Genomic Data Suggests Pathways of Modern White Poplar (Populus alba L.) Range Formation in the Postglacial Era
Abstract
1. Introduction
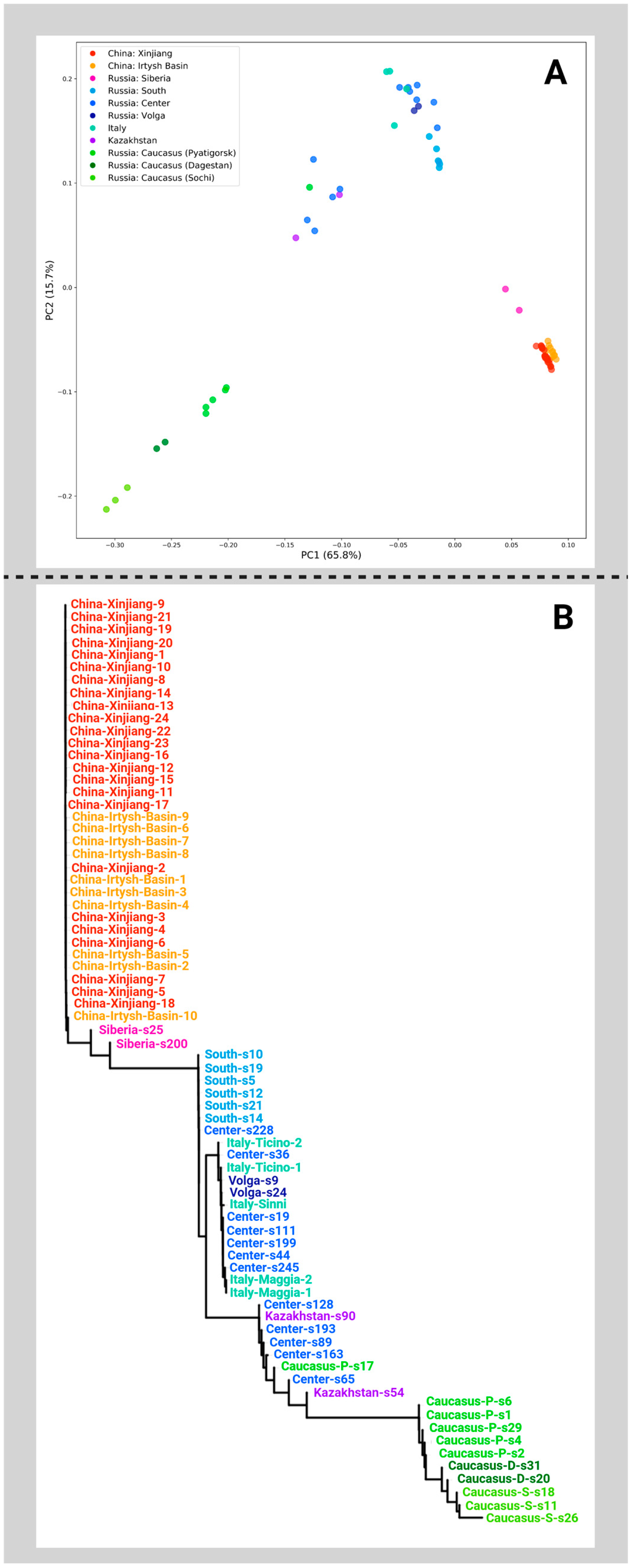
2. Results
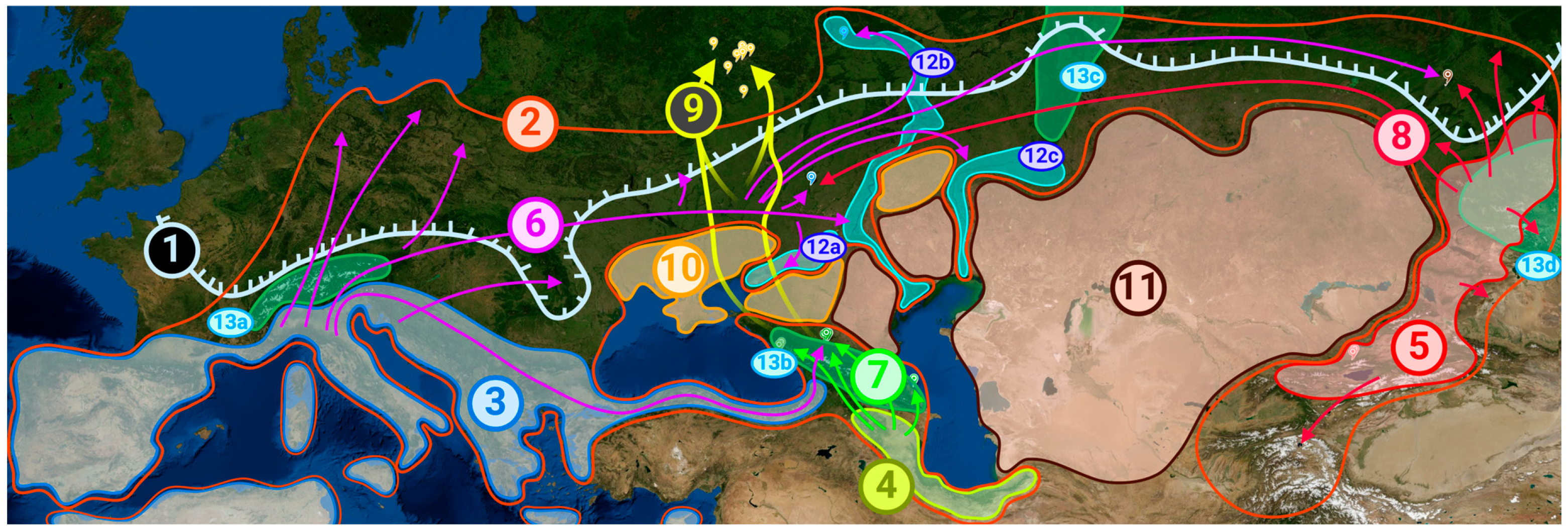
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. DNA Isolation, Library Preparation, and Whole-Genome Sequencing
4.3. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ogutcen, E.; De Lima Ferreira, P.; Wagner, N.D.; Marinček, P.; Vir Leong, J.; Aubona, G.; Cavender-Bares, J.; Michálek, J.; Schroeder, L.; Sedio, B.E.; et al. Phylogenetic Insights into the Salicaceae: The Evolution of Willows and Beyond. Mol. Phylogenet. Evol. 2024, 199, 108161. [Google Scholar] [CrossRef]
- Tuskan, G.A.; DiFazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The Genome of Black Cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef]
- Zhang, Z.-S.; Zeng, Q.-Y.; Liu, Y.-J. Frequent Ploidy Changes in Salicaceae Indicates Widespread Sharing of the Salicoid Whole Genome Duplication by the Relatives of Populus L. and Salix L. BMC Plant Biol. 2021, 21, 535. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, J.; Li, E.; Xu, S.; Zhan, Z.; Zhang, X.; Yang, Z.; Guo, F.; Liu, K.; Liu, D.; et al. Phylogenomics and Biogeography of Populus Based on Comprehensive Sampling Reveal Deep-Level Relationships and Multiple Intercontinental Dispersals. Front. Plant Sci. 2022, 13, 813177. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, Z.; Wang, W.; Huang, Q.; Zeng, Y.; Jin, Y.; Li, H.; Du, S.; Zhang, J. Origin and Evolutionary History of Populus (Salicaceae): Further Insights Based on Time Divergence and Biogeographic Analysis. Front. Plant Sci. 2022, 13, 1031087. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, L.; Zhang, Z.; Li, M.; Wang, D.; Zhang, X.; Xi, Z.; Keefover-Ring, K.; Smart, L.B.; DiFazio, S.P.; et al. Phylogenomics of the Genus Populus Reveals Extensive Interspecific Gene Flow and Balancing Selection. New Phytol. 2020, 225, 1370–1382. [Google Scholar] [CrossRef] [PubMed]
- Gladysh, N.S.; Kovalev, M.A.; Lantsova, M.S.; Popchenko, M.I.; Bolsheva, N.L.; Starkova, A.M.; Bulavkina, E.V.; Karpov, D.S.; Kudryavtsev, A.A.; Kudryavtseva, A.V. The Molecular and Genetic Mechanisms of Sex Determination in Poplar. Mol. Biol. 2024, 58, 178–191. [Google Scholar] [CrossRef]
- Li, Y.; Wang, D.; Wang, W.; Yang, W.; Gao, J.; Zhang, W.; Shan, L.; Kang, M.; Chen, Y.; Ma, T. A Chromosome-level Populus qiongdaoensis Genome Assembly Provides Insights into Tropical Adaptation and a Cryptic Turnover of Sex Determination. Mol. Ecol. 2023, 32, 1366–1380. [Google Scholar] [CrossRef] [PubMed]
- Geraldes, A.; Farzaneh, N.; Grassa, C.J.; McKown, A.D.; Guy, R.D.; Mansfield, S.D.; Douglas, C.J.; Cronk, Q.C.B. LANDSCAPE GENOMICS OF POPULUS TRICHOCARPA: THE ROLE OF HYBRIDIZATION, LIMITED GENE FLOW, AND NATURAL SELECTION IN SHAPING PATTERNS OF POPULATION STRUCTURE. Evolution 2014, 68, 3260–3280. [Google Scholar] [CrossRef]
- McKown, A.D.; Guy, R.D.; Klápště, J.; Geraldes, A.; Friedmann, M.; Cronk, Q.C.B.; El-Kassaby, Y.A.; Mansfield, S.D.; Douglas, C.J. Geographical and Environmental Gradients Shape Phenotypic Trait Variation and Genetic Structure in Populus Trichocarpa. New Phytol. 2014, 201, 1263–1276. [Google Scholar] [CrossRef]
- Keller, S.R.; Olson, M.S.; Silim, S.; Schroeder, W.; Tiffin, P. Genomic Diversity, Population Structure, and Migration Following Rapid Range Expansion in the Balsam Poplar, Populus balsamifera. Mol. Ecol. 2010, 19, 1212–1226. [Google Scholar] [CrossRef]
- Meirmans, P.G.; Godbout, J.; Lamothe, M.; Thompson, S.L.; Isabel, N. History Rather than Hybridization Determines Population Structure and Adaptation in Populus balsamifera. J. Evol. Biol. 2017, 30, 2044–2058. [Google Scholar] [CrossRef]
- Wang, D.; Wang, Z.; Kang, X.; Zhang, J. Genetic Analysis of Admixture and Hybrid Patterns of Populus hopeiensis and P. tomentosa. Sci. Rep. 2019, 9, 4821. [Google Scholar] [CrossRef]
- Cvelev, N.N.; Fëdorov, A.A.; Botaničeskij Institut Imeni, V.L.; Komarova (Eds.) Flora evropejskoj časti SSSR. T. 10: Pokrytosemennye: Dvudolʹnye/pod red. N. N. Cveleva; Izdat. Nauka: Leningrad, Russia, 2001; ISBN 978-5-8085-0122-5. [Google Scholar]
- Chrtek, J. PH Davis (ed.) Flora of Turkey and the East Aegean Islands. Vol. 7.: Edinburgh University Press 1982, 947 Pp., 27 Figs., 116 Maps. Price £ 65.00. Folia Geobot. Phytotax. 1984, 19, 322. [Google Scholar] [CrossRef]
- Fussi, B.; Lexer, C.; Heinze, B. Phylogeography of Populus alba (L.) and Populus tremula (L.) in Central Europe: Secondary Contact and Hybridisation during Recolonisation from Disconnected Refugia. Tree Genet. Genomes 2010, 6, 439–450. [Google Scholar] [CrossRef]
- Brundu, G.; Lupi, R.; Zapelli, I.; Fossati, T.; Patrignani, G.; Camarda, I.; Sala, F.; Castiglione, S. The Origin of Clonal Diversity and Structure of Populus alba in Sardinia: Evidence from Nuclear and Plastid Microsatellite Markers. Ann. Bot. 2008, 102, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization of the United Nations. Overview International Commission on Poplars and Other Fast-Growing Trees Sustaining People and the Environment. Available online: https://www.fao.org/ipc/overview/en/ (accessed on 14 October 2024).
- Liberloo, M.; Calfapietra, C.; Lukac, M.; Godbold, D.; Luo, Z.; Polle, A.; Hoosbeek, M.R.; Kull, O.; Marek, M.; Raines, C.; et al. Woody Biomass Production during the Second Rotation of a Bio-energy Populus Plantation Increases in a Future High CO2 World. Glob. Change Biol. 2006, 12, 1094–1106. [Google Scholar] [CrossRef]
- Macaya-Sanz, D.; Chen, J.; Kalluri, U.C.; Muchero, W.; Tschaplinski, T.J.; Gunter, L.E.; Simon, S.J.; Biswal, A.K.; Bryan, A.C.; Payyavula, R.; et al. Agronomic Performance of Populus Deltoides Trees Engineered for Biofuel Production. Biotechnol. Biofuels 2017, 10, 253. [Google Scholar] [CrossRef]
- Tőzsér, D.; Horváth, R.; Simon, E.; Magura, T. Heavy Metal Uptake by Plant Parts of Populus Species: A Meta-Analysis. Env. Sci. Pollut. Res. 2023, 30, 69416–69430. [Google Scholar] [CrossRef]
- Di Lonardo, S.; Capuana, M.; Arnetoli, M.; Gabbrielli, R.; Gonnelli, C. Exploring the Metal Phytoremediation Potential of Three Populus alba L. Clones Using an in Vitro Screening. Environ. Sci. Pollut. Res. 2011, 18, 82–90. [Google Scholar] [CrossRef] [PubMed]
- EUFORGEN. European Forest Genetic Resources Programme. Available online: https://www.euforgen.org/ (accessed on 15 December 2024).
- White Poplar (Populus alba). Available online: https://www.illinoiswildflowers.info/trees/plants/wh_poplar.html (accessed on 15 December 2024).
- Liu, J.-G.; Han, X.; Yang, T.; Cui, W.-H.; Wu, A.-M.; Fu, C.-X.; Wang, B.-C.; Liu, L.-J. Genome-Wide Transcriptional Adaptation to Salt Stress in Populus. BMC Plant Biol. 2019, 19, 367. [Google Scholar] [CrossRef]
- Rosso, L.; Cantamessa, S.; Bergante, S.; Biselli, C.; Fricano, A.; Chiarabaglio, P.M.; Gennaro, M.; Nervo, G.; Secchi, F.; Carra, A. Responses to Drought Stress in Poplar: What Do We Know and What Can We Learn? Life 2023, 13, 533. [Google Scholar] [CrossRef] [PubMed]
- Castiglione, S.; Cicatelli, A.; Lupi, R.; Patrignani, G.; Fossati, T.; Brundu, G.; Sabatti, M.; Van Loo, M.; Lexer, C. Genetic Structure and Introgression in Riparian Populations of Populus alba L. Plant Biosyst. Int. J. Deal. All Asp. Plant Biol. 2010, 144, 656–668. [Google Scholar] [CrossRef]
- Fussi, B.; Bonello, J.; Calleja, E.; Heinze, B. Combining the Use of Molecular Techniques and Archival Documentary Evidence to Trace the Origin of Populus Alba in a Central Mediterranean Archipelago. Eur. J. For. Res 2012, 131, 347–354. [Google Scholar] [CrossRef]
- Liu, Y.-J.; Wang, X.-R.; Zeng, Q.-Y. De Novo Assembly of White Poplar Genome and Genetic Diversity of White Poplar Population in Irtysh River Basin in China. Sci. China Life Sci. 2019, 62, 609–618. [Google Scholar] [CrossRef]
- Zeng, Y.-F.; Zhang, J.-G.; Duan, A.-G.; Abuduhamiti, B. Genetic Structure of Populus Hybrid Zone along the Irtysh River Provides Insight into Plastid-Nuclear Incompatibility. Sci. Rep. 2016, 6, 28043. [Google Scholar] [CrossRef] [PubMed]
- Xiang, X.; Zhou, X.; Zi, H.; Wei, H.; Cao, D.; Zhang, Y.; Zhang, L.; Hu, J. Populus cathayana Genome and Population Resequencing Provide Insights into Its Evolution and Adaptation. Hortic. Res. 2024, 11, uhad255. [Google Scholar] [CrossRef]
- Ehlers, J.; Gibbard, P.L.; Hughes, P.D. Quaternary Glaciations and Chronology. In Past Glacial Environments; Elsevier: Amsterdam, The Netherlands, 2018; pp. 77–101. ISBN 978-0-08-100524-8. [Google Scholar]
- Böse, M.; Lüthgens, C.; Lee, J.R.; Rose, J. Quaternary Glaciations of Northern Europe. Quat. Sci. Rev. 2012, 44, 1–25. [Google Scholar] [CrossRef]
- Solomina, O.N.; Bradley, R.S.; Hodgson, D.A.; Ivy-Ochs, S.; Jomelli, V.; Mackintosh, A.N.; Nesje, A.; Owen, L.A.; Wanner, H.; Wiles, G.C.; et al. Holocene Glacier Fluctuations. Quat. Sci. Rev. 2015, 111, 9–34. [Google Scholar] [CrossRef]
- Wanner, H.; Solomina, O.; Grosjean, M.; Ritz, S.P.; Jetel, M. Structure and Origin of Holocene Cold Events. Quat. Sci. Rev. 2011, 30, 3109–3123. [Google Scholar] [CrossRef]
- Berger, A.; Loutre, M.F. An Exceptionally Long Interglacial Ahead? Science 2002, 297, 1287–1288. [Google Scholar] [CrossRef]
- Ehlers, J.; Astakhov, V.; Gibbard, P.L.; Mangerud, J.; Svendsen, J.I. GLACIATIONS Middle Pleistocene in Eurasia. In Encyclopedia of Quaternary Science; Elsevier: Amsterdam, The Netherlands, 2013; pp. 172–179. ISBN 978-0-444-53642-6. [Google Scholar]
- Чeтвepтичный Пepиoд. Плeйcтoцeн (800 000–10 300 Лeт Haзaд), Toм 2 @ HAЦИOHAЛЬHЫЙ ATЛAC POCCИИ. Available online: https://nationalatlas.ru/tom2/26-27.html (accessed on 14 October 2024).
- Чeтвepтичныe Oбpaзoвaния, Toм 2 @ HAЦИOHAЛЬHЫЙ ATЛAC POCCИИ. Available online: https://nationalatlas.ru/tom2/60-62.html (accessed on 14 October 2024).
- Budantsev, L.J. (Ed.) Iskopaemye cvetkovye rastenija SSSR. 4: Nyctaginaceae-Salicaceae/L; KMK: Moscow, Russia, 2005; ISBN 978-5-87317-224-5. [Google Scholar]
- Melnikova, N.V.; Pushkova, E.N.; Dvorianinova, E.M.; Beniaminov, A.D.; Novakovskiy, R.O.; Povkhova, L.V.; Bolsheva, N.L.; Snezhkina, A.V.; Kudryavtseva, A.V.; Krasnov, G.S.; et al. Genome Assembly and Sex-Determining Region of Male and Female Populus × Sibirica. Front. Plant Sci. 2021, 12, 625416. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Skotte, L.; Korneliussen, T.S.; Albrechtsen, A. Estimating Individual Admixture Proportions from Next Generation Sequencing Data. Genetics 2013, 195, 693–702. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast Model-Based Estimation of Ancestry in Unrelated Individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Meisner, J.; Albrechtsen, A. Inferring Population Structure and Admixture Proportions in Low-Depth NGS Data. Genetics 2018, 210, 719–731. [Google Scholar] [CrossRef]
- Garrison, E.; Marth, G. Haplotype-Based Variant Detection from Short-Read Sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- Ortiz, E.M. Vcf2phylip v2.0: Convert a VCF Matrix into Several Matrix Formats for Phylogenetic Analysis. Zenodo 2019. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A Fast, Scalable and User-Friendly Tool for Maximum Likelihood Phylogenetic Inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Dylus, D.; Altenhoff, A.; Majidian, S.; Sedlazeck, F.J.; Dessimoz, C. Inference of Phylogenetic Trees Directly from Raw Sequencing Reads Using Read2Tree. Nat. Biotechnol. 2024, 42, 139–147. [Google Scholar] [CrossRef]
- Schneider, A.; Dessimoz, C.; Gonnet, G.H. OMA Browser—Exploring Orthologous Relations across 352 Complete Genomes. Bioinformatics 2007, 23, 2180–2182. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Zdobnov, E.M. BUSCO: Assessing Genomic Data Quality and Beyond. Curr. Protoc. 2021, 1, e323. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. CP Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Müller, N.A.; Kersten, B.; Leite Montalvão, A.P.; Mähler, N.; Bernhardsson, C.; Bräutigam, K.; Carracedo Lorenzo, Z.; Hoenicka, H.; Kumar, V.; Mader, M.; et al. A Single Gene Underlies the Dynamic Evolution of Poplar Sex Determination. Nat. Plants 2020, 6, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Shang, H.; Hess, J.; Pickup, M.; Field, D.L.; Ingvarsson, P.K.; Liu, J.; Lexer, C. Evolution of Strong Reproductive Isolation in Plants: Broad-Scale Patterns and Lessons from a Perennial Model Group. Phil. Trans. R. Soc. B 2020, 375, 20190544. [Google Scholar] [CrossRef]
- Christe, C.; Stölting, K.N.; Paris, M.; Fraïsse, C.; Bierne, N.; Lexer, C. Adaptive Evolution and Segregating Load Contribute to the Genomic Landscape of Divergence in Two Tree Species Connected by Episodic Gene Flow. Mol. Ecol. 2017, 26, 59–76. [Google Scholar] [CrossRef] [PubMed]
- Okumura, S.; Sawada, M.; Park, Y.W.; Hayashi, T.; Shimamura, M.; Takase, H.; Tomizawa, K.-I. Transformation of Poplar (Populus alba) Plastids and Expression of Foreign Proteins in Tree Chloroplasts. Transgenic Res. 2006, 15, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Brenner, W.G.; Mader, M.; Müller, N.A.; Hoenicka, H.; Schroeder, H.; Zorn, I.; Fladung, M.; Kersten, B. High Level of Conservation of Mitochondrial RNA Editing Sites Among Four Populus Species. G3 Genes Genomes Genet. 2019, 9, 709–717. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gladysh, N.S.; Popchenko, M.I.; Kovalev, M.A.; Volodin, V.V.; Krasnov, G.S.; Bogdanova, A.S.; Karpov, D.S.; Bolsheva, N.L.; Kudryavtseva, A.V. Genomic Data Suggests Pathways of Modern White Poplar (Populus alba L.) Range Formation in the Postglacial Era. Plants 2025, 14, 3328. https://doi.org/10.3390/plants14213328
Gladysh NS, Popchenko MI, Kovalev MA, Volodin VV, Krasnov GS, Bogdanova AS, Karpov DS, Bolsheva NL, Kudryavtseva AV. Genomic Data Suggests Pathways of Modern White Poplar (Populus alba L.) Range Formation in the Postglacial Era. Plants. 2025; 14(21):3328. https://doi.org/10.3390/plants14213328
Chicago/Turabian StyleGladysh, Natalya S., Mikhail I. Popchenko, Maxim A. Kovalev, Vsevolod V. Volodin, George S. Krasnov, Alina S. Bogdanova, Dmitry S. Karpov, Nadezhda L. Bolsheva, and Anna V. Kudryavtseva. 2025. "Genomic Data Suggests Pathways of Modern White Poplar (Populus alba L.) Range Formation in the Postglacial Era" Plants 14, no. 21: 3328. https://doi.org/10.3390/plants14213328
APA StyleGladysh, N. S., Popchenko, M. I., Kovalev, M. A., Volodin, V. V., Krasnov, G. S., Bogdanova, A. S., Karpov, D. S., Bolsheva, N. L., & Kudryavtseva, A. V. (2025). Genomic Data Suggests Pathways of Modern White Poplar (Populus alba L.) Range Formation in the Postglacial Era. Plants, 14(21), 3328. https://doi.org/10.3390/plants14213328






