MutMap-Based Cloning of a Soybean Mosaic Virus Resistance Gene
Abstract
1. Introduction
2. Results
2.1. Identification and Evaluation of SCV Resistance in EMS-Induced Mutants
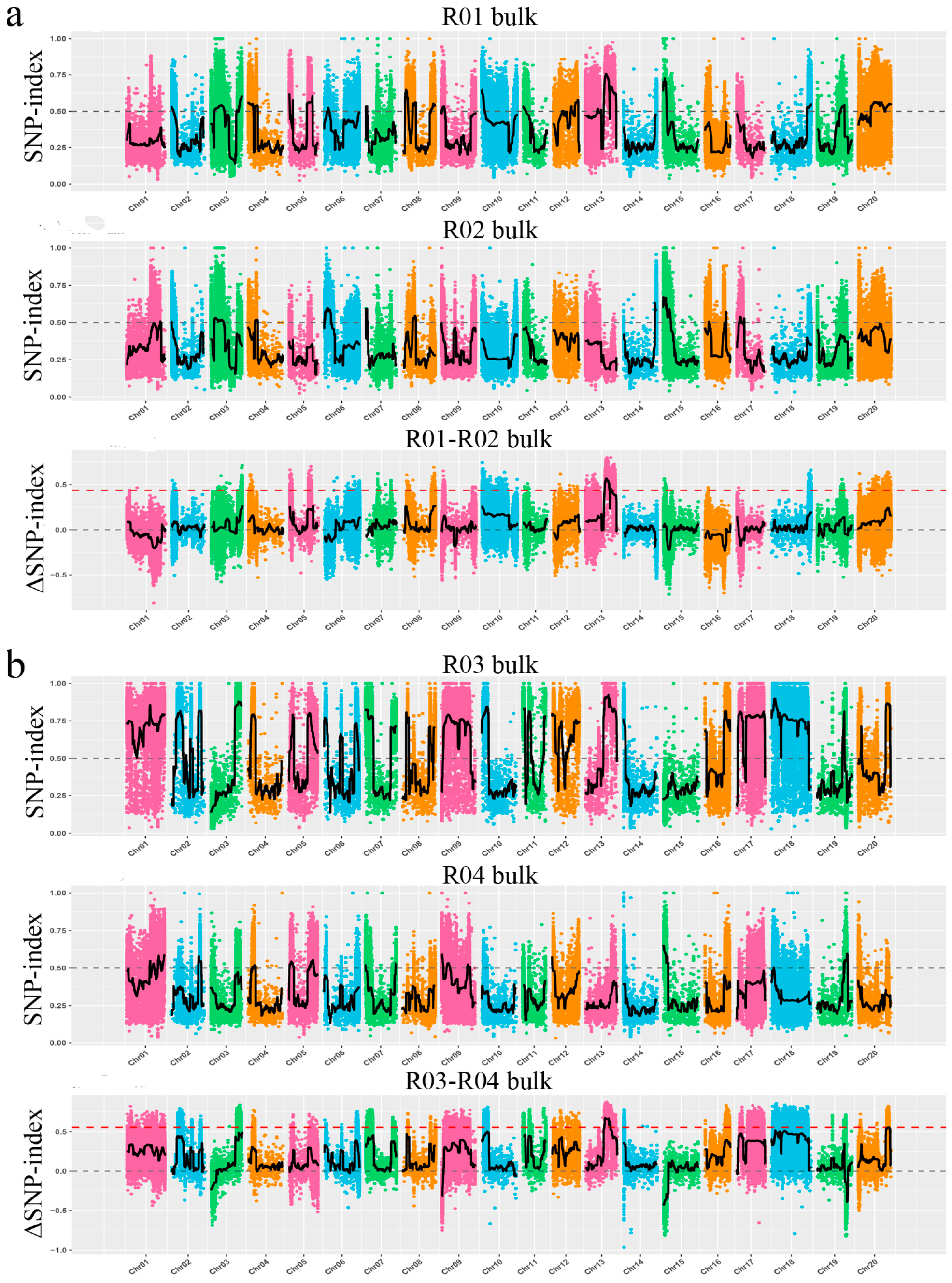
2.2. MutMap-Based Mapping of Resistance Candidate Intervals
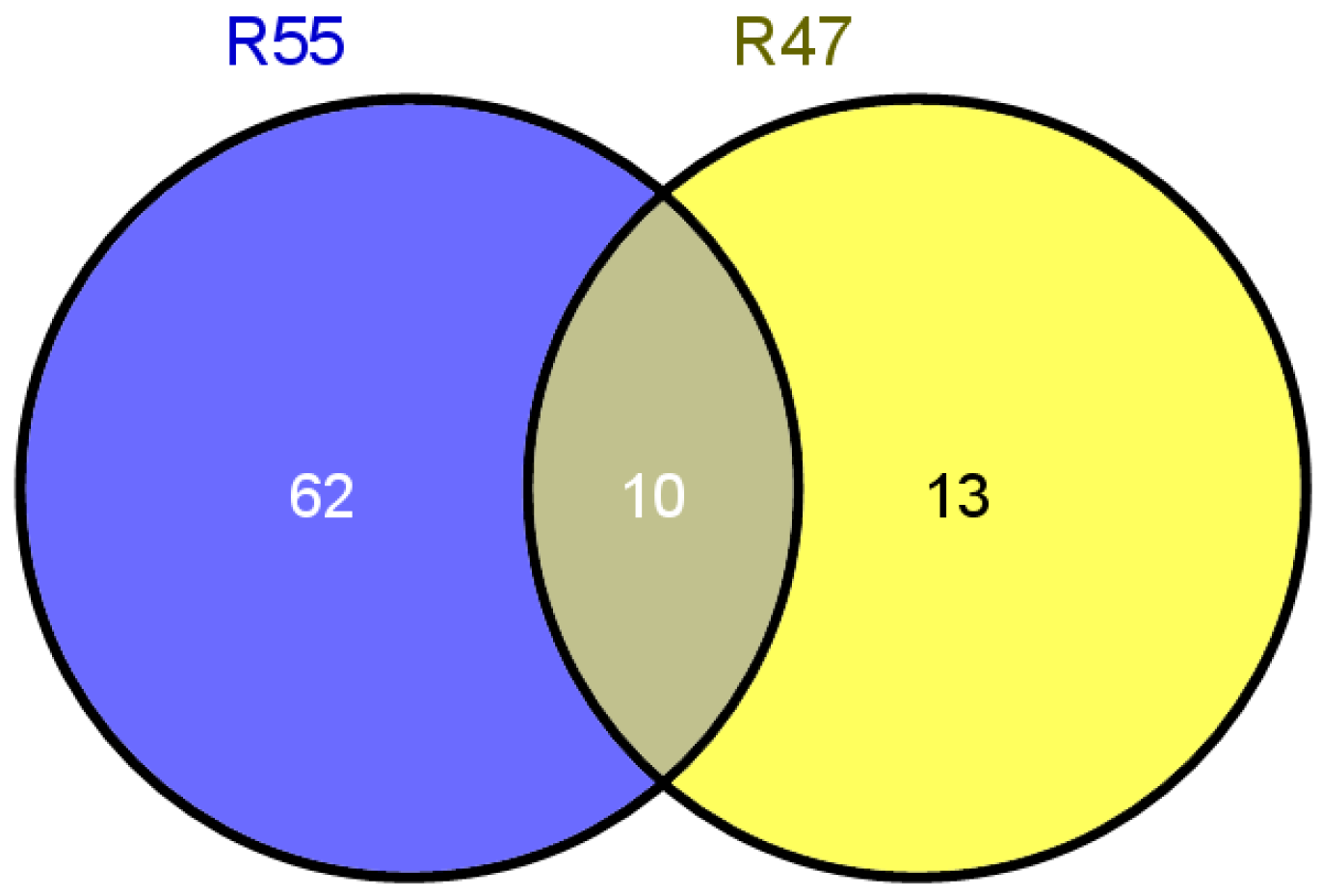
2.3. Screening of Resistance Candidate Genes
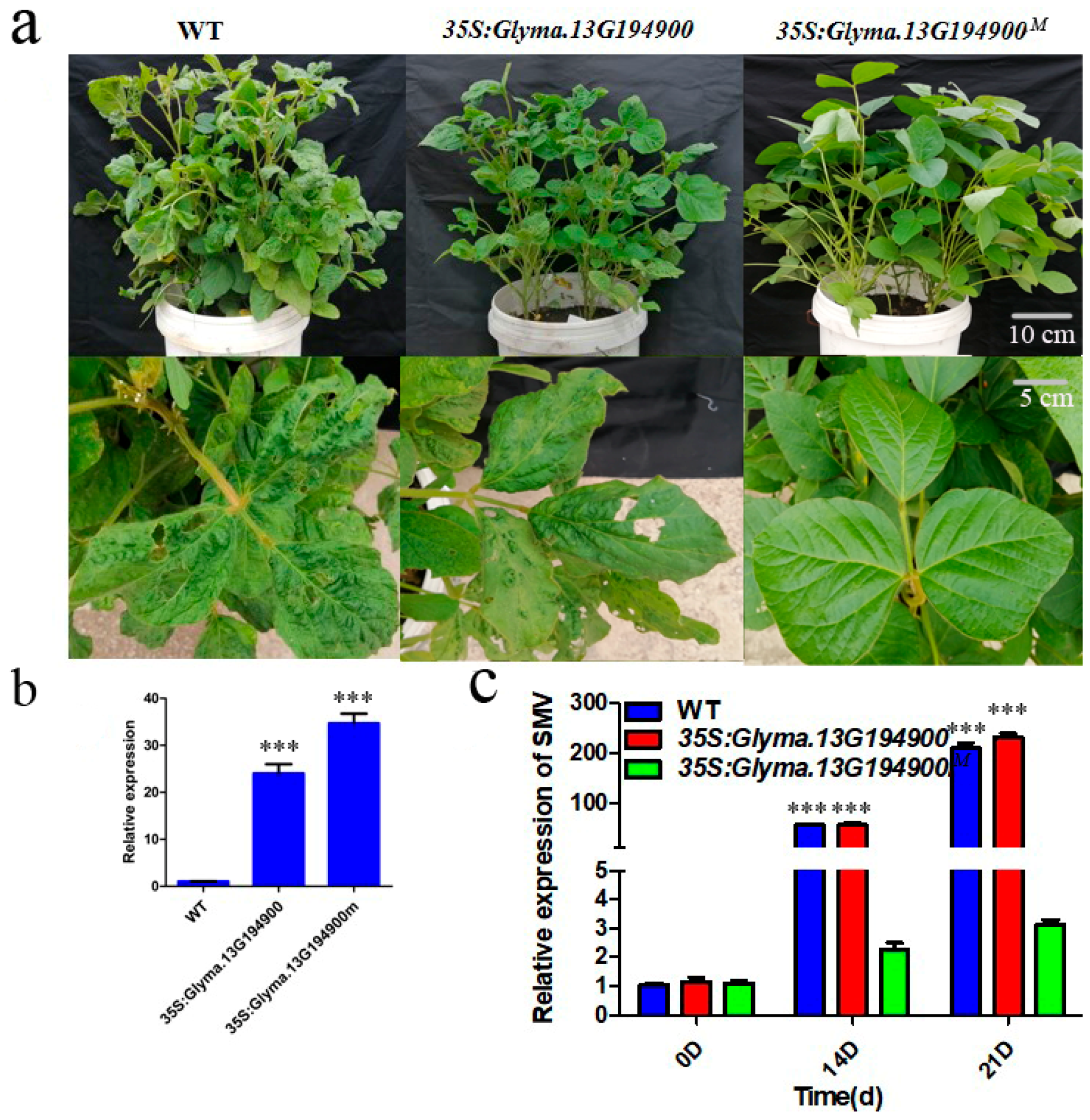
2.4. Transgenic Resistance Assay of the Candidate Resistance Gene
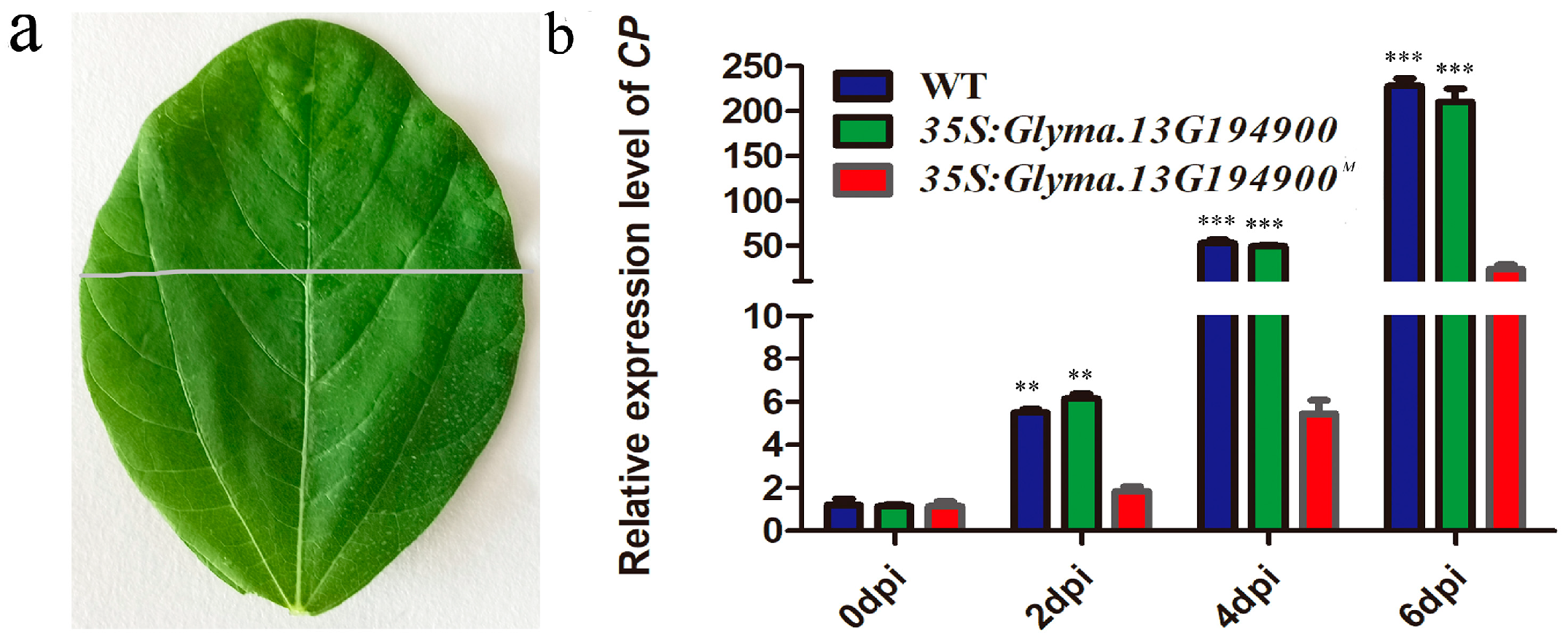
2.5. Overexpression of Glyma.13G194900M Inhibits Viral Replication and/or Cell-to-Cell Movement
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Virus
4.2. Gene Clone and Vector Construction
4.3. Soybean Transformation and Screening of Transgenic Plants
4.4. Genetic Mapping and Bulked Segregant Analysis (BSA)
4.5. Quantitative RT-PCR (qRT-PCR)
4.6. Disease Resistance Assessment in Transgenic Plants
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hartman, G.L.; West, E.D.; Herman, T.K. Crops that feed the world 2. Soybean—Worldwide production, use, and constraints caused by pathogens and pests. Food Secur. 2011, 3, 5–17. [Google Scholar] [CrossRef]
- Cui, X.; Chen, X.; Wang, A. Detection, Understanding and Control of Soybean Mosaic Virus; InTech: Rijeka, Croatia, 2011; pp. 335–354. [Google Scholar]
- Hajimorad, M.R.; Domier, L.L.; Tolin, S.A.; Whitham, S.A.; Saghai Maroof, M.A. Soybean mosaic virus: A successful potyvirus with a wide distribution but restricted natural host range. Mol. Plant Pathol. 2018, 19, 1563–1579. [Google Scholar] [CrossRef]
- Zhou, J.; Tzanetakis, I.E.; French, R.; Ghabrial, S.A. Characterization of Soybean vein necrosis virus, a New Tospovirus Associated with Necrotic Symptoms in Soybean. Virus Genes 2011, 43, 289–294. [Google Scholar] [CrossRef]
- Seo, J.K.; Ohshima, K.; Lee, H.G.; Son, M.; Choi, H.S.; Lee, S.H.; Sohn, S.H.; Kim, K.H. Molecular variability and genetic structure of the population of soybean mosaic virus based on the analysis of complete genome sequences. Virology 2009, 393, 91–103. [Google Scholar] [CrossRef]
- Cho, E.K.; Goodman, R.M. Strains of soybean mosaic virus: Classification based on virulence in resistant soybean cultivars. Phytopathology 1979, 69, 467–470. [Google Scholar] [CrossRef]
- Cho, E.K.; Goodman, R.M. Evaluation of resistance in soybeans to Soybean mosaic virus strains. Crop Sci. 1982, 22, 1133–1136. [Google Scholar] [CrossRef]
- Li, K.; Yang, Q.H.; Zhi, H.J.; Gai, J.Y. Identification and distribution of soybean mosaic virus strains in southern China. Plant Dis. 2010, 94, 351–357. [Google Scholar] [CrossRef]
- Wang, X.; Gai, J.; Pu, Z. Classification and distribution of strain groups of soybean mosaic virus in middle and lower Huang-huai and Changjiang valleys. Soybean Sci. 2003, 22, 102–107, (In Chinese with English Abstract). [Google Scholar]
- Gore, M.A.; Hayes, A.J.; Jeong, S.C.; Yue, Y.G.; Buss, G.R.; Saghai Maroof, M.A. Mapping tightly linked genes controlling potyvirus infection at the Rsv1 and Rpv1 region in soybean. Genome 2002, 45, 592–599. [Google Scholar] [CrossRef]
- Hwang, T.Y.; Moon, J.K.; Yu, S.; Yang, K.; Mohankumar, S.; Yu, Y.H.; Lee, Y.H.; Kim, H.S.; Kim, H.M.; Saghai Maroof, M.A.; et al. Application of comparative genomics in developing molecular markers tightly linked to the virus resistance gene Rsv4 in soybean. Genome 2006, 49, 380–388. [Google Scholar] [CrossRef]
- Jeong, S.C.; Kristipati, S.; Hayes, A.J.; Maughan, P.J.; Noffsinger, S.L.; Gunduz, I.; Buss, G.R.; Saghai Maroof, M.A. Genetic and sequence analysis of markers tightly linked to the resistance gene, Rsv3. Crop Sci. 2002, 42, 265. [Google Scholar]
- Chen, P.; Ma, G.; Buss, G.R.; Gunduz, I.; Roane, C.W.; Tolin, S.A. Inheritance and allelism tests of Raiden soybean for resistance to soybean mosaic virus. J. Hered. 2001, 92, 51–55. [Google Scholar] [CrossRef]
- Gunduz, I.; Buss, G.R.; Chen, P.; Tolin, S.A. Characterization of SMV resistance genes in Tousan 140 and Hourei soybean. Crop Sci. 2002, 42, 90–95. [Google Scholar] [CrossRef]
- Hayes, A.J.; Ma, G.; Buss, G.R.; Saghai Maroof, M.A. Molecular marker mapping of Rsv4, a gene conferring resistance to all known strains of soybean mosaic virus. Crop Sci. 2000, 40, 1434. [Google Scholar] [CrossRef]
- Widyasari, K.; Tran, P.T.; Shin, J.; Son, H.; Kim, K.H. Overexpression of purple acid phosphatase GmPAP2.1 confers resistance to Soybean mosaic virus in a susceptible soybean cultivar. J. Exp. Bot. 2022, 73, 1623–1642. [Google Scholar] [CrossRef]
- Rui, R.; Liu, S.; Karthikeyan, A.; Wang, T.; Niu, H.P.; Yin, J.L.; Yang, Y.H.; Yang, Q.H.; Zhi, H.J.; Li, K. Fine-mapping and identification of a novel locus Rsc15 underlying soybean resistance to Soybean mosaic virus. Theor. Appl. Genet. 2017, 130, 2395–2410. [Google Scholar] [CrossRef]
- Zhou, L.; He, H.L.; Liu, R.F.; Han, Q.; Shou, H.X.; Liu, B. Overexpression of GmAKT2 potassium channel enhances resistance to soybean mosaic virus. BMC Plant Biol. 2014, 14, 154. [Google Scholar] [CrossRef]
- Seo, J.K.; Kwon, S.J.; Cho, W.K.; Choi, H.S.; Kim, K.H. Type 2c protein phosphatase is a key regulator of antiviral extreme resistance limiting virus spread. Sci. Rep. 2014, 4, 5905. [Google Scholar] [CrossRef]
- Zhou, Z.; He, H.L.; Ma, L.P.; Yu, X.Q.; Mi, Q.; Pang, J.S.; Tang, G.X.; Liu, B. Overexpression of a GmCnx1 gene enhanced activity of nitrate reductase and aldehyde oxidase, and boosted mosaic virus resistance in soybean. PLoS ONE 2015, 10, e0124273. [Google Scholar] [CrossRef]
- Luan, H.X.; Shine, M.B.; Cui, X.Y.; Chen, X.; Ma, N.; Kachroo, O.; Zhi, H.J.; Kachroo, A. The potyviral p3 protein targets eEF1a to promote the unfolded protein response and viral pathogenesis. Plant Physiol. 2016, 172, 221–234. [Google Scholar] [CrossRef]
- He, H.L.; Yang, X.D.; Xun, H.W.; Lou, X.; Li, S.Z.; Zhang, Z.B.; Jiang, L.L.; Dong, Y.S.; Wang, S.C.; Liu, B. Over-expression of GmSN1 enhances virus resistance in Arabidopsis and soybean. Plant Cell Rep. 2017, 36, 1441–1455. [Google Scholar] [CrossRef]
- Xun, H.W.; Yang, X.D.; He, H.L.; Wang, M.; Guo, P.; Wang, Y.; Pang, J.S.; Dong, Y.S.; Feng, X.Z.; Wang, S.C.; et al. Over-expression of GmKR3, a TIR-NBS-LRR type R gene, confers resistance to multiple viruses in soybean. Plant Mol. Biol. 2019, 99, 95–111. [Google Scholar] [CrossRef]
- Luan, H.X.; Niu, H.P.; Luo, J.Y.; Zhi, H.J. Soybean cytochrome b5 Is a restriction factor for soybean mosaic virus. Viruses 2019, 11, 546. [Google Scholar] [CrossRef]
- Zhang, P.P.; Du, H.Y.; Wang, J.; Pu, Y.X.; Yang, C.Y.; Yan, R.J.; Yang, H.; Cheng, H.; Yu, D.Y. Multiplex CRISPR/Cas9-mediated metabolic engineering increases soya bean isoflavone content and resistance to soya bean mosaic virus. Plant Biotechnol. J. 2021, 18, 1384–1395. [Google Scholar] [CrossRef]
- Zhao, X.; Yan, J.; Luo, Z.H.; Gao, S.N.; Teng, W.L.; Zhan, Y.H.; Qiu, L.J.; Zheng, H.K.; Li, W.B.; Han, Y.P. GmST1, which encodes a sulfotransferase, confers resistance to soybean mosaic virus strains G2 and G3. Plant Cell Environ. 2021, 44, 2777–2792. [Google Scholar] [CrossRef]
- Yin, J.L.; Wang, L.Q.; Jin, T.T.; Nie, Y.; Liu, H.; Qiu, Y.L.; Yang, Y.H.; Li, B.W.; Zhang, J.J.; Wang, D.G.; et al. A cell wall-localized NLR confers resistance to soybean mosaic virus by recognizing viral-encoded cylindrical inclusion protein. Mol. Plant 2021, 14, 1881–1900. [Google Scholar] [CrossRef]
- Ren, Q.Y.; Jiang, H.; Xiang, W.Y.; Nie, Y.; Xue, S.; Zhi, H.J.; Li, K.; Gai, J. A MADS-box gene is involved in soybean resistance to multiple soybean mosaic virus strains. Crop J. 2022, 10, 802–808. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Du, H.P.; Zhao, T.T.; Liao, C.M.; Feng, T.; Qin, J.; Liu, B.H.; Kong, F.J.; Che, Z.J.; Chen, L.Y. GmTOC1b negatively regulates resistance to Soybean mosaic virus. Crop J. 2023, 11, 1762–1773. [Google Scholar] [CrossRef]
- Song, S.; Wang, J.; Zhou, J.; Cheng, X.; Hu, Y.; Wang, J.; Zou, J.; Zhao, Y.; Liu, C.; Hu, Z.; et al. Single-Cell RNA-Sequencing of Soybean Reveals Transcriptional Changes and Antiviral Functions of GmGSTU23 and GmGSTU24 in Response to Soybean mosaic Virus. Plant Cell Environ. 2024. [Google Scholar] [CrossRef]
- Jin, T.; Yin, J.; Wang, T.; Xue, S.; Li, B.; Zong, T.; Yang, Y.; Liu, H.; Liu, M.; Xu, K.; et al. RSC3K of soybean cv. Kefeng No.1 confers resistance to soybean mosaic virus by interacting with the viral protein P3. J. Integr. Plant Biol. 2023, 65, 838–853. [Google Scholar] [CrossRef]
- Chu, J.H.; Li, W.L.; Shao, Z.Q.; Yang, Z.W.; Xing, X.Z.; Zhang, H.; Tian, R.; Zhang, H.T.; Li, X.H.; Zhang, C.Y. Novel genetic loci and a functional gene Gm18GRSC3 conferring SMV-SC3 resistance in soybean. Crop J. 2025, 13, 1127–1136. [Google Scholar] [CrossRef]
- Li, B.W.; Wang, L.Q.; Qian, X.Y.; Liu, H.; Jin, T.T.; Yin, J.L.; Hu, T.; Liu, M.Z.; Guo, D.Q.; Li, K.; et al. Gma-miR398c/d negatively regulates soybean resistance to Soybean mosaic virus by targeting SOD family genes. Crop J. 2025, 13, 1490–1502. [Google Scholar] [CrossRef]
- Peters, J.L.; Cnudde, F.; Gerats, T. Forward genetics and map-based cloning approaches. Trends Plant Sci. 2003, 8, 484–491. [Google Scholar] [CrossRef]
- Abe, A.; Kosugi, S.; Yoshida, K.; Natsume, S.; Takagi, H.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Mitsuoka, C.; Tamiru, M.; et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 2012, 30, 174–178. [Google Scholar] [CrossRef]
- Sugihara, Y.; Young, L.; Yaegashi, H.; Natsume, S.; Shea, D.J.; Takagi, H.; Booker, H.; Innan, H.; Terauchi, R.; Abe, A. High-performance pipeline for MutMap and QTL-seq. PeerJ 2022, 10, e13170. [Google Scholar] [CrossRef]
- Li, Z.F.; Guo, Y.; Ou, L.; Hong, H.; Wang, J.; Liu, Z.X.; Guo, B.; Zhang, L.; Qiu, L. Identification of the dwarf gene GmDW1 in soybean (Glycine max L.) by combining mapping-by-sequencing and linkage analysis. Theor. Appl. Genet. 2018, 131, 1001–1016. [Google Scholar] [CrossRef]
- Shen, Y.; Zhou, Z.; Wang, Z.; Li, W.; Fang, C.; Wu, M.; Ma, Y.; Liu, T.; Kong, L.A.; Peng, D.L.; et al. Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell 2014, 26, 996–1008. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.B.; Brommonschenkel, S.H.; Chunwongse, J.; Frary, A.; Ganal, M.W.; Spivey, R.; Wu, T.; Earle, E.D.; Tanksley, S.D. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 1993, 262, 1432–1436. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Yang, S.; Tang, K.; Zhang, Y.; Leng, J.; Ma, J.; Wang, Q.; Feng, X. GmPGL1, a Thiamine Thiazole Synthase, Is Required for the Biosynthesis of Thiamine in Soybean. Front. Plant Sci. 2019, 10, 1546. [Google Scholar] [CrossRef]
- Kim, Y.; Schumaker, K.S.; Zhu, J.K. EMS mutagenesis of Arabidopsis. Methods Mol. Biol. 2006, 323, 101–113. [Google Scholar]
- Campbell, B.W.; Mani, D.; Curtin, S.J.; Slattery, R.A.; Michno, J.M.; Ort, D.R.; Schaus, P.J.; Palmer, R.G.; Orf, J.H.; Stupar, R.M. Identical substitutions in magnesium chelatase paralogs result in chlorophyll-deficient soybean mutants. G3 Genes|Genomes|Genet. 2014, 5, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Piau, M.; Schmitt-Keichinger, C. The Hypersensitive Response to Plant Viruses. Viruses 2023, 15, 2000. [Google Scholar] [CrossRef]
- Mundt, C.C. Pyramiding for Resistance Durability: Theory and Practice. Phytopathology 2018, 108, 792–802. [Google Scholar] [CrossRef]
- Chen, L.; Wang, L.; Zhang, L.; Li, Y.G.; Han, S.J. Enhancing Agrobacterium-mediated soybean transformation efficiency with an auxiliary solution. Crop Health 2024, 2, 17. [Google Scholar] [CrossRef]


| Candidate Gene | Functional Annotation | New Leaf Tissue Expression Level (FPKM) |
|---|---|---|
| Glyma.13G162200 | Translocase of chloroplast 90 | 1.7 |
| Glyma.13G178200 | RNA processing and modification | 0.5 |
| Glyma.13G179000 | Signal transduction mechanisms | 2.1 |
| Glyma.13G182200 | Glycosyl transferases group | 0 |
| Glyma.13G182600 | RNA processing and modification | 6.2 |
| Glyma.13G190000 | NB-LRR-ARC domain (R-gene) | 0.3 |
| Glyma.13G191000 | LOB domain-containing protein | 0 |
| Glyma.13G194800 | NB-LRR-ARC domain (R-gene) | 1.3 |
| Glyma.13G194900 | NB-LRR-ARC domain (R-gene) | 4.2 |
| Glyma.13G201800 | DNA Repair protein | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.; Zhong, X.; Yu, D.; Rao, D.; Niu, L.; Xun, H.; Zhu, X.; Yi, L.; Qian, X.; Meng, F. MutMap-Based Cloning of a Soybean Mosaic Virus Resistance Gene. Plants 2025, 14, 3504. https://doi.org/10.3390/plants14223504
Wang B, Zhong X, Yu D, Rao D, Niu L, Xun H, Zhu X, Yi L, Qian X, Meng F. MutMap-Based Cloning of a Soybean Mosaic Virus Resistance Gene. Plants. 2025; 14(22):3504. https://doi.org/10.3390/plants14223504
Chicago/Turabian StyleWang, Bin, Xiaofang Zhong, Debin Yu, Demin Rao, Lu Niu, Hongwei Xun, Xiangyu Zhu, Lu Yi, Xueyan Qian, and Fangang Meng. 2025. "MutMap-Based Cloning of a Soybean Mosaic Virus Resistance Gene" Plants 14, no. 22: 3504. https://doi.org/10.3390/plants14223504
APA StyleWang, B., Zhong, X., Yu, D., Rao, D., Niu, L., Xun, H., Zhu, X., Yi, L., Qian, X., & Meng, F. (2025). MutMap-Based Cloning of a Soybean Mosaic Virus Resistance Gene. Plants, 14(22), 3504. https://doi.org/10.3390/plants14223504







