Killam, A.; Bajjalieh, N.; Li, Z.; Boerma, H. QTL for seed protein and amino acids in the Benning× Danbaekkong soybean population. Theor. Appl. Genet. 2015, 128, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Whiting, R.M.; Torabi, S.; Lukens, L.; Eskandari, M. Genomic regions associated with important seed quality traits in food-grade soybeans. BMC Plant Biol. 2020, 20, 485. [Google Scholar] [CrossRef]
- Clevinger, E.M.; Biyashev, R.; Haak, D.; Song, Q.; Pilot, G.; Saghai Maroof, M. Identification of quantitative trait loci controlling soybean seed protein and oil content. PLoS ONE 2023, 18, e0286329. [Google Scholar] [CrossRef]
- Jamison, D.R.; Chen, P.; Hettiarachchy, N.S.; Miller, D.M.; Shakiba, E. Identification of Quantitative Trait Loci (QTL) for Sucrose and Protein Content in Soybean Seed. Plants 2024, 13, 650. [Google Scholar] [CrossRef]
- Kim, W.J.; Kang, B.H.; Moon, C.Y.; Kang, S.; Shin, S.; Chowdhury, S.; Choi, M.-S.; Park, S.-K.; Moon, J.-K.; Ha, B.-K. Quantitative trait loci (QTL) analysis of seed protein and oil content in wild soybean (Glycine soja). Int. J. Mol. Sci. 2023, 24, 4077. [Google Scholar] [CrossRef]
- Wang, S.; Liu, S.; Wang, J.; Yokosho, K.; Zhou, B.; Yu, Y.-C.; Liu, Z.; Frommer, W.B.; Ma, J.F.; Chen, L.-Q. Simultaneous changes in seed size, oil content and protein content driven by selection of SWEET homologues during soybean domestication. Natl. Sci. Rev. 2020, 7, 1776–1786. [Google Scholar] [CrossRef]
- Duan, Z.; Zhang, M.; Zhang, Z.; Liang, S.; Fan, L.; Yang, X.; Yuan, Y.; Pan, Y.; Zhou, G.; Liu, S.; et al. Natural allelic variation of GmST05 controlling seed size and quality in soybean. Plant Biotechnol. J. 2022, 20, 1807–1818. [Google Scholar] [CrossRef]
- Qi, Z.; Guo, C.; Li, H.; Qiu, H.; Li, H.; Jong, C.; Yu, G.; Zhang, Y.; Hu, L.; Wu, X. Natural variation in Fatty Acid 9 is a determinant of fatty acid and protein content. Plant Biotechnol. J. 2024, 22, 759–773. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, L.; Zuo, H.; Yang, Y.; Hu, D.; Zhang, S.; Yuan, W.; Zhai, X.; He, M.; Xu, M. GmGASA12 coordinates hormonal dynamics to enhance soybean water-soluble protein accumulation and seed size. J. Integr. Plant Biol. 2025, 67, 2401–2415. [Google Scholar] [CrossRef] [PubMed]
- Cao, x.; Liu, B.; Zhang, Y. SEA: A software package of segregation analysis of quantitative traits in plants. J. Nanjing Agric. Univ. 2013, 36, 1–6. (In Chinese) [Google Scholar]
- Liu, J.; Jiang, A.; Ma, R.; Gao, W.; Tan, P.; Li, X.; Du, C.; Zhang, J.; Zhang, X.; Zhang, L. QTL Mapping for seed quality traits under multiple environments in soybean (Glycine max L.). Agronomy 2023, 13, 2382. [Google Scholar] [CrossRef]
- Guo, X.; Jiang, J.; Liu, Y.; Yu, L.; Chang, R.; Guan, R.; Qiu, L. Identification of a novel salt tolerance-related locus in wild soybean (Glycine soja Sieb. & Zucc.). Front. Plant Sci. 2021, 12, 791175. [Google Scholar] [CrossRef]
- Miao, N.; Zhou, J.; Li, M.; Zhang, J.; Hu, Y.; Guo, J.; Zhang, T.; Shi, L. Remodeling and protecting the membrane system to resist phosphorus deficiency in wild soybean (Glycine soja) seedling leaves. Planta 2022, 255, 53. [Google Scholar] [CrossRef]
- Liu, Y.; Cao, L.; Wu, X.; Wang, S.; Zhang, P.; Li, M.; Jiang, J.; Ding, X.; Cao, X. Functional characterization of wild soybean (Glycine soja) GsSnRK1. 1 protein kinase in plant resistance to abiotic stresses. J. Plant Physiol. 2023, 280, 153881. [Google Scholar] [CrossRef]
- Chotekajorn, A.; Hashiguchi, T.; Hashiguchi, M.; Tanaka, H.; Akashi, R. Evaluation of seed amino acid content and its correlation network analysis in wild soybean (Glycine soja) germplasm in Japan. Plant Genet. Resour. 2021, 19, 35–43. [Google Scholar] [CrossRef]
- Dwiyanti, M.S.; Maruyama, S.; Hirono, M.; Sato, M.; Park, E.; Yoon, S.H.; Yamada, T.; Abe, J. Natural diversity of seed α-tocopherol ratio in wild soybean (Glycine soja) germplasm collection. Breed. Sci. 2016, 66, 653–657. [Google Scholar] [CrossRef]
- Jun, T.-H.; Van, K.; Kim, M.Y.; Lee, S.-H.; Walker, D.R. Association analysis using SSR markers to find QTL for seed protein content in soybean. Euphytica 2008, 162, 179–191. [Google Scholar] [CrossRef]
- Sonah, H.; O’Donoughue, L.; Cober, E.; Rajcan, I.; Belzile, F. Identification of loci governing eight agronomic traits using a GBS-GWAS approach and validation by QTL mapping in soya bean. Plant Biotechnol. J. 2015, 13, 211–221. [Google Scholar] [CrossRef]
- Li, X.; Shao, Z.; Tian, R.; Zhang, H.; Du, H.; Kong, Y.; Li, W.; Zhang, C. Mining QTLs and candidate genes for seed protein and oil contents across multiple environments and backgrounds in soybean. Mol. Breed. 2019, 39, 139. [Google Scholar] [CrossRef]
- Wang, Y. Cloning and Expression Analysis of Three WD40 Transcription Factors in Roses. Master’s Thesis, Shandong Agricultural University, Weihai, China, 2019. (In Chinese). [Google Scholar]
- van der Voorn, L.; Ploegh, H.L. The WD-40 repeat. Febs Lett. 1992, 307, 131–134. [Google Scholar] [CrossRef]
- Will, N.; Piserchio, A.; Snyder, I.; Ferguson, S.B.; Giles, D.H.; Dalby, K.N.; Ghose, R. Structure of the C-Terminal helical repeat domain of eukaryotic elongation factor 2 Kinase. Biochemistry 2016, 55, 5377–5386. [Google Scholar] [CrossRef]
- Liu, H.; Xiu, Z.; Yang, H.; Ma, Z.; Yang, D.; Wang, H.; Tan, B.-C. Maize Shrek1 encodes a WD40 protein that regulates pre-rRNA processing in ribosome biogenesis. Plant Cell 2022, 34, 4028–4044. [Google Scholar] [CrossRef]
- Sun, J.; Li, W.; Wei, X.; Shou, H.; Tran, L.S.P.; Feng, X.; Wang, S. Mechanistic roles of GmSWEET10a/b and GmSUT1 in the oil–protein balance in soybean mature seeds at transcriptional and metabolic levels. Plant J. 2025, 123, e70435. [Google Scholar] [CrossRef]
- Zhong, X.; Wang, J.; Shi, X.; Bai, M.; Yuan, C.; Cai, C.; Wang, N.; Zhu, X.; Kuang, H.; Wang, X. Genetically optimizing soybean nodulation improves yield and protein content. Nat. Plants 2024, 10, 736–742. [Google Scholar] [CrossRef]
- Zheng, H.; Feng, X.; Wang, L.; Shao, W.; Guo, S.; Zhao, D.; Li, J.; Yan, L.; Miao, L.; Sun, B. GmSop20 functions as a key coordinator of the oil-to-protein ratio in soybean seeds. Adv. Sci. 2025, 12, e05181. [Google Scholar] [CrossRef] [PubMed]
- Xin, D.; Qi, Z.; Jiang, H.; Hu, Z.; Zhu, R.; Hu, J.; Han, H.; Hu, G.; Liu, C.; Chen, Q. QTL location and epistatic effect analysis of 100-seed weight using wild soybean (Glycine soja Sieb. & Zucc.) chromosome segment substitution lines. PLoS ONE 2016, 11, e0149380. [Google Scholar]
- Doyle, J.; Doyle, J.; Brown, A. Analysis of a polyploid complex in Glycine with chloroplast and nuclear DNA. Aust. Syst. Bot. 1990, 3, 125–136. [Google Scholar] [CrossRef]
- Wang, S.; Basten, C.; Zeng, Z.; Wang, S.C.; Basten, C.J.; Zeng, Z.B. Windows QTL Cartographer 2.5; Department of Statistics, North Carolina State University: Raleigh, NC, USA, 2007. [Google Scholar]
- Zhang, D.; Zhang, H.; Hu, Z.; Chu, S.; Yu, K.; Lv, L.; Yang, Y.; Zhang, X.; Chen, X.; Kan, G. Artificial selection on GmOLEO1 contributes to the increase in seed oil during soybean domestication. PLoS Genet. 2019, 15, e1008267. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Li, X.; Shi, Z.; Gao, J.; Wang, X.; Guo, K. CandiHap: A haplotype analysis toolkit for natural variation study. Mol. Breed. 2023, 43, 21. [Google Scholar] [CrossRef] [PubMed]






| Population | Model | AIC | M-G Var | Heritability (M-G) | P (nW) | P (Dn) |
|---|---|---|---|---|---|---|
| F2 | 1MG-A | 225.7 | 0.091 | 0.094 | 0.884 | 1 |
| Population | Model | AIC | M-G Var | Heritability (M-G) | nW2P (nW) | P (Dn) |
|---|---|---|---|---|---|---|
| F2(R1) | 1MG-A | 275.6 | 0.024 | 0.026 | 0.265 | 1 |
| F2(R2) | 1MG-AD | 542.774 | 0.866 | 0.736 | 0.914 | 1 |
| Gene Name | Homologous Gene | GO Annotation | KEGG Annotation | nr Annotation |
|---|---|---|---|---|
| Glyma.16G164900 | AT1G76730.1 | none | none | NAGB/RpiA/CoA Transferase Superfamily Protein |
| Glyma.16G165000 | AT1G43190.2 | GO:0003676 | K14948 | Polypyrimidine Tract-Binding Protein |
| Glyma.16G165100 | AT5G52820.1 | GO:0005515 | K14855 | WD-40 Repeat Family Protein |
| Glyma.16G165200 | AT2G34430.1 | GO:0016020 | K08912 | Light-Harvesting Chlorophyll Protein Complex II Subunit B1 |
| Glyma.16G165300 | AT1G34040.1 | GO:0016846 | none | Pyridoxal 5′-Phosphate Transferase Superfamily Protein |
| Glyma.16G165400 | AT1G12300.1 | none | K17710 | Tetratricopeptide Repeat Superfamily Protein |
| Glyma.16G165500 | AT2G34430.1 | GO:0016020 | K08912 | Light-Harvesting Chlorophyll Protein Complex II Subunit B1 |
| Glyma.16G165600 | AT1G76740.1 | none | none | none |
| Glyma.16G165700 | AT5G10090.1 | none | none | Tetratricopeptide Repeat Superfamily Protein |
| Maximum Temperature | Minimum Temperature | Average Temperature | Active Accumulated Temperature | Daily Average Active Accumulated Temperature | |
|---|---|---|---|---|---|
| 2017 | 35 °C | −6 °C | 17.66 °C | 3986.5 °C | 26.2 °C |
| 2018 | 35 °C | −3 °C | 17.51 °C | 4129.5 °C | 26.1 °C |
| 2019 | 33 °C | −4 °C | 17.21 °C | 3876.0 °C | 26.5 °C |
| 2020 | 33 °C | −4 °C | 17.43 °C | 4329.0 °C | 26.7 °C |
| Maximum Precipitation | Minimum Precipitation | Average Precipitation | |
|---|---|---|---|
| 2017 | 29.87 mm | 0 mm | 2.82 mm |
| 2018 | 37.50 mm | 0 mm | 3.60 mm |
| 2019 | 46.89 mm | 0 mm | 4.39 mm |
| 2020 | 55.92 mm | 0 mm | 4.20 mm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Xie, J.; Li, G.; Shen, M.; Zheng, Y.; Meng, F.; Fan, X.; Sun, X.; Zhang, Y.; Wang, M.; et al. Fine Mapping of qSPJ_1 and Candidate Gene Identification for Soybean Seed Protein Content. Plants 2025, 14, 3525. https://doi.org/10.3390/plants14223525
Chen J, Xie J, Li G, Shen M, Zheng Y, Meng F, Fan X, Sun X, Zhang Y, Wang M, et al. Fine Mapping of qSPJ_1 and Candidate Gene Identification for Soybean Seed Protein Content. Plants. 2025; 14(22):3525. https://doi.org/10.3390/plants14223525
Chicago/Turabian StyleChen, Jiayuan, Jianguo Xie, Guang Li, Mingzhe Shen, Yuhong Zheng, Fanfan Meng, Xuhong Fan, Xingmiao Sun, Yunfeng Zhang, Mingliang Wang, and et al. 2025. "Fine Mapping of qSPJ_1 and Candidate Gene Identification for Soybean Seed Protein Content" Plants 14, no. 22: 3525. https://doi.org/10.3390/plants14223525
APA StyleChen, J., Xie, J., Li, G., Shen, M., Zheng, Y., Meng, F., Fan, X., Sun, X., Zhang, Y., Wang, M., Yang, Z., Xiong, X., Wang, Q., Wang, S., & Jiang, H. (2025). Fine Mapping of qSPJ_1 and Candidate Gene Identification for Soybean Seed Protein Content. Plants, 14(22), 3525. https://doi.org/10.3390/plants14223525






