Linking Soil Microbial Functional Profiles to Fungal Disease Resistance in Winter Barley Under Different Fertilisation Regimes
Abstract
1. Introduction
2. Results
2.1. Morphological Assessment
2.2. Disease Severity Across Barley Variants
2.3. Effects of Cropping System and Fertilisation on Foliar Disease Severity
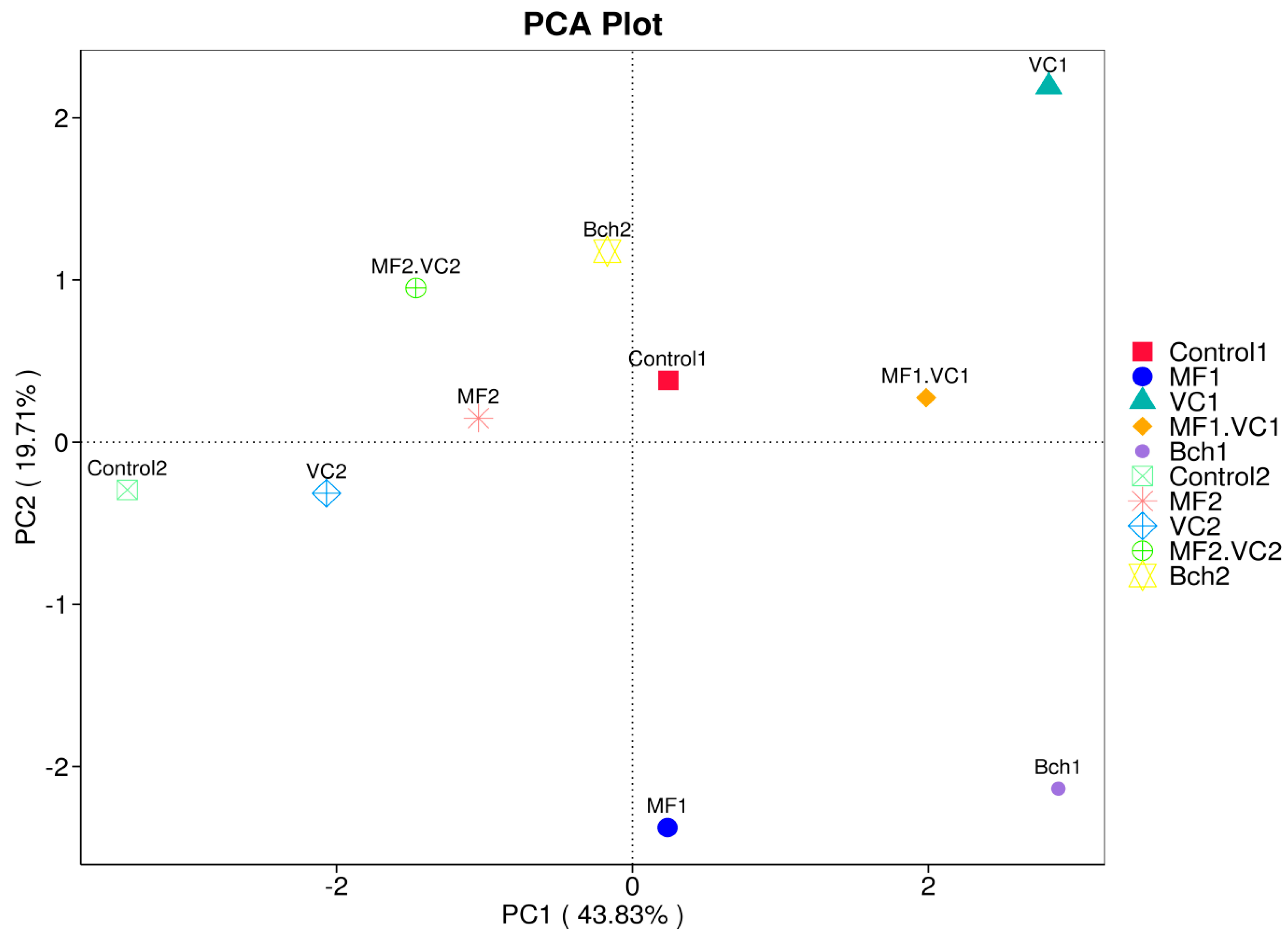
2.4. Principal Component Analysis (PCA) of Disease Pressure and Soil Management
2.4.1. Bacterial Community Shifts Across Treatments
2.4.2. Fungal Community Shifts Across Treatments
2.5. Functional Annotation of Soil Microbial Communities
2.5.1. Main Functions Across Soil Microbial Communities
2.5.2. Heatmaps of Functional Profiles of Bacteria and Fungi
2.5.3. PCA of Bacterial Functional Annotation Profiles
3. Discussion
4. Materials and Methods
4.1. Field Description and Experimental Design
4.2. Phytopathological Assessment
- RI ≥ 80%: High resistance;
- 60% ≤ RI < 80%: Moderate resistance;
- 40% ≤ RI < 60%: Low resistance;
- RI < 40%: High susceptibility.
4.3. Soil DNA Extraction, Amplicon Sequencing, and Bioinformatic Analysis
4.3.1. DNA Extraction
4.3.2. Amplicon Library Preparation
4.3.3. Library Construction and Sequencing
4.4. Functional Annotation of Microbial Communities
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Walters, D.R.; Avrova, A.; Bingham, I.J.; Burnett, F.J.; Fountaine, J.; Havis, N.D.; Hoad, S.P.; Hughes, G.; Looseley, M.; Oxley, S.J.P.; et al. Control of foliar diseases in barley: Towards an integrated approach. Eur. J. Plant Pathol. 2012, 133, 33–73. [Google Scholar] [CrossRef]
- MZH. Field Crop Yields—Harvest 2024 (Preliminary Data). Ministry of Agriculture and Food (BG). 2024. Available online: https://www.mzh.government.bg/media/filer_public/2025/04/01/ra451_publicationcrops2024_preliminarydata.pdf (accessed on 26 September 2025).
- Park, R.F.; Golegaonkar, P.G.; Derevnina, L.; Sandhu, K.S.; Karaoglu, H.; Elmansour, H.M.; Dracatos, P.M.; Singh, D. Leaf rust of cultivated barley: Pathology and control. Annu. Rev. Phytopathol. 2015, 53, 565–589. [Google Scholar] [CrossRef] [PubMed]
- Ogrodowicz, P.; Mikołajczak, K.; Kempa, M.; Mokrzycka, M.; Krajewski, P.; Kuczyńska, A. Genome-wide association study of agronomical and root-related traits in spring barley collection grown under field conditions. Front. Plant Sci. 2023, 14, 1077631. [Google Scholar] [CrossRef] [PubMed]
- Tekauz, A.; McCallum, B.D.; Gilbert, J. Fusarium head blight of barley in western Canada. Can. J. Plant Pathol. 2000, 22, 9–16. [Google Scholar] [CrossRef]
- Mahesha, H.S.; Saini, R.P.; Singh, T.; Singh, A.K.; Srinivasan, R. Potential breeding strategies for developing disease-resistant barley: Progress, challenges, and applications. In Cereal Diseases: Nanobiotechnological Approaches for Diagnosis and Management; Abd-Elsalam, K.A., Mohamed, H.I., Eds.; Springer: Singapore, 2022; pp. 163–181. [Google Scholar] [CrossRef]
- Ren, X.; Wang, C.; Ren, Z.; Wang, J.; Zhang, P.; Zhao, S.; Li, M.; Yuan, M.; Yu, X.; Li, Z.; et al. Genetics of resistance to leaf rust in wheat: An overview in a genome-wide level. Sustainability 2023, 15, 3247. [Google Scholar] [CrossRef]
- Hussain, B.; Mohuiddin, F.A.; Wani, S.H.; Murtaza, I.; Ahmad, S.; Mohammed, R.; Muneeb-ur-Rehman, M.; Rana, A.; Al-Ashkar, I.; Rahman, M.A.; et al. Characterization of barley genotypes and their biochemical responses against leaf rust (Puccinia hordei) disease under cold arid environment. Pol. J. Environ. Stud. 2024, 33, 185–195. [Google Scholar] [CrossRef]
- Langlands-Perry, C.; Pitarch, A.; Lapalu, N.; Cuenin, M.; Bergez, C.; Noly, A.; Amezrou, R.; Gélisse, S.; Barrachina, C.; Parrinello, H.; et al. Quantitative and qualitative plant-pathogen interactions call upon similar pathogenicity genes with a spectrum of effects. Front. Plant Sci. 2023, 14, 1128546. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Schlatter, D.C.; Kinkel, L.L.; Thomashow, L.S.; Weller, D.M.; Paulitz, T.C. Disease suppressive soils: New insights from the soil microbiome. Phytopathology 2017, 107, 1284–1297. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Yang, B.; Balazs, K.R.; Butterfield, B.J.; Laushman, K.M.; Munson, S.M.; Gornish, E.S.; Barberán, A. Does restoration of plant diversity trigger concomitant soil microbiome changes in dryland ecosystems? J. Appl. Ecol. 2022, 59, 560–573. [Google Scholar] [CrossRef]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef] [PubMed]
- Urbanová, M.; Šnajdr, J.; Baldrian, P. Composition of fungal and bacterial communities in forest litter and soil is largely determined by dominant trees. Soil Biol. Biochem. 2015, 84, 53–64. [Google Scholar] [CrossRef]
- Kuypers, M.M.; Marchant, H.K.; Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 2018, 16, 263–276. [Google Scholar] [CrossRef]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Bahram, M.; Zobel, M. How mycorrhizal associations drive plant population and community biology. Science 2020, 367, eaba1223. [Google Scholar] [CrossRef]
- Bziuk, N.; Maccario, L.; Sørensen, S.J.; Schikora, A.; Smalla, K. Barley rhizosphere microbiome transplantation—A strategy to decrease susceptibility of barley grown in soils with low microbial diversity to powdery mildew. Front. Microbiol. 2022, 13, 830905. [Google Scholar] [CrossRef]
- Dreiseitl, A. Specific resistance of barley to powdery mildew, its use and beyond: A concise critical review. Genes 2020, 11, 971. [Google Scholar] [CrossRef]
- Czembor, J.H.; Czembor, E. Barley sources of resistance to the net form of net blotch (Pyrenophora teres f. teres). Biol. Life Sci. Forum 2023, 27, 9. [Google Scholar] [CrossRef]
- Tini, F.; Covarelli, L.; Ricci, G.; Balducci, E.; Orfei, M.; Beccari, G. Management of Pyrenophora teres f. teres, the causal agent of net form net blotch of barley, in a two-year field experiment in central Italy. Pathogens 2022, 11, 291. [Google Scholar] [CrossRef]
- Singh, R.P.; Hodson, D.P.; Huerta-Espino, J.; Bhavani, S.; Njau, P.; Herrera-Foessel, S.A.; Singh, P.K. Progress in breeding for resistance to rusts in wheat. Crop Pasture Sci. 2021, 72, 96–109. [Google Scholar]
- Lehmann, S.; Serrano, M.; L’Haridon, F.; Tjamos, S.E.; Metraux, J.P. Reactive oxygen species and plant resistance to fungal pathogens. Phytochemistry 2015, 112, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wright, S.; Shen, Y.; Du, L. Bioactive natural products from Lysobacter. Nat. Prod. Rep. 2012, 29, 1277–1287. [Google Scholar] [CrossRef]
- Xu, H.J.; Wang, X.H.; Li, H.; Yao, H.Y.; Su, J.Q.; Zhu, Y.G. Biochar impacts soil microbial community composition and nitrogen cycling in an acidic soil planted with rape. Environ. Sci. Technol. 2016, 50, 9437–9446. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Zuo, J.; Dong, H. Changes in soil properties and bacterial community composition with biochar amendment after six years. Agronomy 2020, 10, 746. [Google Scholar] [CrossRef]
- Albers, S.V.; Jarrell, K.F. The archaellum: An update on the unique archaeal motility structure. Trends Microbiol. 2018, 26, 351–362. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, Q.; Ju, M.; Yan, S.; Zhang, Q.; Gu, P. The endophytic fungi diversity, community structure, and ecological function prediction of Sophora alopecuroides in Ningxia, China. Microorganisms 2022, 10, 2099. [Google Scholar] [CrossRef]
- Chehri, K.; Salleh, B.; Yli-Mattila, T.; Reddy, K.R.N.; Abbasi, S. Molecular characterization of pathogenic Fusarium species in cucurbit plants from Kermanshah province, Iran. Saudi J. Biol. Sci. 2011, 18, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Mouchacca, J. Thermophilic fungi: Biodiversity and taxonomic status. Cryptogam. Mycol. 1997, 18, 19–70. [Google Scholar] [CrossRef]
- Nicoletti, R.; Trincone, A. Bioactive compounds produced by strains of Penicillium and Talaromyces of marine origin. Mar. Drugs 2016, 14, 37. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Baquerizo, M.; Reich, P.B.; Khachane, A.N.; Campbell, C.D.; Thomas, N.; Freitag, T.E.; Abu Al-Soud, W.; Sørensen, S.; Bardgett, R.D.; Singh, B.K. It is elemental: Soil nutrient stoichiometry drives bacterial diversity. Environ. Microbiol. 2017, 19, 1176–1188. [Google Scholar] [CrossRef] [PubMed]
- Escudero-Martinez, C.; Coulter, M.; Alegria Terrazas, R.; Foito, A.; Kapadia, R.; Pietrangelo, L.; Maver, M.; Sharma, R.; Aprile, A.; Morris, J.; et al. Identifying plant genes shaping microbiota composition in the barley rhizosphere. Nat. Commun. 2022, 13, 3443. [Google Scholar] [CrossRef]
- Joshi, D.; Kaushik, A.; Kumar, R.; Arya, A.; Santoyo, G.; Singh, V.K.; Kashyap, N.; Solanki, M.K.; Kumari, M.; Bhardwaj, N.; et al. Improving Plant Performance Through Microbiome Manipulation: The Potential Role of Current Bioengineering Approaches. Bacteria 2025, 4, 12. [Google Scholar] [CrossRef]
- Pokharel, S.S.; Ali, Z.; Wang, C.; Jiang, X.; Chen, F. Leguminous Cover Crops Promote Microbial Community Diversity in the Rhizosphere Soil of Tea Plants: Insights from 16S rRNA Microbiome Analysis. Agronomy 2025, 15, 2217. [Google Scholar] [CrossRef]
- Weerasinghe, V.; Bakker, M.G.; Tucker, J.R.; Fernando, W.D.; Sura, S.; Badea, A.; Wijekoon, C. Comparison of Bacterial Endophytes in Barley Grains Infected and Non-Infected with Fusarium Head Blight Using Metabarcoding. Plant Pathol. 2025, 74, 2241–2255. [Google Scholar] [CrossRef]
- Shilev, S.; Mitkov, A.; Popova, V.; Neykova, I.; Minev, N.; Szulc, W.; Yordanov, Y.; Yanev, M. Fertilization type differentially affects barley grain yield and nutrient content, soil and microbial properties. Microorganisms 2024, 12, 1447. [Google Scholar] [CrossRef]
- Savary, S.; Zadoks, J.C. Analysis of crop loss in the multiple pathosystem groundnut–rust–late leaf spot. II. Study of the interactions between diseases and crop intensification in factorial experiments. Crop Prot. 1992, 11, 110–120. [Google Scholar] [CrossRef]
- James, W.C. An illustrated series of assessment keys for plant diseases: Their preparation and usage. Can. Plant Dis. Surv. 1971, 51, 39–65. Available online: https://phytopath.ca/wp-content/uploads/cpds-archive/vol51/CPDS_Vol_51_No_2_%2839-65%291971.pdf (accessed on 20 August 2025).
- Kirse, A.; Bourlat, S.J.; Langen, K.; Fonseca, V.G. Unearthing the potential of soil eDNA metabarcoding—Towards best practice advice for invertebrate biodiversity assessment. Front. Ecol. Evol. 2021, 9, 630560. [Google Scholar] [CrossRef]
- Petkova, M.; Shilev, S.; Popova, V.; Neykova, I.; Minev, N. Intercropping of oats with vetch conducts to improve soil bacteriome diversity and structure. Microorganisms 2025, 13, 977. [Google Scholar] [CrossRef]
- Adkins, N.L.; Hall, J.A.; Georgel, P.T. The use of quantitative agarose gel electrophoresis for rapid analysis of the integrity of protein–DNA complexes. J. Biochem. Biophys. Methods 2007, 70, 721–726. [Google Scholar] [CrossRef]
- Maretto, L.; Deb, S.; Ravi, S.; Chiodi, C.; Manfredi, P.; Squartini, A.; Concheri, G.; Renella, G.; Stevanato, P. Microbial diversity of reconstituted, degraded, and agricultural soils assessed by 16S rDNA multi-amplicon sequencing. Front. Environ. Sci. 2022, 9, 807889. [Google Scholar] [CrossRef]
- Abellan-Schneyder, I.; Matchado, M.S.; Reitmeier, S.; Sommer, A.; Sewald, Z.; Baumbach, J.; List, M.; Neuhaus, K. Primer, pipelines, parameters: Issues in 16S rRNA gene sequencing. mSphere 2021, 6, e01202-20. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [PubMed]
- Sansupa, C.; Wahdan, S.F.M.; Hossen, S.; Disayathanoowat, T.; Wubet, T.; Purahong, W. Can we use functional annotation of prokaryotic taxa (FAPROTAX) to assign the ecological functions of soil bacteria? Appl. Sci. 2021, 11, 688. [Google Scholar] [CrossRef]
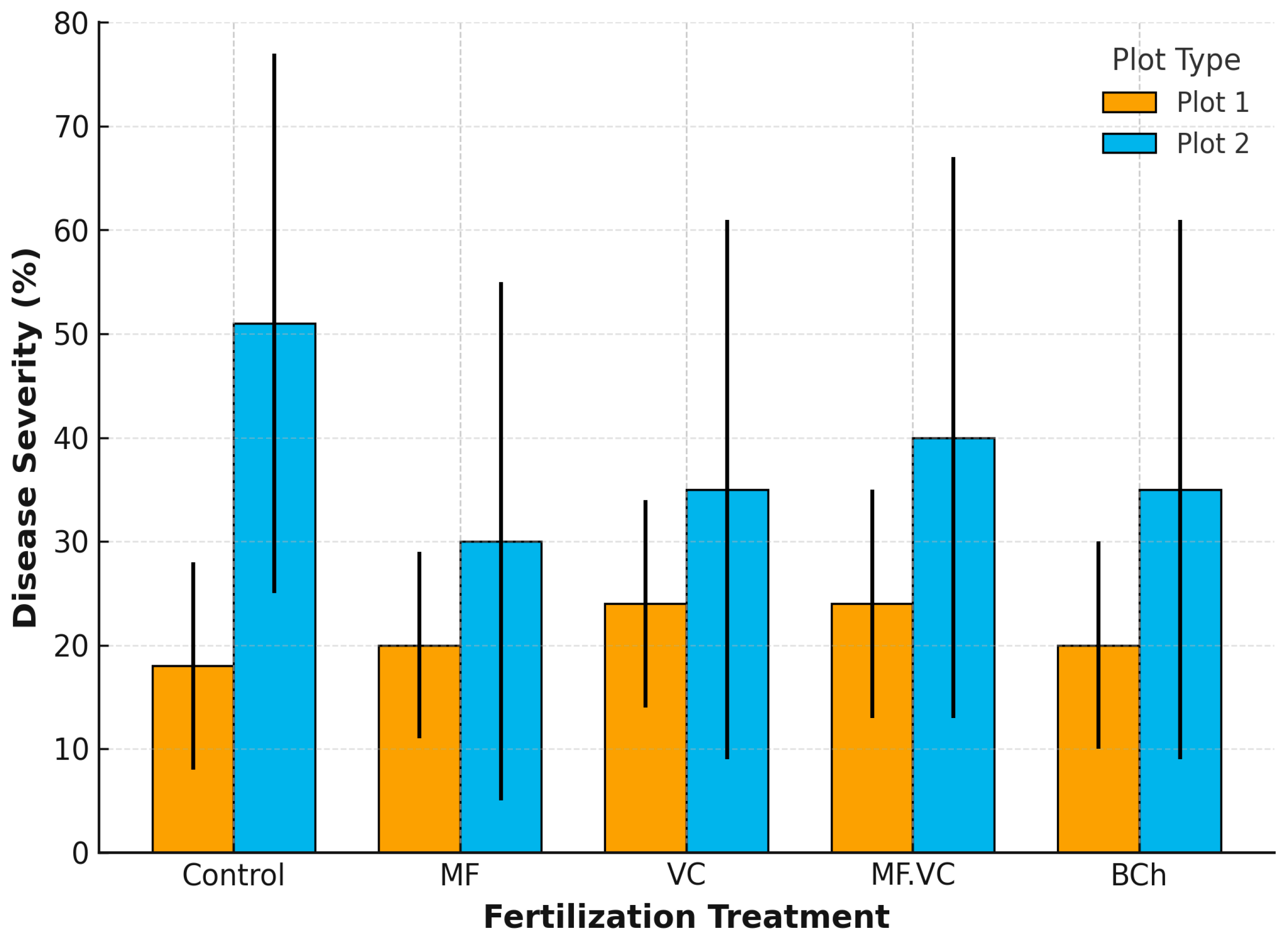


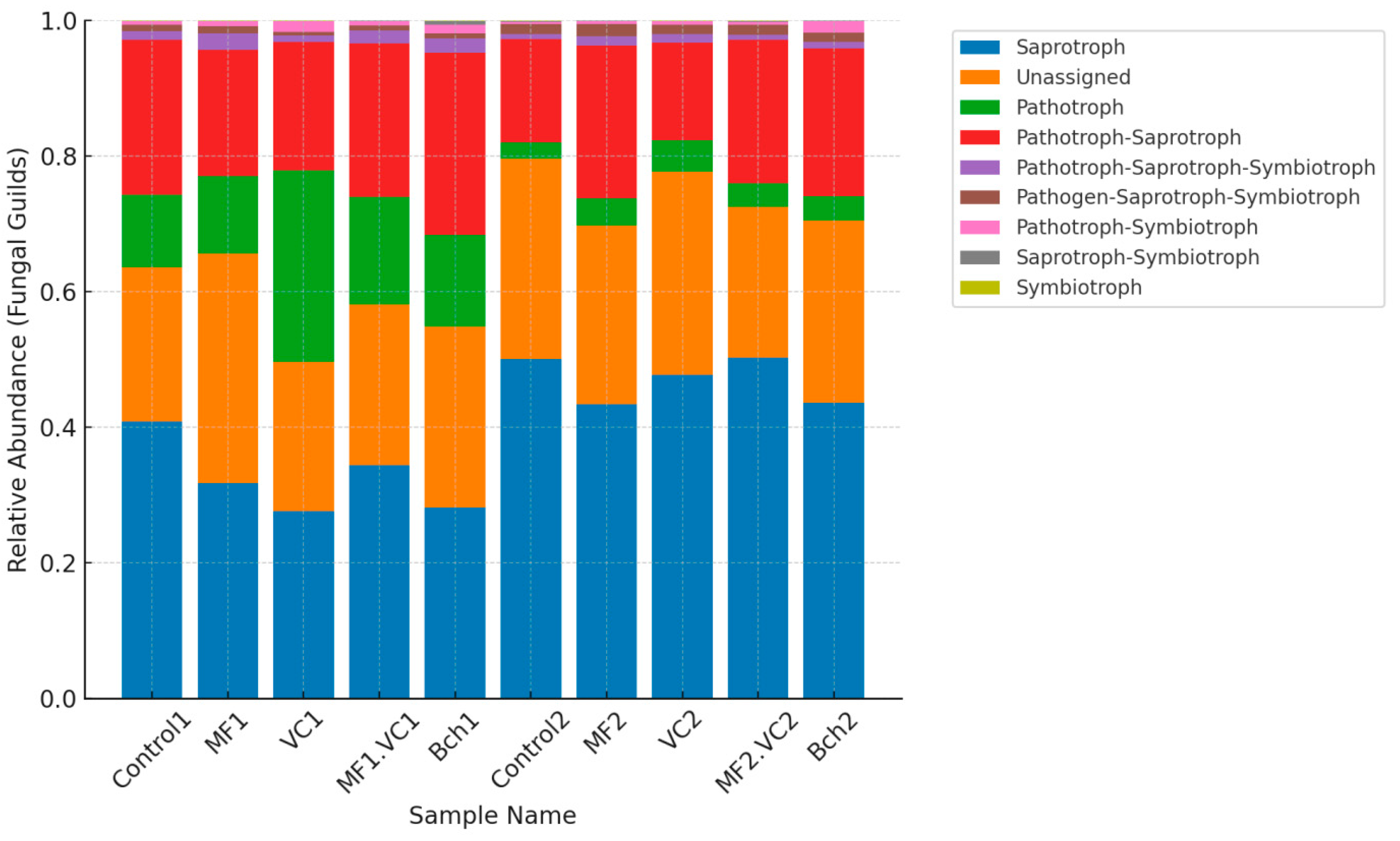
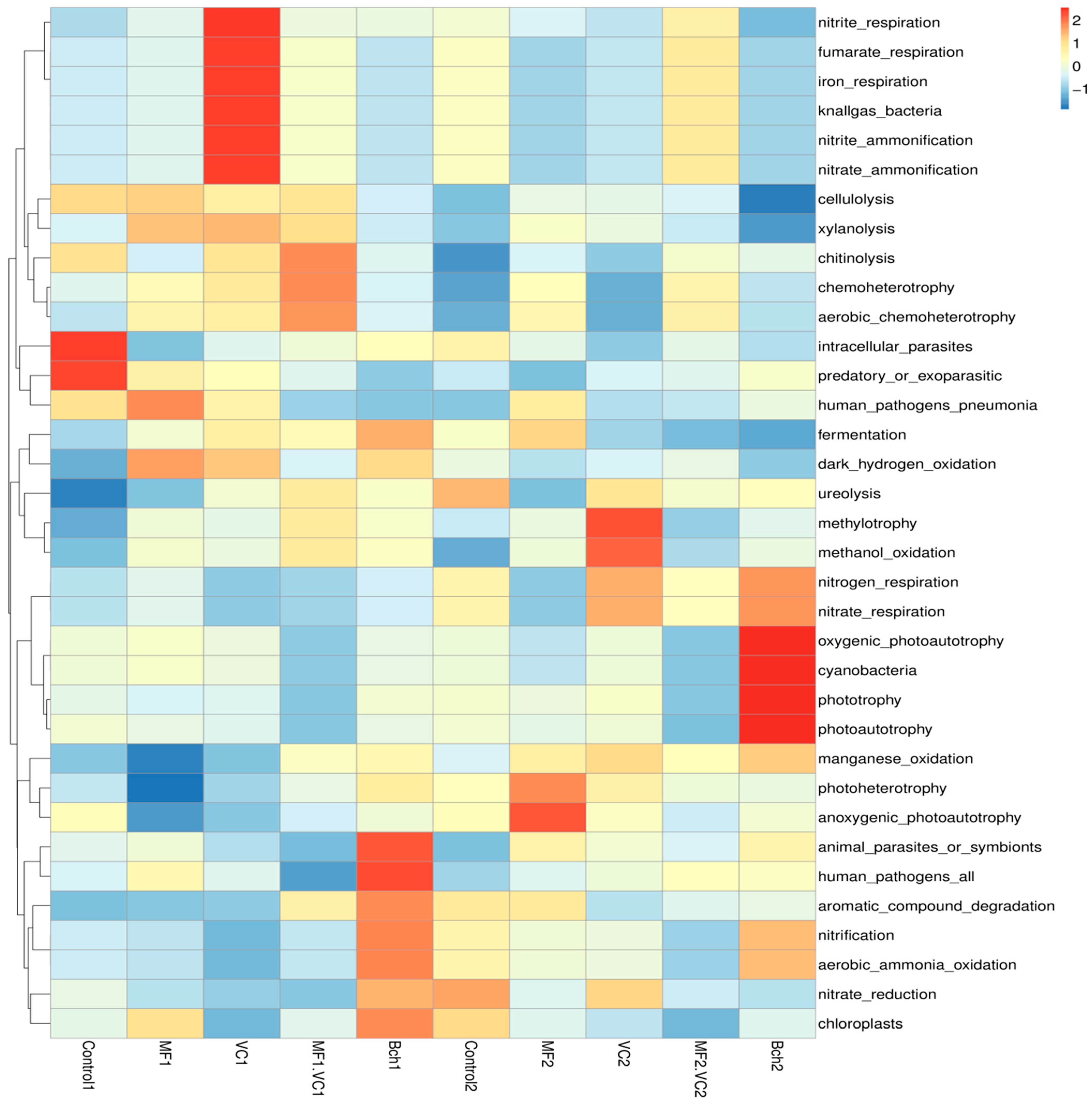
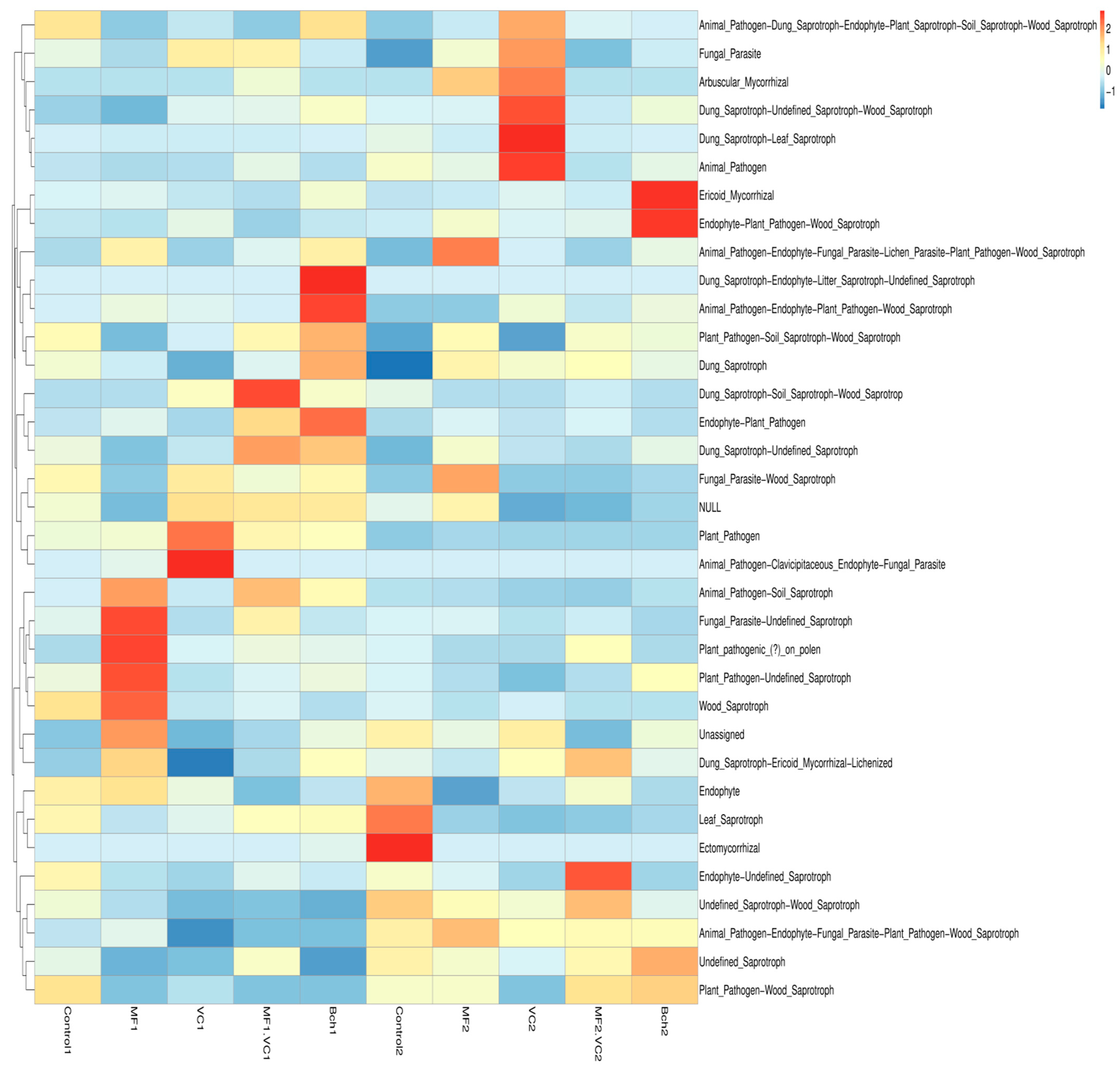

| Plot | Treatment | Disease | Severity (%) | RI (%) | Resistance Category |
|---|---|---|---|---|---|
| Plot 1.1 | Control | Powdery mildew | 10 | 90 | High resistance |
| Plot 1.1 | Control | Net blotch | 33 | 67 | Moderate resistance |
| Plot 1.1 | Control | Brown rust | 13 | 87 | High resistance |
| Plot 1.2 | MF | Powdery mildew | 12 | 88 | High resistance |
| Plot 1.2 | MF | Net blotch | 20 | 80 | High resistance |
| Plot 1.2 | MF | Brown rust | 31 | 69 | Moderate resistance |
| Plot 1.3 | VC | Powdery mildew | 35 | 65 | Moderate resistance |
| Plot 1.3 | VC | Net blotch | 25 | 75 | Moderate resistance |
| Plot 1.3 | VC | Brown rust | 12 | 88 | High resistance |
| Plot 1.4 | MF.VC | Powdery mildew | 15 | 85 | High resistance |
| Plot 1.4 | MF.VC | Net blotch | 38 | 62 | Moderate resistance |
| Plot 1.4 | MF.VC | Brown rust | 20 | 80 | High resistance |
| Plot 1.5 | BCh | Powdery mildew | 18 | 82 | High resistance |
| Plot 1.5 | BCh | Net blotch | 11 | 89 | High resistance |
| Plot 1.5 | BCh | Brown rust | 32 | 68 | Moderate resistance |
| Plot 2.1 | Control | Powdery mildew | 55 | 45 | Low resistance |
| Plot 2.1 | Control | Net blotch | 20 | 80 | High resistance |
| Plot 2.1 | Control | Brown rust | 80 | 20 | High susceptibility |
| Plot 2.2 | MF | Powdery mildew | 65 | 35 | High susceptibility |
| Plot 2.2 | MF | Net blotch | 20 | 80 | High resistance |
| Plot 2.2 | MF | Brown rust | 5 | 95 | High resistance |
| Plot 2.3 | VC | Powdery mildew | 10 | 90 | High resistance |
| Plot 2.3 | VC | Net blotch | 70 | 30 | High susceptibility |
| Plot 2.3 | VC | Brown rust | 25 | 75 | Moderate resistance |
| Plot 2.4 | MF.VC | Powdery mildew | 30 | 70 | Moderate resistance |
| Plot 2.4 | MF.VC | Net blotch | 15 | 85 | High resistance |
| Plot 2.4 | MF.VC | Brown rust | 76 | 24 | High susceptibility |
| Plot 2.5 | BCh | Powdery mildew | 72 | 28 | High susceptibility |
| Plot 2.5 | BCh | Net blotch | 25 | 75 | Moderate resistance |
| Plot 2.5 | BCh | Brown rust | 10 | 90 | High resistance |
| Plot Type | F-Statistic | p-Value | Significance (p < 0.05) |
|---|---|---|---|
| Plot 1 (After green manure) | 0.139 | 0.964 | No |
| Plot 2 (After Fallow) | 0.205 | 0.93 | No |
| Taxonomy | Lysobacter dokdonensis | Arenimicrobium luteum | Gemmatimonadetes bacterium LX87 | Gemmatimonadetes bacterium LP81 | Jahnella thaxteri | Agromyces ramosus | Steroidobacter sp. | Nitrospira japonica | Aquimonas sp. | |
|---|---|---|---|---|---|---|---|---|---|---|
| Control1 | 0.9 | 0.41 | 0.79 | 0.32 | 0.15 | 0.15 | 0.22 | 0.2 | 0.19 | 0.17 |
| MF1 | 0.4 | 0.82 | 0.54 | 0.29 | 0.06 | 0.1 | 0.18 | 0.27 | 0.16 | 0.3 |
| VC1 | 1.01 | 0.55 | 0.53 | 0.45 | 0.01 | 0.12 | 0.29 | 0.16 | 0.17 | 0.28 |
| MF1.VC1 | 1.32 | 0.11 | 0.52 | 0.32 | 0.55 | 0.38 | 0.22 | 0.08 | 0.15 | 0.19 |
| Bch1 | 0.85 | 0.23 | 0.4 | 0.34 | 0.03 | 0.5 | 0.28 | 0.12 | 0.22 | 0.08 |
| Control2 | 0.32 | 0.19 | 0.21 | 0.32 | 0 | 0.26 | 0.17 | 0.33 | 0.32 | 0.11 |
| MF2 | 0.56 | 0.26 | 0.42 | 0.37 | 0.02 | 0.3 | 0.23 | 0.28 | 0.29 | 0.17 |
| VC2 | 0.54 | 0.17 | 0.45 | 0.4 | 0.04 | 0.22 | 0.29 | 0.2 | 0.18 | 0.08 |
| MF2.VC2 | 0.79 | 0.14 | 0.66 | 0.56 | 0.05 | 0.24 | 0.35 | 0.13 | 0.21 | 0.17 |
| Bch2 | 0.91 | 0.11 | 0.72 | 0.47 | 0 | 0.27 | 0.39 | 0.29 | 0.17 | 0.09 |
| Taxonomy | Acrophialophora jodhpurensis | Fusarium equiseti | Humicola nigrescens | Chrysosporium lobatum | Stachybotrys chartarum | Humicola fuscoatra | Fungi sp. | Penicillium polonicum |
|---|---|---|---|---|---|---|---|---|
| Control1 | 5.73 | 10.41 | 0.74 | 11.04 | 3.72 | 0.9 | 1.8 | 0.02 |
| MF1 | 6.88 | 5.64 | 1.48 | 5.42 | 2.02 | 0.8 | 3.93 | 0.01 |
| VC1 | 24.52 | 8.47 | 0.48 | 4.28 | 3.93 | 0.38 | 1.85 | 0.03 |
| MF1.VC1 | 7.15 | 10.26 | 1.25 | 3.81 | 5.2 | 0.45 | 2.41 | 0 |
| Bch1 | 7.31 | 14.63 | 0.34 | 3.46 | 3.16 | 0.31 | 3.15 | 0.22 |
| Control2 | 0.24 | 4.59 | 11.08 | 5 | 4.85 | 3.97 | 2.32 | 0.15 |
| MF2 | 0.23 | 8.39 | 10.1 | 1.92 | 4.77 | 2.92 | 2.14 | 0.13 |
| VC2 | 0.19 | 5.62 | 3.02 | 5.3 | 6 | 4.4 | 2.07 | 0.07 |
| MF2.VC2 | 0.05 | 8.43 | 13.95 | 4.96 | 6.3 | 2.15 | 1.42 | 2.72 |
| Bch2 | 0.62 | 8.69 | 2.16 | 3.04 | 7.22 | 5.08 | 2.2 | 3.29 |
| Plot | Treatment | Description of Treatment |
|---|---|---|
| 1 | Control1 | No fertilisation (reference treatment) |
| 1 | MF1 | Mineral fertiliser NPK (15:15:15) at 0.1 t ha−1 + 50 kg N ha−1 (NH4NO3 at tillering) |
| 1 | VC1 | Vermicompost 12 t ha−1 |
| 1 | MF.VC1 | Combined mineral fertiliser 0.05 t ha−1 + vermicompost 6 t ha−1 |
| 1 | BCh1 | Biochar 10 t ha−1 |
| 2 | Control2 | No fertilisation (reference treatment) |
| 2 | MF2 | Mineral fertiliser NPK (15:15:15) at 0.1 t ha−1 + 50 kg N ha−1 (NH4NO3 at tillering) |
| 2 | VC2 | Vermicompost 12 t ha−1 |
| 2 | MF.VC2 | Combined mineral fertiliser 0.05 t ha−1 + vermicompost 6 t ha−1 |
| 2 | BCh2 | Biochar 10 t ha−1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petkova, M.; Chavdarov, P.; Shilev, S. Linking Soil Microbial Functional Profiles to Fungal Disease Resistance in Winter Barley Under Different Fertilisation Regimes. Plants 2025, 14, 3199. https://doi.org/10.3390/plants14203199
Petkova M, Chavdarov P, Shilev S. Linking Soil Microbial Functional Profiles to Fungal Disease Resistance in Winter Barley Under Different Fertilisation Regimes. Plants. 2025; 14(20):3199. https://doi.org/10.3390/plants14203199
Chicago/Turabian StylePetkova, Mariana, Petar Chavdarov, and Stefan Shilev. 2025. "Linking Soil Microbial Functional Profiles to Fungal Disease Resistance in Winter Barley Under Different Fertilisation Regimes" Plants 14, no. 20: 3199. https://doi.org/10.3390/plants14203199
APA StylePetkova, M., Chavdarov, P., & Shilev, S. (2025). Linking Soil Microbial Functional Profiles to Fungal Disease Resistance in Winter Barley Under Different Fertilisation Regimes. Plants, 14(20), 3199. https://doi.org/10.3390/plants14203199







