Transcriptomic Profiling Unravels the Molecular Mechanisms of GmCML-Mediated Resistance to Fusarium oxysporum in Soybean
Abstract
1. Introduction
2. Results
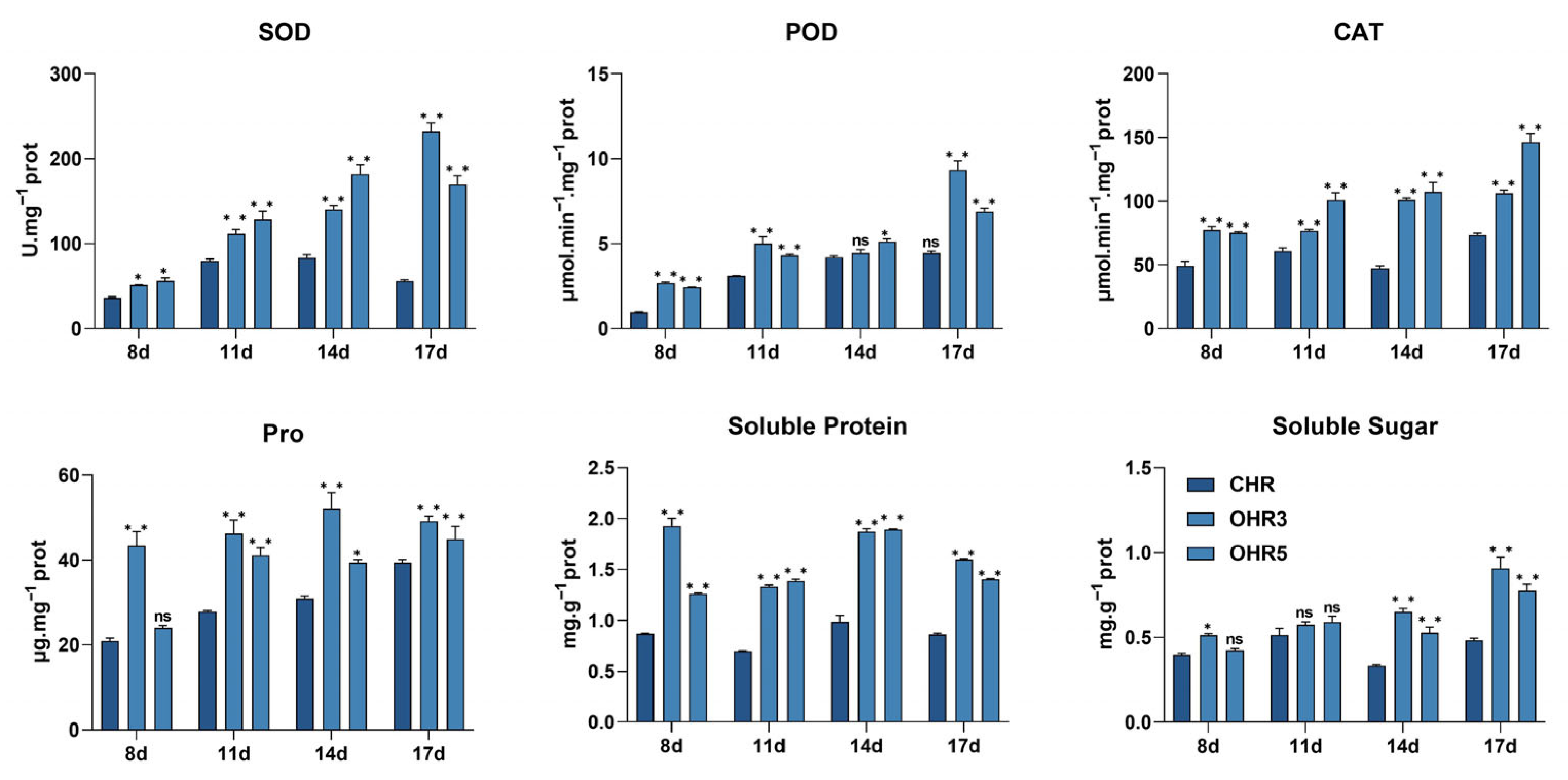
2.1. Contrasting Disease Phenotypes and Physiological Responses Between Resistant and Susceptible Soybean Accessions
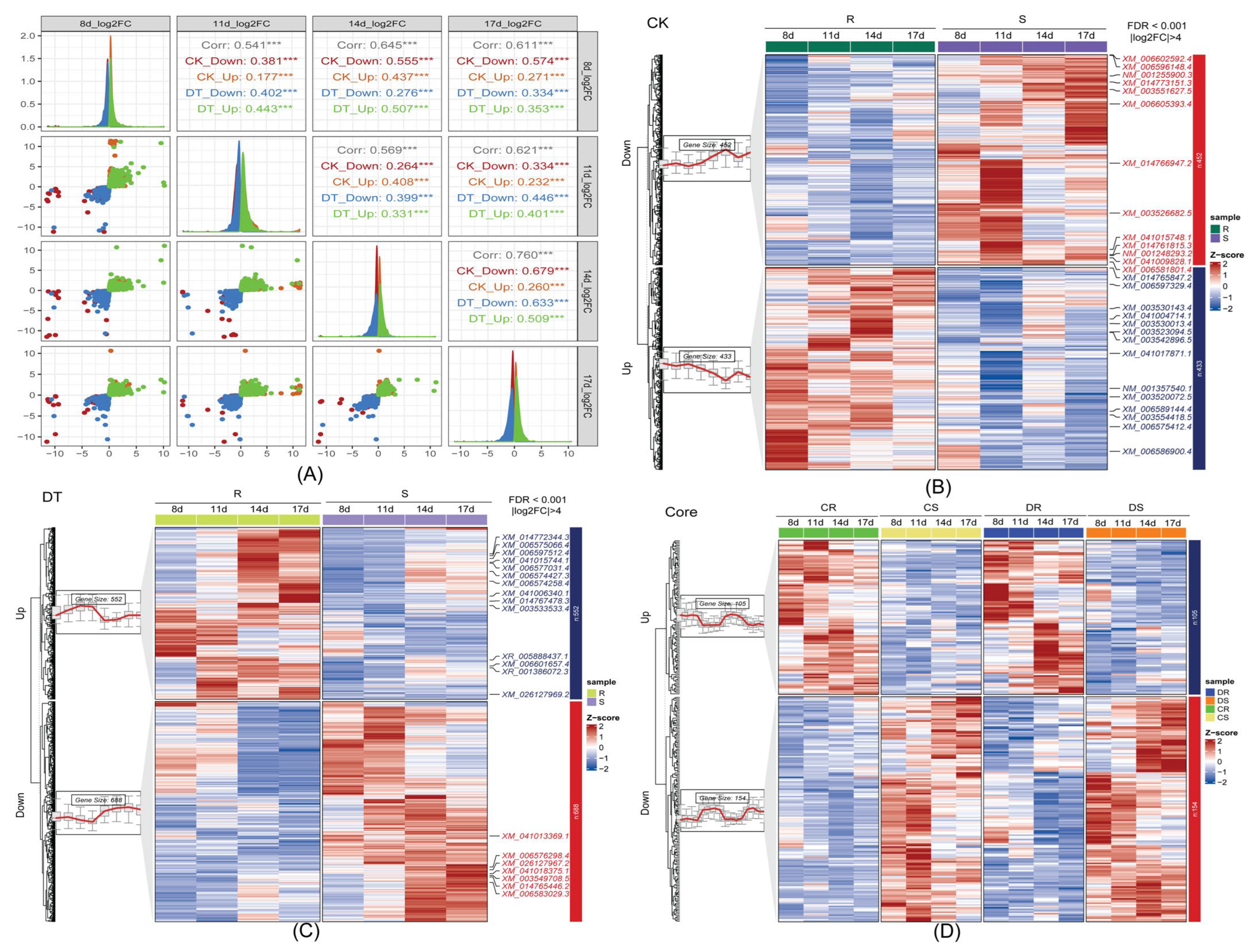
2.2. Comprehensive Transcriptomic Profiling Reveals Defense-Associated Gene Expression Networks
2.3. Expression Pattern Analysis Reveals Coordinated Transcriptional Responses
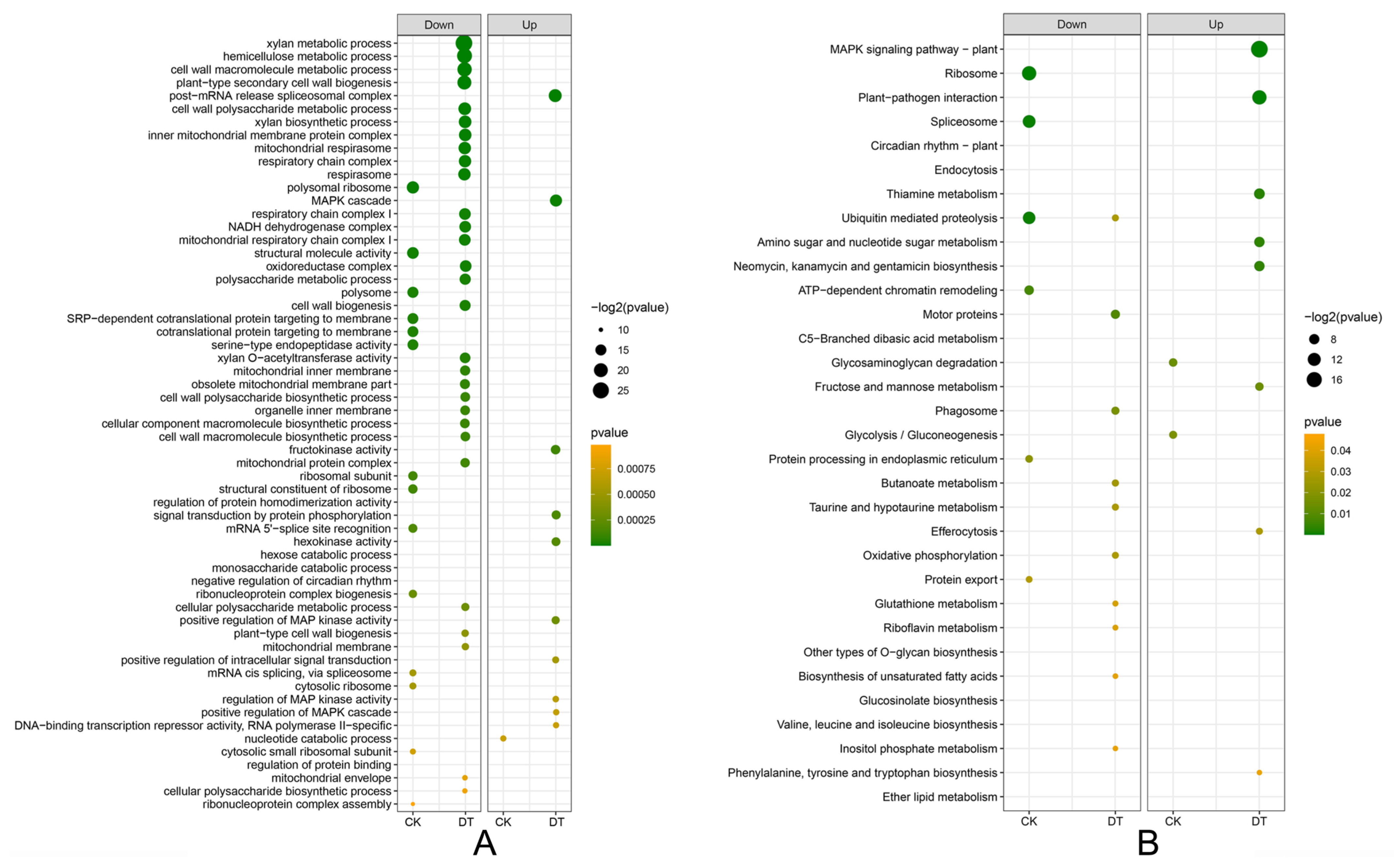
2.4. Functional Enrichment Analysis Identifies Key Defense Pathways
2.5. Validation and Identification of GmCML as a Key Resistance Gene
2.6. Functional Validation Demonstrates GmCML’s Role in Disease Resistance
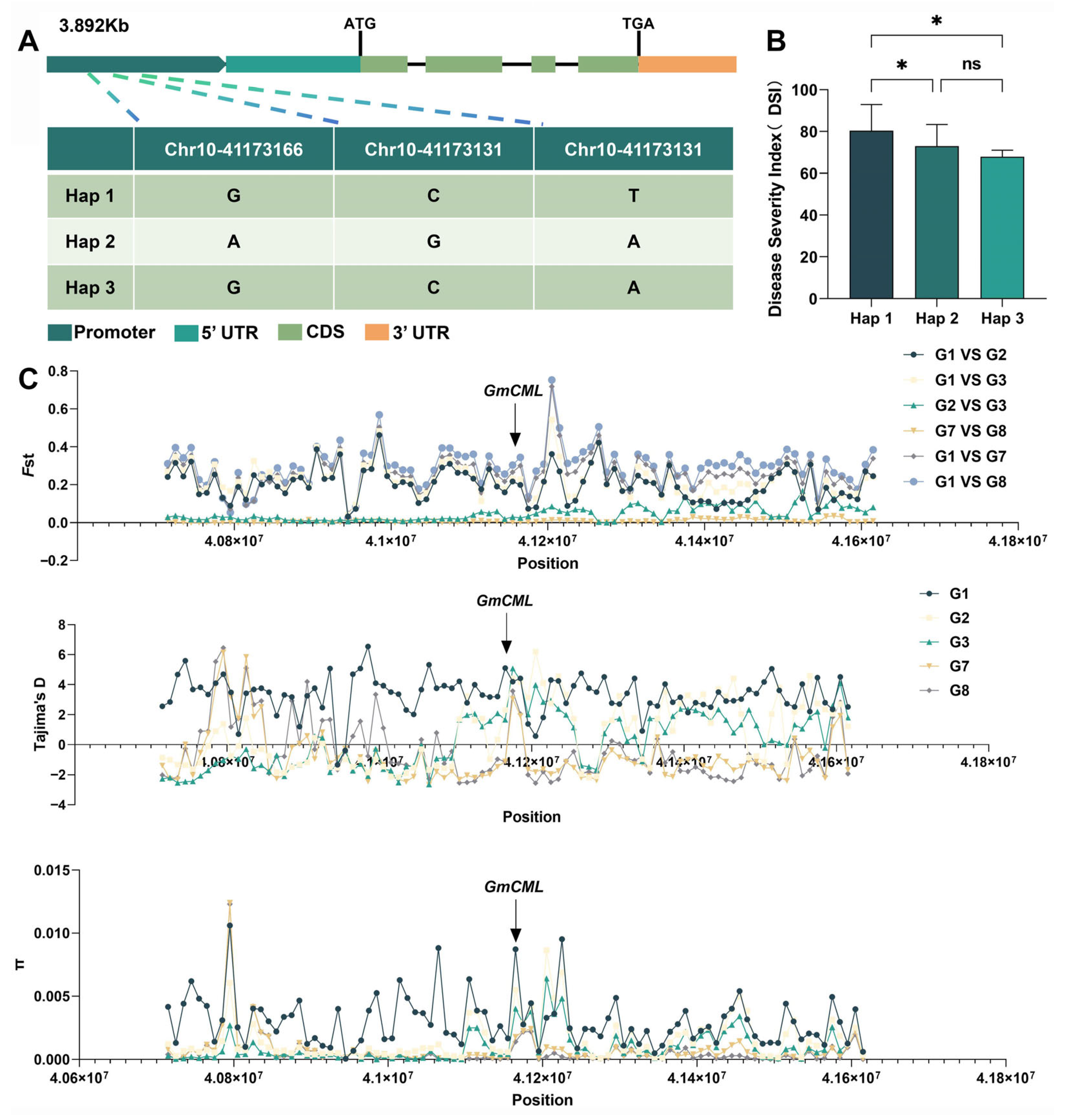
2.7. Population Genetic Analysis Reveals Natural Variation in GmCML
2.8. Evolutionary Analysis Indicates Selection During Domestication
3. Discussion
3.1. GmCML Functions as a Master Regulator of Calcium-Mediated Defense Responses
3.2. Integration with Plant Defense Signaling Networks
3.3. Physiological Integration of Defense Responses
3.4. Broader Implications for Plant Immunity Research
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Pathogen Culture and Inoculum Preparation
4.3. Disease Resistance Evaluation
4.4. Physiological and Biochemical Parameter Analysis
4.4.1. Antioxidant Enzyme Activity Assays
4.4.2. Osmoregulatory Compound Analysis
4.5. RNA Extraction and Quality Assessment
4.6. RNA Sequencing and Data Processing
4.7. Differential Expression Analysis
4.8. Functional Enrichment Analysis
4.9. Quantitative Real-Time PCR Validation
4.10. GmCML Functional Characterization
4.11. Hairy Root Transformation
4.12. Disease Resistance Assay in Hairy Roots
4.13. Population Genetic Analysis
4.13.1. Haplotype Analysis
4.13.2. Analysis of Selective Sweep in the GmCML Gene During Soybean Domestication
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lin, F.; Chhapekar, S.S.; Vieira, C.C.; Da Silva, M.P.; Rojas, A.; Lee, D.; Liu, N.; Pardo, E.M.; Lee, Y.-C.; Dong, Z.; et al. Breeding for disease resistance in soybean: A global perspective. Theor. Appl. Genet. 2022, 135, 3773–3872. [Google Scholar] [CrossRef] [PubMed]
- Whitham, S.A.; Qi, M.; Innes, R.W.; Ma, W.; Lopes-Caitar, V.; Hewezi, T. Molecular Soybean-Pathogen Interactions. Annu. Rev. Phytopathol. 2016, 54, 443–468. [Google Scholar] [CrossRef] [PubMed]
- Sang, Y.; Liu, X.; Yuan, C.; Yao, T.; Li, Y.; Wang, D.; Zhao, H.; Wang, Y. Genome-wide association study on resistance of cultivated soybean to Fusarium oxysporum root rot in Northeast China. BMC Plant Biol. 2023, 23, 625. [Google Scholar] [CrossRef]
- Ellis, M.L.; Jimenez, D.R.C.; Leandro, L.F.; Munkvold, G.P. Genotypic and Phenotypic Characterization of Fungi in the Fusarium oxysporum Species Complex from Soybean Roots. Phytopathology 2014, 104, 1329–1339. [Google Scholar] [CrossRef]
- Chang, X.; Dai, H.; Wang, D.; Zhou, H.; He, W.; Fu, Y.; Ibrahim, F.; Zhou, Y.; Gong, G.; Shang, J.; et al. Identification of Fusarium species associated with soybean root rot in Sichuan Province, China. Eur. J. Plant Pathol. 2018, 151, 563–577. [Google Scholar] [CrossRef]
- Chang, X.; Yan, L.; Naeem, M.; Khaskheli, M.I.; Zhang, H.; Gong, G.; Zhang, M.; Song, C.; Yang, W.; Liu, T. Maize/soybean relay strip intercropping reduces the occurrence of Fusarium root rot and changes the diversity of the pathogenic Fusarium species. Pathogens 2020, 9, 211. [Google Scholar] [CrossRef]
- Bradley, C.A.; Allen, T.W.; Sisson, A.J.; Bergstrom, G.C.; Bissonnette, K.M.; Bond, J.; Byamukama, E.; Chilvers, M.I.; Collins, A.A.; Damicone, J.P. Soybean yield loss estimates due to diseases in the United States and Ontario, Canada, from 2015 to 2019. Plant Health Prog. 2021, 22, 483–495. [Google Scholar] [CrossRef]
- Klein, H.S.; Luna, F.V. The growth of the soybean frontier in South America: The case of Brazil and Argentina. Rev. Hist. Econ.-J. Iber. Lat. Am. Econ. Hist. 2021, 39, 427–468. [Google Scholar] [CrossRef]
- Ninkuu, V.; Han, T.; Dakora, F.D. Development of the soybean industry in Africa: Safeguarding food security in Africa and China—A perspective. Engineering 2025, 49, 272–278. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, S.; Zhang, Y.; Yu, W.; Li, J.; Zhou, H. Study on regional characteristics of soybean production in China from a temporal and spatial perspective. J. Phys. Conf. Ser. 2020, 1592, 012088. [Google Scholar] [CrossRef]
- Wang, T.; Ma, Y.; Luo, S. Spatiotemporal evolution and influencing factors of soybean production in Heilongjiang Province, China. Land 2023, 12, 2090. [Google Scholar] [CrossRef]
- Krupinsky, J.M.; Bailey, K.L.; McMullen, M.P.; Gossen, B.D.; Turkington, T.K. Managing plant disease risk in diversified cropping systems. Agron. J. 2002, 94, 198–209. [Google Scholar] [CrossRef]
- Li, X. Economic Impact of China’s Retaliatory Soybean Tariff on US Soybean Farmers. arXiv 2025, arXiv:2503.10715. [Google Scholar]
- Panwar, V. Investigation of Seed Health in Pathology and Disease Management; Academic University Press: New Delhi, India, 2023; p. 55. [Google Scholar]
- Ekwomadu, T.I.; Mwanza, M. Fusarium fungi pathogens, identification, adverse effects, disease management, and global food security: A review of the latest research. Agriculture 2023, 13, 1810. [Google Scholar] [CrossRef]
- Coleman, J.J. The Fusarium solani species complex: Ubiquitous pathogens of agricultural importance. Mol. Plant Pathol. 2016, 17, 146–158. [Google Scholar] [CrossRef]
- Xu, X.; Shen, G.; Teng, H.; Zhao, J.; Xiao, J.; Guo, L.; Gao, Y.; Chen, J.; Wang, X.; Xiang, W. Unravelling species diversity and pathogenicity of Fusarium spp. associated with soybean leaf and root in Heilongjiang Province, China. Plant Dis. 2024, 108, 852–856. [Google Scholar] [CrossRef]
- Sun, X.; Liu, Y.; He, L.; Kuang, Z.; Dai, S.; Hua, L.; Jiang, Q.; Wei, T.; Ye, P.; Zeng, H. Response of Yields, Soil Physiochemical Characteristics, and the Rhizosphere Microbiome to the Occurrence of Root Rot Caused by Fusarium solani in Ligusticum chuanxiong Hort. Microorganisms 2024, 12, 2350. [Google Scholar] [CrossRef]
- Wang, Y.; Han, J.; Meng, X.; Sun, M.; Qu, S.; Liu, Y.; Li, Y.; Zhan, Y.; Teng, W.; Li, H.; et al. Genome-Wide Association Study and Marker Development for Fusarium oxysporum Root Rot Resistance in Soybean. Int. J. Mol. Sci. 2024, 25, 12573. [Google Scholar] [CrossRef]
- De Sain, M.; Rep, M. The Role of Pathogen-Secreted Proteins in Fungal Vascular Wilt Diseases. Int. J. Mol. Sci. 2015, 16, 23970–23993. [Google Scholar] [CrossRef]
- Naqvi, S.A.H.; Farhan, M.; Ahmad, M.; Kiran, R.; Shahbaz, M.; Abbas, A.; Hakim, F.; Shabbir, M.; Tan, Y.S.; Sathiya Seelan, J.S. Fungicide resistance in Fusarium species: Exploring environmental impacts and sustainable management strategies. Arch. Microbiol. 2025, 207, 31. [Google Scholar] [CrossRef]
- Foroud, N.A.; Chatterton, S.; Reid, L.M.; Turkington, T.K.; Tittlemier, S.A.; Gräfenhan, T. Fusarium diseases of Canadian grain crops: Impact and disease management strategies. In Future Challenges in Crop Protection Against Fungal Pathogens; Springer: Berlin/Heidelberg, Germany, 2014; pp. 267–316. [Google Scholar]
- Sharma, S.; Mandal, S.; Cramer, C.S. Recent Advances in Understanding and Controlling Fusarium Diseases of Alliums. Horticulturae 2024, 10, 527. [Google Scholar] [CrossRef]
- Shah, L.; Ali, A.; Yahya, M.; Zhu, Y.; Wang, S.; Si, H.; Rahman, H.; Ma, C. Integrated control of fusarium head blight and deoxynivalenol mycotoxin in wheat. Plant Pathol. 2018, 67, 532–548. [Google Scholar] [CrossRef]
- Brown, J.K. Durable resistance of crops to disease: A Darwinian perspective. Annu. Rev. Phytopathol. 2015, 53, 513–539. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.A.; Ellsworth, P.C.; Faria, J.C.; Head, G.P.; Owen, M.D.; Pilcher, C.D.; Shelton, A.M.; Meissle, M. Genetically engineered crops: Importance of diversified integrated pest management for agricultural sustainability. Front. Bioeng. Biotechnol. 2019, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Hao, Q.; Yang, H.; Chen, S.; Qu, Y.; Zhang, C.; Chen, L.; Cao, D.; Yuan, S.; Guo, W.; Yang, Z.; et al. RNA-Seq and Comparative Transcriptomic Analyses of Asian Soybean Rust Resistant and Susceptible Soybean Genotypes Provide Insights into Identifying Disease Resistance Genes. Int. J. Mol. Sci. 2023, 24, 13450. [Google Scholar] [CrossRef]
- Naveed, Z.A.; Ali, G.S. Comparative Transcriptome Analysis Between a Resistant and a Susceptible Wild Tomato Accession in Response to Phytophthora parasitica. Int. J. Mol. Sci. 2018, 19, 3735. [Google Scholar] [CrossRef]
- Matić, S.; Bagnaresi, P.; Biselli, C.; Orru’, L.; Amaral Carneiro, G.; Siciliano, I.; Valé, G.; Gullino, M.L.; Spadaro, D. Comparative transcriptome profiling of resistant and susceptible rice genotypes in response to the seedborne pathogen Fusarium fujikuroi. BMC Genom. 2016, 17, 608. [Google Scholar] [CrossRef]
- Lanubile, A.; Muppirala, U.K.; Severin, A.J.; Marocco, A.; Munkvold, G.P. Transcriptome profiling of soybean (Glycine max) roots challenged with pathogenic and non-pathogenic isolates of Fusarium oxysporum. BMC Genom. 2015, 16, 1089. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, J.; Zhang, R.; Li, J.; Li, J.; Zhang, D.; Li, Y.; Wang, J. Transcriptomic and Metabolomic Explanation of the Interaction Between Soybean and Root Rot Caused by Fusarium tricinctum. J. Agric. Food Chem. 2025, 73, 19944–19957. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, Q.; Yan, H.; Cang, X.; Li, W.; He, J.; Zhang, M.; Lou, L.; Wang, R.; Chang, M. Lighting-up wars: Stories of Ca2+ signaling in plant immunity. New Crops 2024, 1, 100027. [Google Scholar] [CrossRef]
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.-M.; He, S.Y.; Xin, X.-F. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 2021, 592, 105–109. [Google Scholar] [CrossRef]
- Grant, M.; Brown, I.; Adams, S.; Knight, M.; Ainslie, A.; Mansfield, J. The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J. 2000, 23, 441–450. [Google Scholar] [CrossRef]
- Chiasson, D.; Ekengren, S.K.; Martin, G.B.; Dobney, S.L.; Snedden, W.A. Calmodulin-like Proteins from Arabidopsis and Tomato are Involved in Host Defense Against Pseudomonas syringae pv. tomato. Plant Mol. Biol. 2005, 58, 887–897. [Google Scholar] [CrossRef] [PubMed]
- Leba, L.-J.; Cheval, C.; Ortiz-Martín, I.; Ranty, B.; Beuzón, C.R.; Galaud, J.-P.; Aldon, D. CML9, an Arabidopsis calmodulin-like protein, contributes to plant innate immunity through a flagellin-dependent signalling pathway. Plant J. 2012, 71, 976–989. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Bheri, M.; Pandey, G.K. Delineating calcium signaling machinery in plants: Tapping the potential through functional genomics. Curr. Genom. 2021, 22, 404–439. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Zhang, Y.; Zhang, X.; Pi, E.; Zhu, Y. Analysis of EF-hand proteins in soybean genome suggests their potential roles in environmental and nutritional stress signaling. Front. Plant Sci. 2017, 8, 877. [Google Scholar] [CrossRef]
- Yadav, M.; Pandey, J.; Chakraborty, A.; Hassan, M.I.; Kundu, J.K.; Roy, A.; Singh, I.K.; Singh, A. A comprehensive analysis of calmodulin-like proteins of glycine max indicates their role in calcium signaling and plant defense against insect attack. Front. Plant Sci. 2022, 13, 817950. [Google Scholar] [CrossRef]
- Yadav, R.; Gupta, P.; Chhabra, R.; Thakur, K.; Dhar, H. Transcriptomics of Host–Pathogen Interaction. In Biotechnological Advances for Disease Tolerance in Plants; Springer: Berlin/Heidelberg, Germany, 2024; pp. 377–397. [Google Scholar]
- Chaitra, G.; Sharma, P.S.; Parameswari, B.; Anbazhagan, P.; Rajashree, A.; Chalam, V.C.; Siddarthan, V.K. Bioinformatics approaches in molecular plant pathology: From genomic data to disease diagnosis. Indian J. Plant Prot. 2025, 53, 59–72. [Google Scholar]
- Zhao, Y.; Qu, X.; Zha, B.; Sun, L.; Zhou, R.; Wang, Y.; Ren, H. Identification of Soybean Germplasm Resources Resistant to Fusarium oxysporum Root Rot Disease. Soybean Sci. 2025, 2, 8–13. [Google Scholar]
- Delplace, F.; Huard-Chauveau, C.; Berthomé, R.; Roby, D. Network organization of the plant immune system: From pathogen perception to robust defense induction. Plant J. 2022, 109, 447–470. [Google Scholar] [CrossRef]
- Rezapour, M.; Walker, S.J.; Ornelles, D.A.; McNutt, P.M.; Atala, A.; Gurcan, M.N. Analysis of gene expression dynamics and differential expression in viral infections using generalized linear models and quasi-likelihood methods. Front. Microbiol. 2024, 15, 1342328. [Google Scholar] [CrossRef]
- Liao, H.; Liu, F.; Wang, X.; Huang, H.; Huang, Q.; Wang, N.; Wei, C. Comparative Transcriptome Analysis of Susceptible and Resistant Rutaceae Plants to Huanglongbing. Agronomy 2025, 15, 1218. [Google Scholar] [CrossRef]
- Ghosh, S.K. Nature and Mechanisms of Abiotic and Biotic Stress Responses and Signaling in the Pinus nigra-Diplodia spp. Pathosystem; The Ohio State University: Columbus, OH, USA, 2023. [Google Scholar]
- Meng, X.; Zhang, S. MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 2013, 51, 245–266. [Google Scholar] [CrossRef]
- Saharan, G.S.; Mehta, N.K.; Meena, P.D. Host Resistance Signaling Network System to Multiple Stresses. In Genomics of Crucifer’s Host-Resistance; Springer: Berlin/Heidelberg, Germany, 2022; pp. 359–463. [Google Scholar]
- Wang, D.; Liang, X.; Bao, Y.; Yang, S.; Zhang, X.; Yu, H.; Zhang, Q.; Xu, G.; Feng, X.; Dou, D. A malectin-like receptor kinase regulates cell death and pattern-triggered immunity in soybean. EMBO Rep. 2020, 21, e50442. [Google Scholar] [CrossRef] [PubMed]
- Künstler, A.; Bacsó, R.; Hafez, Y.M.; Király, L. Reactive oxygen species and plant disease resistance. In Reactive Oxygen Species and Oxidative Damage in Plants Under Stress; Springer: Berlin/Heidelberg, Germany, 2015; pp. 269–303. [Google Scholar]
- Knaus, U.G. Reactive Oxygen Species. In Inflammation: Fundamental Mechanisms; World Scientific Publishing Co., Pte Ltd.: Singapore, 2018; pp. 125–169. [Google Scholar]
- Jing, M.; Ma, H.; Li, H.; Guo, B.; Zhang, X.; Ye, W.; Wang, H.; Wang, Q.; Wang, Y. Differential regulation of defense-related proteins in soybean during compatible and incompatible interactions between Phytophthora sojae and soybean by comparative proteomic analysis. Plant Cell Rep. 2015, 34, 1263–1280. [Google Scholar] [CrossRef] [PubMed]
- Paliwal, K.; Jajoo, A.; Tomar, R.S.; Prakash, A.; Syed, A.; Bright, J.P.; Sayyed, R. Enhancing biotic stress tolerance in soybean affected by Rhizoctonia solani root rot through an integrated approach of biocontrol agent and fungicide. Curr. Microbiol. 2023, 80, 304. [Google Scholar] [CrossRef]
- Glubb, D.M.; Innocenti, F. Mechanisms of genetic regulation in gene expression: Examples from drug metabolizing enzymes and transporters. Wiley Interdiscip. Rev. Syst. Biol. Med. 2011, 3, 299–313. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, Y.; Zhang, R.; Yuan, R.; Zhao, K.; Liu, X.; Wang, X.; Zhang, F.; Lamlom, S.F.; Zhang, B. Comprehensively characterize the soybean CAM/CML gene family, as it provides resistance against both the soybean mosaic virus and Cercospora sojina pathogens. Front. Plant Sci. 2025, 16, 1633325. [Google Scholar] [CrossRef]
- Trivedi, L.; Rathi, Y. Detection of seed mycoflora from chickpea wilt complex seed borne Fusarium oxysporum f. sp. ciceri diseased seeds. World J. Pharm. Pharm. Sci. 2015, 44, 1242–1249. [Google Scholar]
- Hohenfeld, C.S.; de Oliveira, S.A.S.; Ferreira, C.F.; Mello, V.H.; Margarido, G.R.A.; Passos, A.R.; de Oliveira, E.J. Comparative analysis of infected cassava root transcriptomics reveals candidate genes for root rot disease resistance. Sci. Rep. 2024, 14, 10587. [Google Scholar] [CrossRef]
- Ferreira, J.; Zwinderman, A. On the benjamini–hochberg method. Ann. Stat. 2006, 34, 1827–1849. [Google Scholar] [CrossRef]
- Ruihong, H.; Xinshi, L.; Guijuan, G. Analysis of the principal components and the subordinate function of alfalfa drought resistance. Acta Agrestia Sin. 2006, 14, 142–146. [Google Scholar]
- Yang, Z.; Luo, C.; Pei, X.; Wang, S.; Huang, Y.; Li, J.; Liu, B.; Kong, F.; Yang, Q.-Y.; Fang, C. SoyMD: A platform combining multi-omics data with various tools for soybean research and breeding. Nucleic Acids Res. 2023, 52, D1639–D1650. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, R.; You, J.; Li, J.; Qu, X.; Shang, Y.; Ren, H.; Wang, J. Transcriptomic Profiling Unravels the Molecular Mechanisms of GmCML-Mediated Resistance to Fusarium oxysporum in Soybean. Plants 2025, 14, 3222. https://doi.org/10.3390/plants14203222
Zhou R, You J, Li J, Qu X, Shang Y, Ren H, Wang J. Transcriptomic Profiling Unravels the Molecular Mechanisms of GmCML-Mediated Resistance to Fusarium oxysporum in Soybean. Plants. 2025; 14(20):3222. https://doi.org/10.3390/plants14203222
Chicago/Turabian StyleZhou, Runnan, Jia You, Jinrong Li, Xue Qu, Yuxin Shang, Honglei Ren, and Jiajun Wang. 2025. "Transcriptomic Profiling Unravels the Molecular Mechanisms of GmCML-Mediated Resistance to Fusarium oxysporum in Soybean" Plants 14, no. 20: 3222. https://doi.org/10.3390/plants14203222
APA StyleZhou, R., You, J., Li, J., Qu, X., Shang, Y., Ren, H., & Wang, J. (2025). Transcriptomic Profiling Unravels the Molecular Mechanisms of GmCML-Mediated Resistance to Fusarium oxysporum in Soybean. Plants, 14(20), 3222. https://doi.org/10.3390/plants14203222





