Microbial Consortium of Streptomyces spp. from Mining Environments Enhances Phytoremediation Potential of Lemna minor L.
Abstract
1. Introduction
2. Results
2.1. DNA Extraction and 16S rRNA Metabarcoding
2.2. Actinomycetes Isolation and In Vitro Heavy Metals Tolerance
2.3. Plant Growth-Promoting Activities
2.4. In Planta Experiment on Lemna minor L.
3. Discussion
4. Materials and Methods
4.1. Sampling Area
4.2. DNA Extraction and 16S rRNA Gene Metabarcoding
4.3. Isolation and Identification of Actinomycetes Strains
4.4. In Vitro Heavy Metal Tolerance
4.5. Plant Growth-Promoting (PGP) Activities
4.5.1. Phosphate Solubilisation
4.5.2. Indoles Production
4.5.3. Siderophore Production
4.5.4. Ammonia and HCN Production
4.5.5. The 1-Aminocyclopropane-1-Carboxylate (ACC) Deaminase Activity
4.6. Molecular Identification and Phylogenetic Analysis
4.7. In Planta Experiment on Lemna minor L.
- CNT (without PGPB and heavy metals)
- PGPB (with PGPB and without heavy metals)
- CNT-HM (without PGPB and with heavy metals)
- PGPB-HM (with PGPB and heavy metals)
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Han, L.; Zhou, W.; Li, W.; Qian, Y. Urbanization Strategy and Environmental Changes: An Insight with Relationship between Population Change and Fine Particulate Pollution. Sci. Total Environ. 2018, 642, 789–799. [Google Scholar] [CrossRef]
- Bokhari, S.H.; Ahmad, I.; Mahmood-Ul-Hassan, M.; Mohammad, A. Phytoremediation Potential of Lemna minor L. for Heavy Metals. Int. J. Phytoremediat. 2016, 18, 25–32. [Google Scholar] [CrossRef]
- Arantza, S.-J.; Hiram, M.-R.; Erika, K.; Chávez-Avilés, M.N.; Valiente-Banuet, J.I.; Fierros-Romero, G. Bio- and Phytoremediation: Plants and Microbes to the Rescue of Heavy Metal Polluted Soils. SN Appl. Sci. 2022, 4, 59. [Google Scholar] [CrossRef]
- Mishra, S.; Bharagava, R.N.; More, N.; Yadav, A.; Zainith, S.; Mani, S.; Chowdhary, P. Heavy Metal Contamination: An Alarming Threat to Environment and Human Health. In Environmental Biotechnology: For Sustainable Future; Springer: Singapore, 2019; pp. 103–125. [Google Scholar]
- Li, C.; Zhou, K.; Qin, W.; Tian, C.; Qi, M.; Yan, X.; Han, W. A Review on Heavy Metals Contamination in Soil: Effects, Sources, and Remediation Techniques. Soil. Sediment. Contam. Int. J. 2019, 28, 380–394. [Google Scholar] [CrossRef]
- Rascio, N.; Navari-Izzo, F. Heavy Metal Hyperaccumulating Plants: How and Why Do They Do It? And What Makes Them so Interesting? Plant Sci. 2011, 180, 169–181. [Google Scholar] [CrossRef]
- Aithani, D.; Kushawaha, J. Heavy Metals Contamination in Environment. In Remediation of Heavy Metals; Wiley: Hoboken, NJ, USA, 2024; pp. 15–30. [Google Scholar]
- Khan, F.I.; Husain, T.; Hejazi, R. An Overview and Analysis of Site Remediation Technologies. J. Environ. Manag. 2004, 71, 95–122. [Google Scholar] [CrossRef] [PubMed]
- Brar, S.K.; Verma, M.; Surampalli, R.Y.; Misra, K.; Tyagi, R.D.; Meunier, N.; Blais, J.F. Bioremediation of Hazardous Wastes—A Review. Pract. Period. Hazard. Toxic. Radioact. Waste Manag. 2006, 10, 59–72. [Google Scholar] [CrossRef]
- Fulekar, M.H. Bioremediation Technology for Hazardous Wastes—Recent Advances. In Bioremediation Technology; Springer: Dordrecht, The Netherlands, 2010; pp. 135–166. [Google Scholar]
- Ramírez-García, R.; Gohil, N.; Singh, V. Recent Advances, Challenges, and Opportunities in Bioremediation of Hazardous Materials. In Phytomanagement of Polluted Sites; Elsevier: Amsterdam, The Netherlands, 2019; pp. 517–568. [Google Scholar]
- Ekperusi, A.O.; Sikoki, F.D.; Nwachukwu, E.O. Application of Common Duckweed (Lemna minor) in Phytoremediation of Chemicals in the Environment: State and Future Perspective. Chemosphere 2019, 223, 285–309. [Google Scholar] [CrossRef] [PubMed]
- Hillman, W.S.; Culley, D.D. The Uses of Duckweed: The Rapid Growth, Nutritional Value, and High Biomass Productivity of These Floating Plants Suggest Their Use in Water Treatment, as Feed Crops, and in Energy-Efficient Farming. Am. Sci. 1978, 66, 442–451. [Google Scholar]
- Basile, A.; Sorbo, S.; Conte, B.; Cobianchi, R.C.; Trinchella, F.; Capasso, C.; Carginale, V. Toxicity, Accumulation, and Removal of Heavy Metals by Three Aquatic Macrophytes. Int. J. Phytoremediat. 2012, 14, 374–387. [Google Scholar] [CrossRef] [PubMed]
- Ohlbaum, M.; Wadgaonkar, S.L.; van Bruggen, J.J.A.; Nancharaiah, Y.V.; Lens, P.N.L. Phytoremediation of Seleniferous Soil Leachate Using the Aquatic Plants Lemna minor and Egeria densa. Ecol. Eng. 2018, 120, 321–328. [Google Scholar] [CrossRef]
- Stout, L.M.; Dodova, E.N.; Tyson, J.F.; Nüsslein, K. Phytoprotective Influence of Bacteria on Growth and Cadmium Accumulation in the Aquatic Plant Lemna Minor. Water Res. 2010, 44, 4970–4979. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Van Le, Q.; Peng, W.; Yang, Y.; Yang, H.; Gu, H.; Lam, S.S.; Sonne, C. A Review on Phytoremediation of Contaminants in Air, Water and Soil. J. Hazard. Mater. 2021, 403, 123658. [Google Scholar] [CrossRef]
- Andrews, J.H.; Harris, R.F. The Ecology and Biogeography of Microorganisms on Plant Surfaces. Annu. Rev. Phytopathol. 2000, 38, 145–180. [Google Scholar] [CrossRef]
- Boonmak, C.; Kettongruang, S.; Buranathong, B.; Morikawa, M.; Duangmal, K. Duckweed-Associated Bacteria as Plant Growth-Promotor to Enhance Growth of Spirodela Polyrhiza in Wastewater Effluent from a Poultry Farm. Arch. Microbiol. 2024, 206, 43. [Google Scholar] [CrossRef]
- Kristanti, R.A.; Kanbe, M.; Hadibarata, T.; Toyama, T.; Tanaka, Y.; Mori, K. Isolation and Characterization of 3-Nitrophenol-Degrading Bacteria Associated with Rhizosphere of Spirodela polyrrhiza. Environ. Sci. Pollut. Res. 2012, 19, 1852–1858. [Google Scholar] [CrossRef]
- Yamaga, F.; Washio, K.; Morikawa, M. Sustainable Biodegradation of Phenol by Acinetobacter calcoaceticus P23 Isolated from the Rhizosphere of Duckweed Lemna aoukikusa. Environ. Sci. Technol. 2010, 44, 6470–6474. [Google Scholar] [CrossRef]
- Kuiper, I.; Lagendijk, E.L.; Bloemberg, G.V.; Lugtenberg, B.J.J. Rhizoremediation: A Beneficial Plant-Microbe Interaction. Mol. Plant-Microbe Interact. 2004, 17, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Verma, J.P.; Jaiswal, D.K. Book Review: Advances in Biodegradation and Bioremediation of Industrial Waste. Front. Microbiol. 2016, 6, 175774. [Google Scholar] [CrossRef]
- Singh, A.; Ward, O.P. Biotechnology and Bioremediation—An Overview; Springer: Berlin/Heidelberg, Germany, 2004; pp. 1–17. [Google Scholar]
- Abhilash, P.C.; Powell, J.R.; Singh, H.B.; Singh, B.K. Plant–Microbe Interactions: Novel Applications for Exploitation in Multipurpose Remediation Technologies. Trends Biotechnol. 2012, 30, 416–420. [Google Scholar] [CrossRef]
- Sarwar, N.; Imran, M.; Shaheen, M.R.; Ishaque, W.; Kamran, M.A.; Matloob, A.; Rehim, A.; Hussain, S. Phytoremediation Strategies for Soils Contaminated with Heavy Metals: Modifications and Future Perspectives. Chemosphere 2017, 171, 710–721. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; van Themaat, E.V.L.; Schulze-Lefert, P. Structure and Functions of the Bacterial Microbiota of Plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef]
- Taj, Z.Z.; Rajkumar, M. Perspectives of Plant Growth-Promoting Actinomycetes in Heavy Metal Phytoremediation. In Plant Growth Promoting Actinobacteria; Springer: Singapore, 2016; pp. 213–231. [Google Scholar]
- Farda, B.; Djebaili, R.; Vaccarelli, I.; Del Gallo, M.; Pellegrini, M. Actinomycetes from Caves: An Overview of Their Diversity, Biotechnological Properties, and Insights for Their Use in Soil Environments. Microorganisms 2022, 10, 453. [Google Scholar] [CrossRef]
- Djebaili, R.; Pellegrini, M.; Smati, M.; Del Gallo, M.; Kitouni, M. Actinomycete Strains Isolated from Saline Soils: Plant-Growth-Promoting Traits and Inoculation Effects on Solanum lycopersicum. Sustainability 2020, 12, 4617. [Google Scholar] [CrossRef]
- Djebaili, R.; Pellegrini, M.; Rossi, M.; Forni, C.; Smati, M.; Del Gallo, M.; Kitouni, M. Characterization of Plant Growth-Promoting Traits and Inoculation Effects on Triticum Durum of Actinomycetes Isolates under Salt Stress Conditions. Soil Syst. 2021, 5, 26. [Google Scholar] [CrossRef]
- Goodfellow, M.; Williams, S.T. Ecology of Actinomycetes. Annu. Rev. Microbiol. 1983, 37, 189–216. [Google Scholar] [CrossRef]
- Ball, A.S.; Betts, W.B.; McCarthy, A.J. Degradation of Lignin-Related Compounds by Actinomycetes. Appl. Environ. Microbiol. 1989, 55, 1642–1644. [Google Scholar] [CrossRef]
- Gupta, R.S. The Phylogeny of Proteobacteria: Relationships to Other Eubacterial Phyla and Eukaryotes. FEMS Microbiol. Rev. 2000, 24, 367–402. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Lee, S.H.; Jo, H.Y.; Finneran, K.T.; Kwon, M.J. Diversity and Composition of Soil Acidobacteria and Proteobacteria Communities as a Bacterial Indicator of Past Land-Use Change from Forest to Farmland. Sci. Total Environ. 2021, 797, 148944. [Google Scholar] [CrossRef]
- Sharma, V.; Vashishtha, A.; Jos, A.L.M.; Khosla, A.; Basu, N.; Yadav, R.; Bhatt, A.; Gulani, A.; Singh, P.; Lakhera, S.; et al. Phylogenomics of the Phylum Proteobacteria: Resolving the Complex Relationships. Curr. Microbiol. 2022, 79, 224. [Google Scholar] [CrossRef] [PubMed]
- Stackebrandt, E.; Murray, R.G.E.; Truper, H.G. Proteobacteria Classis Nov., a Name for the Phylogenetic Taxon That Includes the “Purple Bacteria and Their Relatives. Int. J. Syst. Bacteriol. 1988, 38, 321–325. [Google Scholar] [CrossRef]
- Woese, C.R. Bacterial Evolution. Microbiol. Rev. 1987, 51, 221–271. [Google Scholar] [CrossRef]
- Williams, K.P.; Kelly, D.P. Proposal for a New Class within the Phylum Proteobacteria, Acidithiobacillia Classis Nov., with the Type Order Acidithiobacillales, and Emended Description of the Class Gammaproteobacteria. Int. J. Syst. Evol. Microbiol. 2013, 63, 2901–2906. [Google Scholar] [CrossRef]
- Garrity, G.M.; Bell, J.A.; Lilburn, T. Proteobacteria Phyl. Nov. In Bergey’s Manual of Systematics of Archaea and Bacteria; Springer: New York, NY, USA, 2005; Volume 2, pp. 607–624. [Google Scholar]
- McAllister, S.M.; Davis, R.E.; McBeth, J.M.; Tebo, B.M.; Emerson, D.; Moyer, C.L. Biodiversity and Emerging Biogeography of the Neutrophilic Iron-Oxidizing Zetaproteobacteria. Appl. Environ. Microbiol. 2011, 77, 5445–5457. [Google Scholar] [CrossRef]
- Ludwig, W.; Klenk, H.-P. Overview: A Phylogenetic Backbone and Taxonomic Framework for Procaryotic Systematics. In Bergey’s Manual of Systematic Bacteriology; Springer: New York, NY, USA, 2005; pp. 49–66. [Google Scholar] [CrossRef]
- Vingadassalom, D.; Kolb, A.; Mayer, C.; Rybkine, T.; Collatz, E.; Podglajen, I. An Unusual Primary Sigma Factor in the Bacteroidetes Phylum. Mol. Microbiol. 2005, 56, 888–902. [Google Scholar] [CrossRef]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The Rhizosphere Microbiome: Significance of Plant Beneficial, Plant Pathogenic, and Human Pathogenic Microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef] [PubMed]
- Thomas, F.; Hehemann, J.H.; Rebuffet, E.; Czjzek, M.; Michel, G. Environmental and Gut Bacteroidetes: The Food Connection. Front. Microbiol. 2011, 2, 9588. [Google Scholar] [CrossRef] [PubMed]
- Borsetto, C.; Amos, G.C.A.; Da Rocha, U.N.; Mitchell, A.L.; Finn, R.D.; Laidi, R.F.; Vallin, C.; Pearce, D.A.; Newsham, K.K.; Wellington, E.M.H. Microbial Community Drivers of PK/NRP Gene Diversity in Selected Global Soils. Microbiome 2019, 7, 78. [Google Scholar] [CrossRef]
- Brinkmann, S.; Kurz, M.; Patras, M.A.; Hartwig, C.; Marner, M.; Leis, B.; Billion, A.; Kleiner, Y.; Bauer, A.; Toti, L.; et al. Genomic and Chemical Decryption of the Bacteroidetes Phylum for Its Potential to Biosynthesize Natural Products. Microbiol. Spectr. 2022, 10, e02479-21. [Google Scholar] [CrossRef] [PubMed]
- Ventura, M.; Canchaya, C.; Tauch, A.; Chandra, G.; Fitzgerald, G.F.; Chater, K.F.; van Sinderen, D. Genomics of Actinobacteria: Tracing the Evolutionary History of an Ancient Phylum. Microbiol. Mol. Biol. Rev. 2007, 71, 495–548. [Google Scholar] [CrossRef]
- Salam, N.; Jiao, J.Y.; Zhang, X.T.; Li, W.J. Update on the Classification of Higher Ranks in the Phylum Actinobacteria. Int. J. Syst. Evol. Microbiol. 2020, 70, 1331–1355. [Google Scholar] [CrossRef] [PubMed]
- Al-Shaibani, M.M.; Mohamed, R.M.S.R.; Sidik, N.M.; Enshasy, H.A.E.; Al-Gheethi, A.; Noman, E.; Al-Mekhlafi, N.A.; Zin, N.M. Biodiversity of Secondary Metabolites Compounds Isolated from Phylum Actinobacteria and Its Therapeutic Applications. Molecules 2021, 26, 4504. [Google Scholar] [CrossRef] [PubMed]
- Atlas, R.M. Principles of Microbiology; McGraw-Hill: Columbus, OH, USA, 1997. [Google Scholar]
- Schrempf, H. Recognition and Degradation of Chitin by Streptomycetes. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2001, 79, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Lechevalier, H.A.; Lechevalier, M.P. Biology of Actinomycetes. Annu. Rev. Microbiol. 1967, 21, 71–100. [Google Scholar] [CrossRef]
- Passari, A.K.; Leo, V.V.; Chandra, P.; Kumar, B.; Nayak, C.; Hashem, A.; Abd Allah, E.F.; Alqarawi, A.A.; Singh, B.P. Bioprospection of Actinobacteria Derived from Freshwater Sediments for Their Potential to Produce Antimicrobial Compounds. Microb. Cell Fact. 2018, 17, 68. [Google Scholar] [CrossRef]
- Kishimoto, N.; Kosako, Y.; Tano, T. Acidobacterium Capsulatum Gen. Nov., Sp. Nov.: An Acidophilic Chemoorganotrophic Bacterium Containing Menaquinone from Acidic Mineral Environment. Curr. Microbiol. 1991, 22, 1–7. [Google Scholar] [CrossRef]
- Lee, S.-H.; Ka, J.-O.; Cho, J.-C. Members of the Phylum Acidobacteria Are Dominant and Metabolically Active in Rhizosphere Soil. FEMS Microbiol. Lett. 2008, 285, 263–269. [Google Scholar] [CrossRef]
- Conradie, T.A.; Jacobs, K. Distribution Patterns of Acidobacteriota in Different Fynbos Soils. PLoS ONE 2021, 16, e0248913. [Google Scholar] [CrossRef]
- Ludwig, W.; Bauer, S.H.; Bauer, M.; Held, I.; Kirchhof, G.; Schulze, R.; Huber, I.; Spring, S.; Hartmann, A.; Schleifer, K.H. Detection and in Situ Identification of Representatives of a Widely Distributed New Bacterial Phylum. FEMS Microbiol. Lett. 2006, 153, 181–190. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Yilmaz, P. Refining the Taxonomic Structure of the Phylum Acidobacteria. Int. J. Syst. Evol. Microbiol. 2018, 68, 3796–3806. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Oliverio, A.M.; Brewer, T.E.; Benavent-González, A.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K.; Fierer, N. A Global Atlas of the Dominant Bacteria Found in Soil. Science (1979) 2018, 359, 320–325. [Google Scholar] [CrossRef]
- Barns, S.M.; Takala, S.L.; Kuske, C.R. Wide Distribution and Diversity of Members of the Bacterial Kingdom Acidobacterium in the Environment. Appl. Environ. Microbiol. 1999, 65, 1731–1737. [Google Scholar] [CrossRef]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic Identification and in Situ Detection of Individual Microbial Cells without Cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, Y.; Zhou, Q.; Huang, S.; Ning, K.; Xu, J.; Kalin, R.M.; Rolfe, S.; Huang, W.E. A Culture-Independent Approach to Unravel Uncultured Bacteria and Functional Genes in a Complex Microbial Community. PLoS ONE 2012, 7, e47530. [Google Scholar] [CrossRef] [PubMed]
- Rappé, M.S.; Giovannoni, S.J. The Uncultured Microbial Majority. Annu. Rev. Microbiol. 2003, 57, 369–394. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Mishra, A. Functional Gene Diversity and Metabolic Potential of Uncultured Bacteria. In Microbial Diversity in the Genomic Era; Elsevier: Amsterdam, The Netherlands, 2024; pp. 481–491. [Google Scholar]
- Almeida, A.; Nayfach, S.; Boland, M.; Strozzi, F.; Beracochea, M.; Shi, Z.J.; Pollard, K.S.; Sakharova, E.; Parks, D.H.; Hugenholtz, P.; et al. A Unified Catalog of 204,938 Reference Genomes from the Human Gut Microbiome. Nat. Biotechnol. 2020, 39, 105–114. [Google Scholar] [CrossRef]
- Nayfach, S.; Roux, S.; Seshadri, R.; Udwary, D.; Varghese, N.; Schulz, F.; Wu, D.; Paez-Espino, D.; Chen, I.M.; Huntemann, M.; et al. A Genomic Catalog of Earth’s Microbiomes. Nat. Biotechnol. 2020, 39, 499–509. [Google Scholar] [CrossRef]
- Rodríguez del Río, Á.; Giner-Lamia, J.; Cantalapiedra, C.P.; Botas, J.; Deng, Z.; Hernández-Plaza, A.; Munar-Palmer, M.; Santamaría-Hernando, S.; Rodríguez-Herva, J.J.; Ruscheweyh, H.-J.; et al. Functional and Evolutionary Significance of Unknown Genes from Uncultivated Taxa. Nature 2024, 626, 377–384. [Google Scholar] [CrossRef]
- Liu, S.; Moon, C.D.; Zheng, N.; Huws, S.; Zhao, S.; Wang, J. Opportunities and Challenges of Using Metagenomic Data to Bring Uncultured Microbes into Cultivation. Microbiome 2022, 10, 76. [Google Scholar] [CrossRef]
- Lloyd, K.G.; Steen, A.D.; Ladau, J.; Yin, J.; Crosby, L. Phylogenetically Novel Uncultured Microbial Cells Dominate Earth Microbiomes. mSystems 2018, 3, e00055-18. [Google Scholar] [CrossRef]
- Farda, B.; Djebaili, R.; Bernardi, M.; Pace, L.; Del Gallo, M.; Pellegrini, M. Bacterial Microbiota and Soil Fertility of Crocus Sativus L. Rhizosphere in the Presence and Absence of Fusarium spp. Land 2022, 11, 2048. [Google Scholar] [CrossRef]
- Bodor, A.; Bounedjoum, N.; Vincze, G.E.; Erdeiné Kis, Á.; Laczi, K.; Bende, G.; Szilágyi, Á.; Kovács, T.; Perei, K.; Rákhely, G. Challenges of Unculturable Bacteria: Environmental Perspectives. Rev. Environ. Sci. Biotechnol. 2020, 19, 1–22. [Google Scholar] [CrossRef]
- dos-Santos, C.M.; Nascimento, W.B.A.; Cesar, M.J.S.C.; Baldani, J.I.; Schwab, S. Diversity of Bacteria of the Genus Sphingomonas Associated with Sugarcane (Saccharum spp.) Culm Apoplast Fluid and Their Agrotechnological Potential. World J. Microbiol. Biotechnol. 2024, 40, 304. [Google Scholar] [CrossRef]
- White, D.C.; Sutton, S.D.; Ringelberg, D.B. The Genus Sphingomonas: Physiology and Ecology. Curr. Opin. Biotechnol. 1996, 7, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, C.L.; Andrewes, A.G.; McQuade, T.J.; Starr, M.P. The Pigment of Pseudomonas Paucimobilis Is a Carotenoid (Nostoxanthin), Rather than a Brominated Aryl-Polyene (Xanthomonadin). Curr. Microbiol. 1979, 3, 1–4. [Google Scholar] [CrossRef]
- Siddaramappa, S.; Viswanathan, V.; Thiyagarajan, S.; Narjala, A. Genomewide Characterisation of the Genetic Diversity of Carotenogenesis in Bacteria of the Order Sphingomonadales. Microb. Genom. 2018, 4, e000172. [Google Scholar] [CrossRef]
- Halo, B.A.; Khan, A.L.; Waqas, M.; Al-Harrasi, A.; Hussain, J.; Ali, L.; Adnan, M.; Lee, I.J. Endophytic Bacteria (Sphingomonas sp. LK11) and Gibberellin Can Improve Solanum lycopersicum Growth and Oxidative Stress under Salinity. J. Plant Interact. 2015, 10, 117–125. [Google Scholar] [CrossRef]
- Asaf, S.; Numan, M.; Khan, A.L.; Al-Harrasi, A. Sphingomonas: From Diversity and Genomics to Functional Role in Environmental Remediation and Plant Growth. Crit. Rev. Biotechnol. 2020, 40, 138–152. [Google Scholar] [CrossRef]
- Yu, F.B.; Shan, S.D.; Luo, L.P.; Guan, L.B.; Qin, H. Isolation and Characterization of a Sphingomonas sp. Strain F-7 Degrading Fenvalerate and Its Use in Bioremediation of Contaminated Soil. J. Environ. Sci. Health Part. B 2013, 48, 198–207. [Google Scholar] [CrossRef]
- Christensen, P.; Cook, F.D. Lysobacter, a New Genus of Nonfruiting, Gliding Bacteria with a High Base Ratio. Int. J. Syst. Bacteriol. 1978, 28, 367–393. [Google Scholar] [CrossRef]
- Park, J.H.; Kim, R.; Aslam, Z.; Jeon, C.O.; Chung, Y.R. Lysobacter capsici sp. Nov., with Antimicrobial Activity, Isolated from the Rhizosphere of Pepper, and Emended Description of the Genus Lysobacter. Int. J. Syst. Evol. Microbiol. 2008, 58, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Niu, B.-Y.; Ren, D.-J.; Zhang, F.-B.; Zhu, H.-T.; Wei, H.-L.; Ma, M.-C.; Gao, M. Lysobacter changpingensis sp. Nov., a Novel Species of the Genus Lysobacter Isolated from a Rhizosphere Soil of Strawberry in China. Folia Microbiol 2023, 68, 991–998. [Google Scholar] [CrossRef]
- Choi, H.; Im, W.-T.; Park, J.-S. Lysobacter spongiae sp. Nov., Isolated from Spongin. J. Microbiol. 2018, 56, 97–103. [Google Scholar] [CrossRef]
- Chhetri, G.; Kim, J.; Kim, I.; Seo, T. Lysobacter caseinilyticus sp. Nov., a Casein Hydrolyzing Bacterium Isolated from Sea Water. Antonie Van Leeuwenhoek 2019, 112, 1349–1356. [Google Scholar] [CrossRef]
- Singh, H.; Du, J.; Ngo, H.T.T.; Won, K.; Yang, J.-E.; Kim, K.-Y.; Yi, T.-H. Lysobacter fragariae sp. Nov. and Lysobacter rhizosphaerae Sp. Nov. Isolated from Rhizosphere of Strawberry Plant. Antonie Van Leeuwenhoek 2015, 107, 1437–1444. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhao, Y.-L.; Cheng, J.; Zhou, X.-K.; Salam, N.; Fang, B.-Z.; Li, Q.-Q.; Hozzein, W.N.; Li, W.-J. Lysobacter cavernae sp. Nov., a Novel Bacterium Isolated from a Cave Sample. Antonie Van Leeuwenhoek 2016, 109, 1047–1053. [Google Scholar] [CrossRef]
- Huo, Y.; Kang, J.-P.; Hurh, J.; Han, Y.; Ahn, J.-C.; Mathiyalagan, R.; Piao, C.; Yang, D.-C. Lysobacter panacihumi sp. Nov., Isolated from Ginseng Cultivated Soil. J. Microbiol. 2018, 56, 748–752. [Google Scholar] [CrossRef]
- Im, W.-T.; Siddiqi, M.Z.; Kim, S.-Y.; Huq, M.d.A.; Lee, J.H.; Choi, K.D. Lysobacter lacus Sp. Nov., Isolated from from Lake Sediment. Int. J. Syst. Evol. Microbiol. 2020, 70, 2211–2216. [Google Scholar] [CrossRef]
- Xu, J.; Sheng, M.; Yang, Z.; Qiu, J.; Zhang, J.; Zhang, L.; He, J. Lysobacter gilvus sp. Nov., Isolated from Activated Sludge. Arch. Microbiol. 2021, 203, 7–11. [Google Scholar] [CrossRef]
- Friedman, B.A.; Dugan, P.R. Identification of Zoogloea Species and the Relationship to Zoogloeal Matrix and Floc Formation. J. Bacteriol. 1968, 95, 1903–1909. [Google Scholar] [CrossRef] [PubMed]
- Hiraishi, A.; Shin, Y.K.; Sugiyama, J. Proposal to Reclassify Zoogloea Ramigera IAM 12670 (P. R. Dugan 115) as Duganella zoogloeoides gen. Nov., sp. Nov. Int. J. Syst. Bacteriol. 1997, 47, 1249–1252. [Google Scholar] [CrossRef]
- Kämpfer, P.; Irgang, R.; Busse, H.-J.; Poblete-Morales, M.; Kleinhagauer, T.; Glaeser, S.P.; Avendaño-Herrera, R. Pseudoduganella Danionis Sp. Nov., Isolated from Zebrafish (Danio rerio). Int. J. Syst. Evol. Microbiol. 2016, 66, 4671–4675. [Google Scholar] [CrossRef]
- Jeon, D.; Kim, I.S.; Choe, H.; Kim, J.-S.; Lee, S.D. Duganella aceris Sp. Nov., Isolated from Tree Sap and Proposal to Transfer of Rugamonas aquatica and Rugamonas rivuli to the Genus Duganella as Duganella aquatica Comb. Nov., with the Emended Description of the Genus Rugamonas. Arch. Microbiol. 2021, 203, 2843–2852. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Nupur; Wu, N.; Madsen, A.M.; Chen, X.; Gardiner, A.T.; Koblížek, M. Gemmatimonas groenlandica sp. Nov. Is an Aerobic Anoxygenic Phototroph in the Phylum Gemmatimonadetes. Front. Microbiol. 2021, 11, 606612. [Google Scholar] [CrossRef]
- Zeng, Y.; Baumbach, J.; Barbosa, E.G.V.; Azevedo, V.; Zhang, C.; Koblížek, M. Metagenomic Evidence for the Presence of Phototrophic Emmatimonadetes Bacteria in Diverse Environments. Environ. Microbiol. Rep. 2016, 8, 139–149. [Google Scholar] [CrossRef]
- Janssen, P.H. Identifying the Dominant Soil Bacterial Taxa in Libraries of 16S RRNA and 16S RRNA Genes. Appl. Environ. Microbiol. 2006, 72, 1719–1728. [Google Scholar] [CrossRef]
- Youssef, N.H.; Elshahed, M.S. Diversity Rankings among Bacterial Lineages in Soil. ISME J. 2009, 3, 305–313. [Google Scholar] [CrossRef]
- DeBruyn, J.M.; Nixon, L.T.; Fawaz, M.N.; Johnson, A.M.; Radosevich, M. Global Biogeography and Quantitative Seasonal Dynamics of Gemmatimonadetes in Soil. Appl. Environ. Microbiol. 2011, 77, 6295–6300. [Google Scholar] [CrossRef] [PubMed]
- Yoneda, Y.; Yamamoto, K.; Makino, A.; Tanaka, Y.; Meng, X.Y.; Hashimoto, J.; Shin-Ya, K.; Satoh, N.; Fujie, M.; Toyama, T.; et al. Novel Plant-Associated Acidobacteria Promotes Growth of Common Floating Aquatic Plants, Duckweeds. Microorganisms 2021, 9, 1133. [Google Scholar] [CrossRef]
- Radić, S.; Stipaničev, D.; Cvjetko, P.; Mikelić, I.L.; Rajčić, M.M.; Širac, S.; Pevalek-Kozlina, B.; Pavlica, M. Ecotoxicological Assessment of Industrial Effluent Using Duckweed (Lemna minor L.) as a Test Organism. Ecotoxicology 2010, 19, 216–222. [Google Scholar] [CrossRef] [PubMed]
- Daud, M.K.; Ali, S.; Abbas, Z.; Zaheer, I.E.; Riaz, M.A.; Malik, A.; Hussain, A.; Rizwan, M.; Zia-ur-Rehman, M.; Zhu, S.J. Potential of Duckweed (Lemna minor) for the Phytoremediation of Landfill Leachate. J. Chem. 2018, 2018, 3951540. [Google Scholar] [CrossRef]
- Piotrowska, A.; Bajguz, A.; Godlewska-Żyłkiewicz, B.; Czerpak, R.; Kamińska, M. Jasmonic Acid as Modulator of Lead Toxicity in Aquatic Plant Wolffia Arrhiza (Lemnaceae). Environ. Exp. Bot. 2009, 66, 507–513. [Google Scholar] [CrossRef]
- Chen, M.; Zhang, L.-L.; Li, J.; He, X.-J.; Cai, J.-C. Bioaccumulation and Tolerance Characteristics of a Submerged Plant (Ceratophyllum demersum L.) Exposed to Toxic Metal Lead. Ecotoxicol. Environ. Saf. 2015, 122, 313–321. [Google Scholar] [CrossRef]
- Sasmaz, M.; Obek, E.; Sasmaz, A. Bioaccumulation of Uranium and Thorium by Lemna minor and Lemna gibba in Pb-Zn-Ag Tailing Water. Bull. Environ. Contam. Toxicol. 2016, 97, 832–837. [Google Scholar] [CrossRef] [PubMed]
- Saimee, Y.; Butdee, W.; Boonmak, C.; Duangmal, K. Actinomycetospora Lemnae sp. Nov., A Novel Actinobacterium Isolated from Lemna aequinoctialis Able to Enhance Duckweed Growth. Curr. Microbiol. 2024, 81, 92. [Google Scholar] [CrossRef]
- Takahashi, Y.; Nakashima, T. Actinomycetes, an Inexhaustible Source of Naturally Occurring Antibiotics. Antibiotics 2018, 7, 45. [Google Scholar] [CrossRef] [PubMed]
- Behera, S.; Das, S. Potential and Prospects of Actinobacteria in the Bioremediation of Environmental Pollutants: Cellular Mechanisms and Genetic Regulations. Microbiol. Res. 2023, 273, 127399. [Google Scholar] [CrossRef]
- Chaturvedi, S.; Khurana, S.M.P. Importance of Actinobacteria for Bioremediation. In Plant Biotechnology: Progress in Genomic Era; Springer: Singapore, 2019; pp. 277–307. [Google Scholar]
- Schmidt, A.; Haferburg, G.; Sineriz, M.; Merten, D.; Büchel, G.; Kothe, E. Heavy Metal Resistance Mechanisms in Actinobacteria for Survival in AMD Contaminated Soils. Geochemistry 2005, 65, 131–144. [Google Scholar] [CrossRef]
- El Baz, S.; Baz, M.; Barakate, M.; Hassani, L.; El Gharmali, A.; Imziln, B. Resistance to and Accumulation of Heavy Metals by Actinobacteria Isolated from Abandoned Mining Areas. Sci. World J. 2015, 2015, 761834. [Google Scholar] [CrossRef]
- Jin, Y.; Luan, Y.; Ning, Y.; Wang, L. Effects and Mechanisms of Microbial Remediation of Heavy Metals in Soil: A Critical Review. Appl. Sci. 2018, 8, 1336. [Google Scholar] [CrossRef]
- Makarani, N.; Kaushal, R.S. Advances in Actinobacteria-Based Bioremediation: Mechanistic Insights, Genetic Regulation, and Emerging Technologies. Biodegradation 2025, 36, 24. [Google Scholar] [CrossRef]
- Majewska, M.; Słomka, A.; Hanaka, A. Siderophore-Producing Bacteria from Spitsbergen Soils—Novel Agents Assisted in Bioremediation of the Metal-Polluted Soils. Environ. Sci. Pollut. Res. 2024, 31, 32371–32381. [Google Scholar] [CrossRef]
- Timková, I.; Sedláková-Kaduková, J.; Pristaš, P. Biosorption and Bioaccumulation Abilities of Actinomycetes/Streptomycetes Isolated from Metal Contaminated Sites. Separations 2018, 5, 54. [Google Scholar] [CrossRef]
- Hesse, E.; O’Brien, S.; Tromas, N.; Bayer, F.; Luján, A.M.; van Veen, E.M.; Hodgson, D.J.; Buckling, A. Ecological Selection of Siderophore-producing Microbial Taxa in Response to Heavy Metal Contamination. Ecol. Lett. 2018, 21, 117–127. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Wirth, S.J.; Alqarawi, A.A.; Abd_Allah, E.F.; Hashem, A. Phytohormones and Beneficial Microbes: Essential Components for Plants to Balance Stress and Fitness. Front. Microbiol. 2017, 8, 278255. [Google Scholar] [CrossRef] [PubMed]
- Babu, A.G.; Kim, J.-D.; Oh, B.-T. Enhancement of Heavy Metal Phytoremediation by Alnus firma with Endophytic Bacillus thuringiensis GDB-1. J. Hazard. Mater. 2013, 250–251, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, S.; Liu, S.; Peng, J.; Zhang, H.; Zhao, Q.; Zheng, L.; Chen, Y.; Shen, Z.; Xu, X.; et al. Enhancing the Phytoremediation of Heavy Metals by Combining Hyperaccumulator and Heavy Metal-Resistant Plant Growth-Promoting Bacteria. Front. Plant Sci. 2022, 13, 912350. [Google Scholar] [CrossRef]
- Makino, A.; Nakai, R.; Yoneda, Y.; Toyama, T.; Tanaka, Y.; Meng, X.-Y.; Mori, K.; Ike, M.; Morikawa, M.; Kamagata, Y.; et al. Isolation of Aquatic Plant Growth-Promoting Bacteria for the Floating Plant Duckweed (Lemna minor). Microorganisms 2022, 10, 1564. [Google Scholar] [CrossRef]
- Mahbub, K.R.; Krishnan, K.; Andrews, S.; Venter, H.; Naidu, R.; Megharaj, M. Bio-Augmentation and Nutrient Amendment Decrease Concentration of Mercury in Contaminated Soil. Sci. Total Environ. 2017, 576, 303–309. [Google Scholar] [CrossRef]
- Ibarrolaza, A.; Coppotelli, B.M.; Del Panno, M.T.; Donati, E.R.; Morelli, I.S. Application of the Knowledge-Based Approach to Strain Selection for a Bioaugmentation Process of Phenanthrene- and Cr (VI)-Contaminated Soil. J. Appl. Microbiol. 2011, 111, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Polti, M.A.; Aparicio, J.D.; Benimeli, C.S.; Amoroso, M.J. Simultaneous Bioremediation of Cr (VI) and Lindane in Soil by Actinobacteria. Int. Biodeterior. Biodegrad. 2014, 88, 48–55. [Google Scholar] [CrossRef]
- Campanella, M.; Chiavaroli, F.; Cocco, R.; Di Croce, L.; Lena, V.; Lombardi, A.; Mancinelli, G.; Palestini, S.; Coordinamento, E.S. Abruzzo, Rapporto Sullo Stato Dell’ambiente 2018 Responsabili Del Progetto; Arpa Abruzzo: Pescara, Italy, 2018. [Google Scholar]
- Mizrahi-Man, O.; Davenport, E.R.; Gilad, Y. Taxonomic Classification of Bacterial 16S RRNA Genes Using Short Sequencing Reads: Evaluation of Effective Study Designs. PLoS ONE 2013, 8, e53608. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Shirling, E.B.; Gottlieb, D. Methods for Characterization of Streptomyces Species. Int. J. Syst. Bacteriol. 1966, 16, 313–340. [Google Scholar] [CrossRef]
- Cappuccino, J.G.; Sherman, N. Microbiology—A Laboratory Manual; Cummings Publishing Co.: Boston, MA, USA, 1996. [Google Scholar]
- Remy, E.; Duque, P. Assessing Tolerance to Heavy-Metal Stress in Arabidopsis Thaliana Seedlings. In Environmental Responses in Plants: Methods and Protocols; Springer: New York, NY, USA, 2016; pp. 197–208. [Google Scholar]
- Farda, B.; Djebaili, R.; Del Gallo, M.; Ercole, C.; Bellatreccia, F.; Pellegrini, M. The “Infernaccio” Gorges: Microbial Diversity of Black Deposits and Isolation of Manganese-Solubilizing Bacteria. Biology 2022, 11, 1204. [Google Scholar] [CrossRef]
- Nautiyal, C.S. An Efficient Microbiological Growth Medium for Screening Phosphate Solubilizing Microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef]
- Cherif-Silini, H.; Silini, A.; Yahiaoui, B.; Ouzari, I.; Boudabous, A. Phylogenetic and Plant-Growth-Promoting Characteristics of Bacillus Isolated from the Wheat Rhizosphere. Ann. Microbiol. 2016, 66, 1087–1097. [Google Scholar] [CrossRef]
- Olsen, S.R.; Sommers, L.E.; Page, A.L. Methods of Soil Analysis: Part 2 Chemical and Microbiological Properties; American Society of Agronomy: Madison, WI, USA, 1982; Volume 2, pp. 403–430. [Google Scholar]
- Gordon, S.A.; Weber, R.P. Colorimetric Estimation of Indoleacetic Acid. Plant Physiol. 1951, 26, 192–195. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J.B. Universal Chemical Assay for the Detection and Determination of Siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Farda, B.; Mattedi, A.; Djebaili, R.; Pace, L.; Del Gallo, M.; Pellegrini, M. Microbial Community Investigation of Wild Brambles with Root Nodulation from a Calcareous Nitrogen-Deficient Soil. Soil Syst. 2022, 6, 96. [Google Scholar] [CrossRef]
- Donate-Correa, J.; León-Barrios, M.; Pérez-Galdona, R. Screening for Plant Growth-Promoting Rhizobacteria in Chamaecytisus proliferus (Tagasaste), a Forage Tree-Shrub Legume Endemic to the Canary Islands. Plant Soil 2005, 266, 261–272. [Google Scholar] [CrossRef]
- Brígido, C.; Duan, J.; Glick, B.R. Methods to Study 1-Aminocyclopropane-1-Carboxylate (ACC) Deaminase in Plant Growth-Promoting Bacteria. In Handbook for Azospirillum; Springer International Publishing: Cham, Switzerland, 2015; pp. 287–305. [Google Scholar]
- Dworkin, M.; Foster, J.W. Experiments with some microorganisms which utilize ethane and hydrogen. J. Bacteriol. 1958, 75, 592–603. [Google Scholar] [CrossRef] [PubMed]
- Jain, D.; Kour, R.; Bhojiya, A.A.; Meena, R.H.; Singh, A.; Mohanty, S.R.; Rajpurohit, D.; Ameta, K.D. Zinc Tolerant Plant Growth Promoting Bacteria Alleviates Phytotoxic Effects of Zinc on Maize through Zinc Immobilization. Sci. Rep. 2020, 10, 13865. [Google Scholar] [CrossRef] [PubMed]
- Arnon, D.I. Copper enzymes in isolated chloroplasts, Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef]
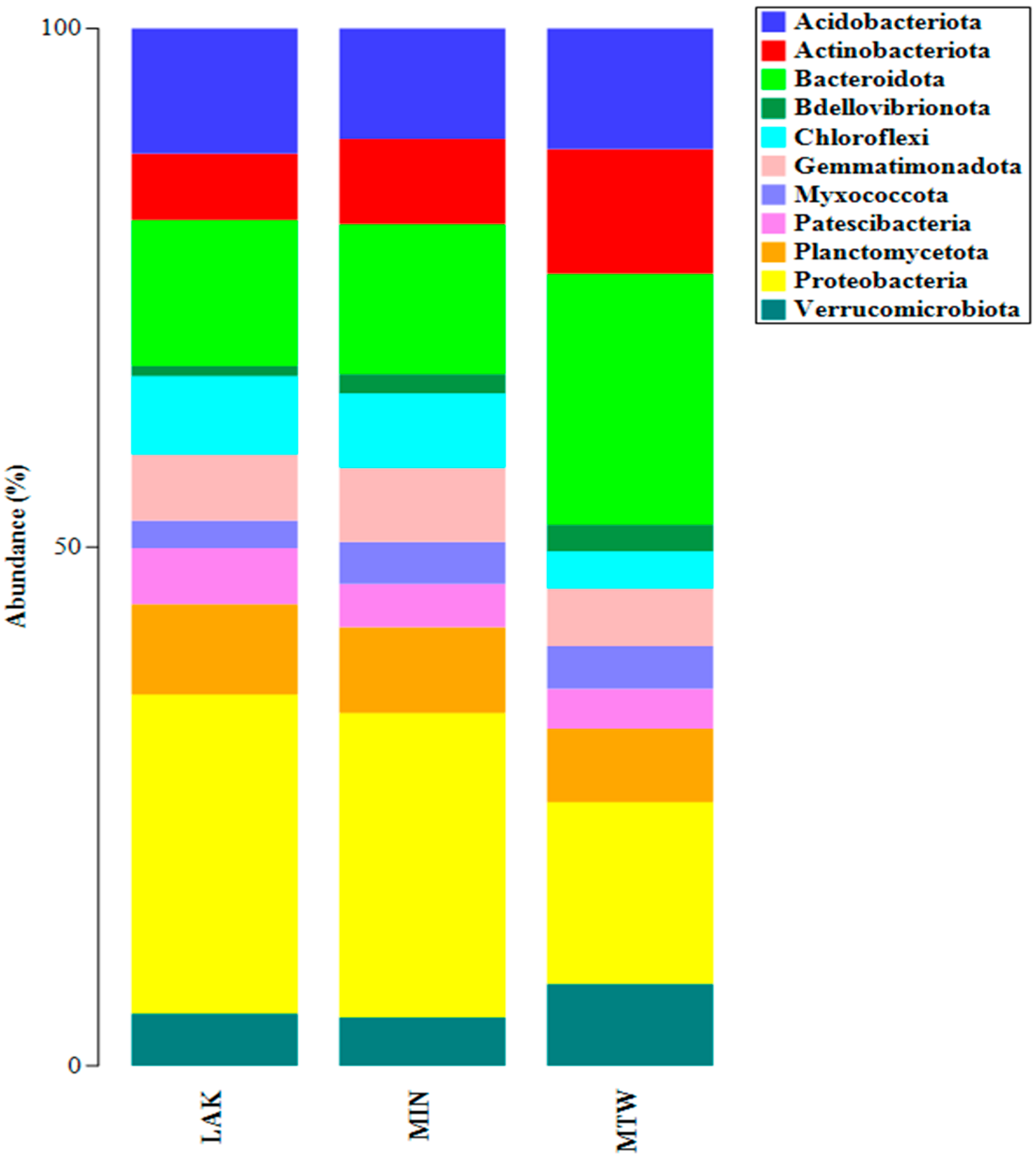
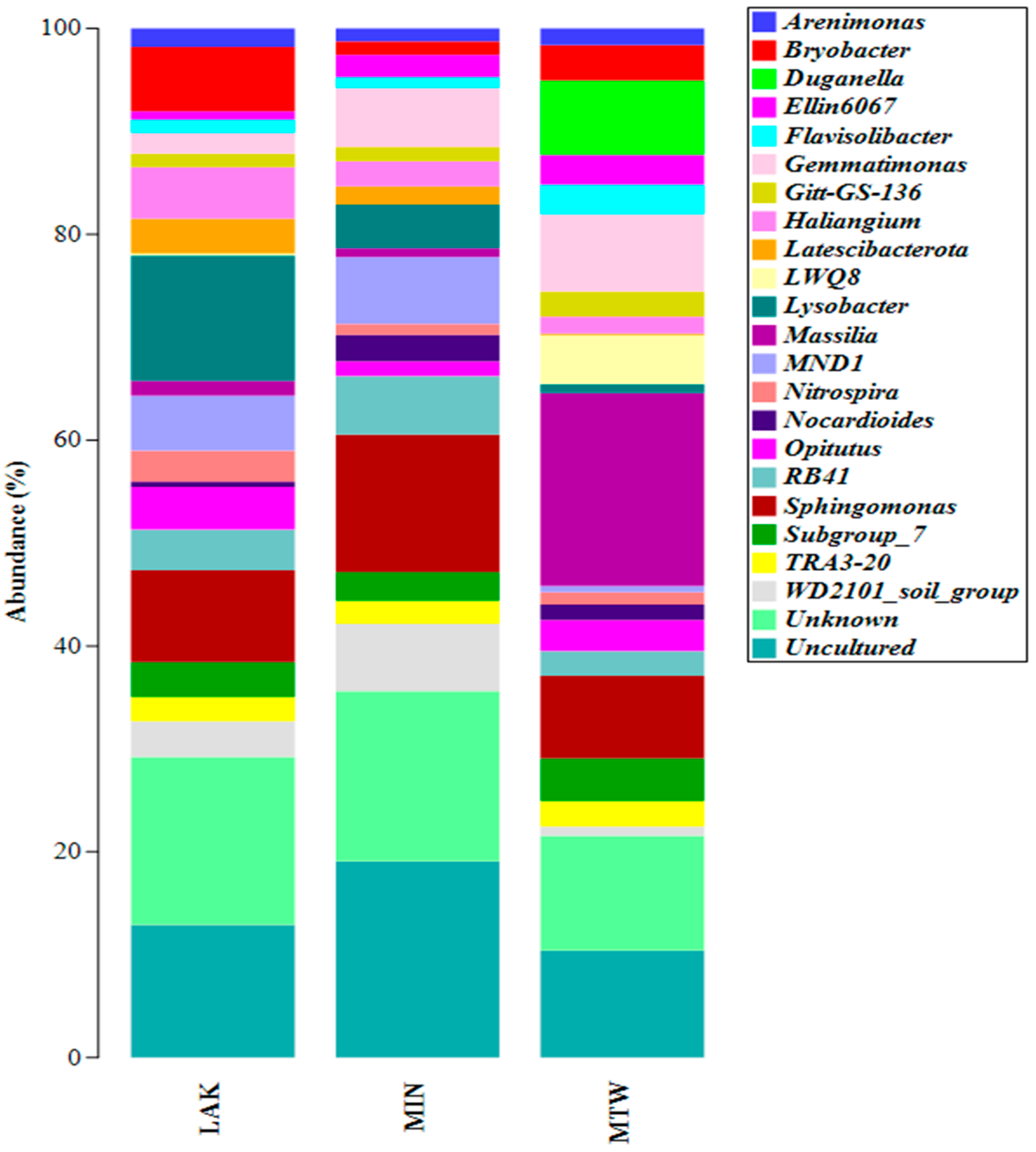
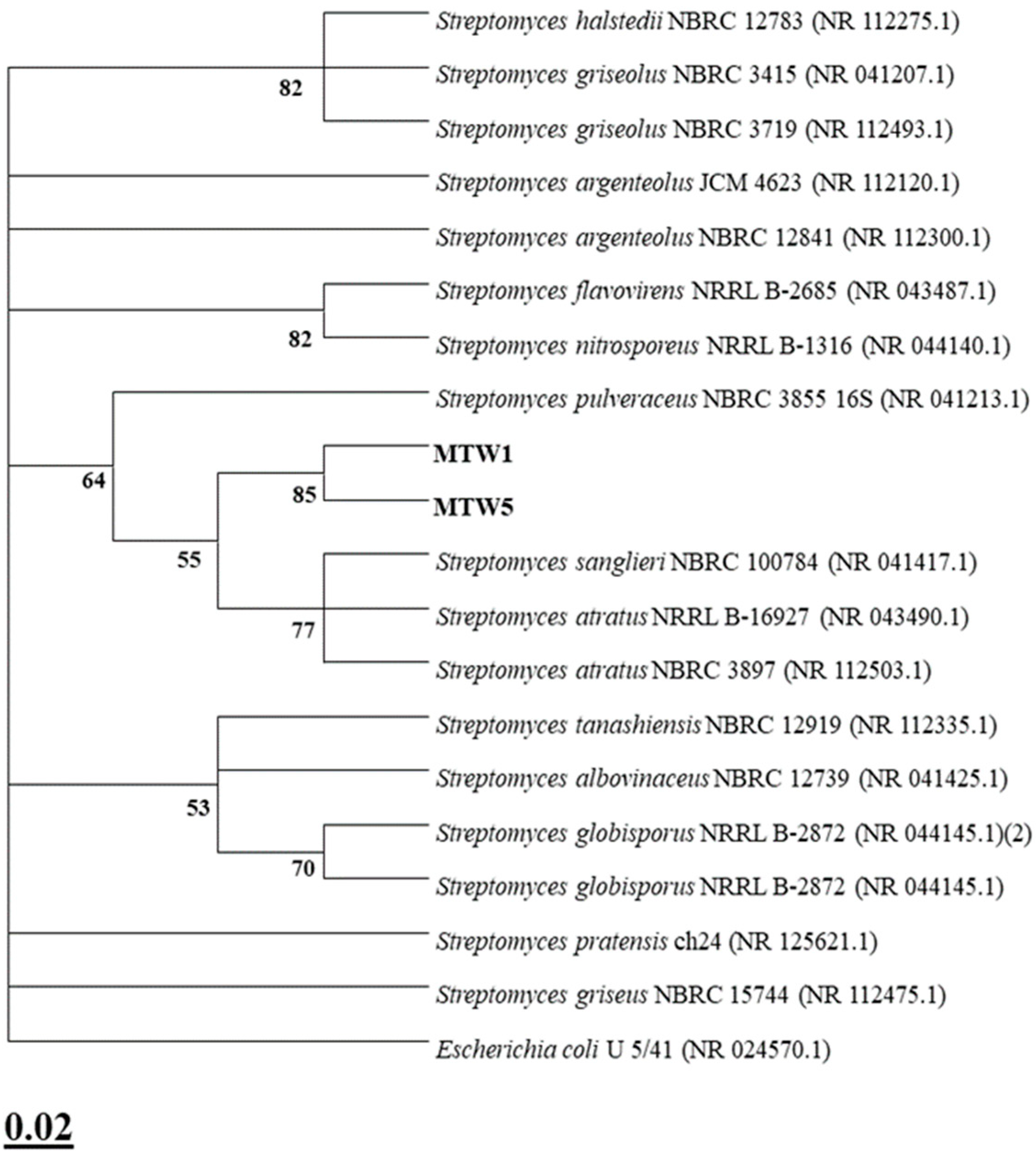
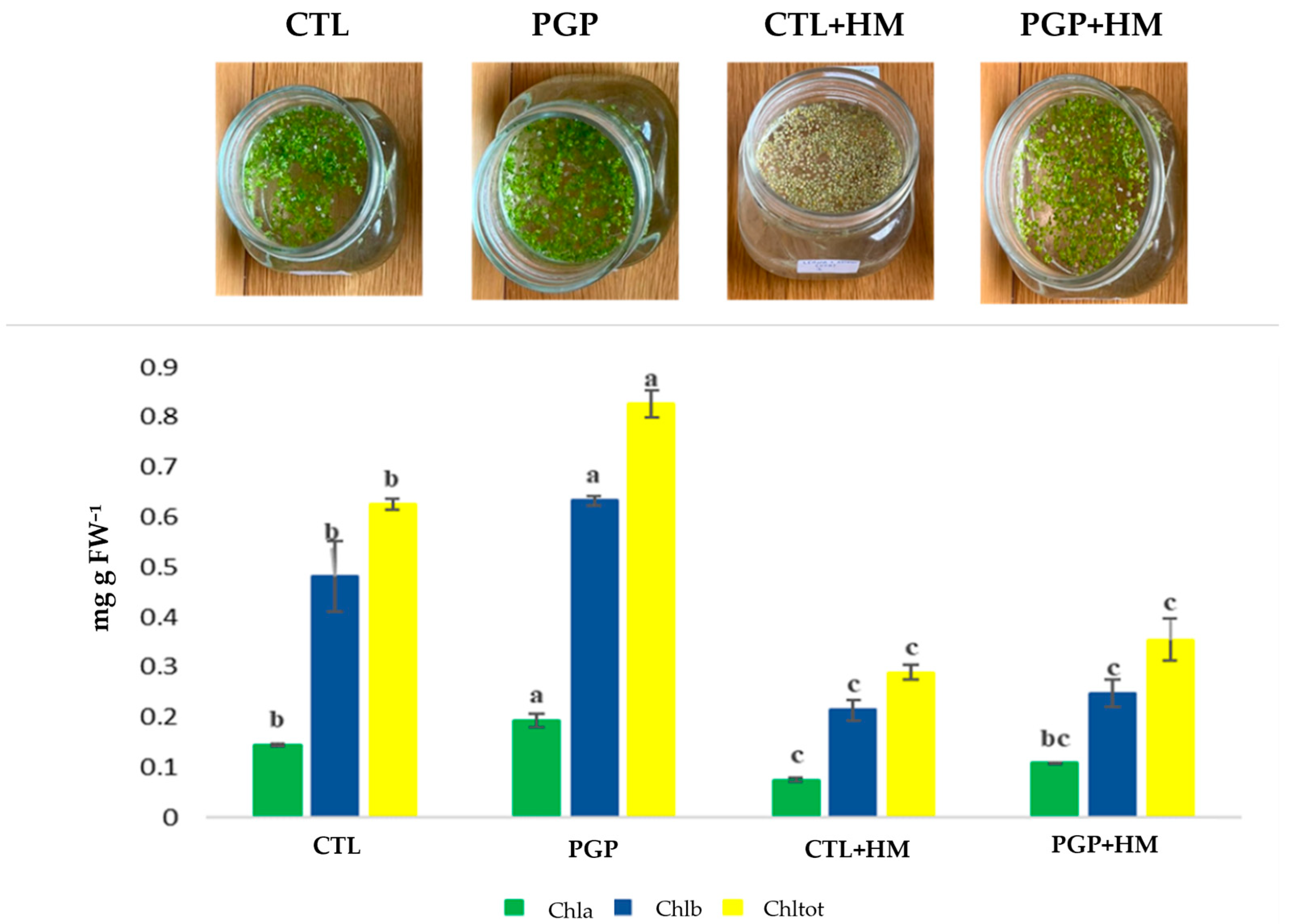

| LAK | MIN | MTW | |
|---|---|---|---|
| Taxa_S | 1273 | 1036 | 740 |
| Individuals | 28,505 | 18,070 | 13,029 |
| Simpson_1-D | 0.9978 | 0.998 | 0.9962 |
| Shannon_H | 6.635 | 6.577 | 6.134 |
| Evenness_eH/S | 0.5981 | 0.6935 | 0.6233 |
| Chao-1 | 1274 | 1038 | 741.6 |
| Al | Cd | Cu | Mn 0.5% | Mn 1% | Ni | Pb | Zn | |
|---|---|---|---|---|---|---|---|---|
| MIN 1 | + | - | ++ | - | - | +++ | ++ | - |
| MIN 2 | + | - | +++ | - | - | +++ | ++ | +++ |
| MTW 1 | + | + | +++ | +++ | +++ | +++ | ++ | + |
| MTW 2 | + | + | ++ | +++ | ++ | +++ | +++ | + |
| MTW 3 | + | + | - | - | - | + | + | - |
| MTW 4 | - | - | - | - | - | ++ | + | - |
| MTW 5 | ++ | ++ | ++ | +++ | ++ | +++ | ++ | + |
| MTW 6 | + | ++ | + | ++ | ++ | ++ | ++ | ++ |
| MTW 7 | + | - | ++ | - | - | +++ | + | - |
| MTW 8 | + | + | ++ | - | - | + | ++ | +++ |
| P µg mL−1 | Ind µg mL−1 | Sid % | HCN | NH3 | ACCd | |
|---|---|---|---|---|---|---|
| MIN 1 | 1.89 ± 0.00 a | - | - | + | +++ | - |
| MIN 2 | - | - | 13.15 ± 0.96 a | + | + | - |
| MTW 1 | 13.37 ± 0.52 b | 2.85 ± 0.06 | 21.52 ± 2.16 a | ++ | ++ | - |
| MTW 2 | 1.89 ± 0.00 a | - | 12.82 ± 0.76 a | + | + | - |
| MTW 3 | - | - | 24.95 ± 1.25 a | + | + | - |
| MTW 4 | - | - | 15.92 ± 2.30 a | + | + | - |
| MTW 5 | - | - | 22.87 ± 3.06 a | + | +++ | - |
| MTW 6 | - | - | 16.70 ± 1.00 a | +++ | ++ | - |
| MTW 7 | 1.89 ± 0.00 a | - | - | ++ | +++ | - |
| MTW 8 | - | - | - | ++ | ++ | - |
| Survival Rate (%) | Plant Biomass (%) | Root Lengths (cm) | |
|---|---|---|---|
| CTL | 100 ± 0 b | 2.26 ± 0.70 a | 2.37 ± 0.44 ab |
| PGP | 100 ± 0 b | 3.63 ± 0.33 a | 2.82 ± 0.08 ab |
| CTL + HM | 46.00 ± 0.82 ab | 1.87 ± 0.09 b | 1.45 ± 0.48 b |
| PGP + HM | 97.67 ± 0.47 ab | 2.33 ± 0.12 b | 1.77 ± 0.37 b |
| Heavy Metals | Final Concentration |
|---|---|
| Al2(SO4)3 | 2 mM |
| CdSO4·8H2O | 0.1 mM |
| CuCl2·2H2O | 0.15 mM |
| Pb(NO3)2 | 1 mM |
| NiCl2·6H2O | 0.2 mM |
| ZnSO4·7H2O | 1 mM |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Djebaili, R.; Farda, B.; Gialdini, O.; Vaccarelli, I.; Rezaee Danesh, Y.; Pellegrini, M. Microbial Consortium of Streptomyces spp. from Mining Environments Enhances Phytoremediation Potential of Lemna minor L. Plants 2025, 14, 3467. https://doi.org/10.3390/plants14223467
Djebaili R, Farda B, Gialdini O, Vaccarelli I, Rezaee Danesh Y, Pellegrini M. Microbial Consortium of Streptomyces spp. from Mining Environments Enhances Phytoremediation Potential of Lemna minor L. Plants. 2025; 14(22):3467. https://doi.org/10.3390/plants14223467
Chicago/Turabian StyleDjebaili, Rihab, Beatrice Farda, Oscar Gialdini, Ilaria Vaccarelli, Younes Rezaee Danesh, and Marika Pellegrini. 2025. "Microbial Consortium of Streptomyces spp. from Mining Environments Enhances Phytoremediation Potential of Lemna minor L." Plants 14, no. 22: 3467. https://doi.org/10.3390/plants14223467
APA StyleDjebaili, R., Farda, B., Gialdini, O., Vaccarelli, I., Rezaee Danesh, Y., & Pellegrini, M. (2025). Microbial Consortium of Streptomyces spp. from Mining Environments Enhances Phytoremediation Potential of Lemna minor L. Plants, 14(22), 3467. https://doi.org/10.3390/plants14223467










