Genomic-Wide Association Markers and Candidate Genes for the High-Protein Trait in Storage Roots of Cassava (Manihot esculenta)
Abstract
1. Introduction
2. Results
2.1. Statistical Analysis of Phenotypic Traits and Screening of High-Protein Varieties
2.2. Distribution and Detection of Genomic Variation Sites
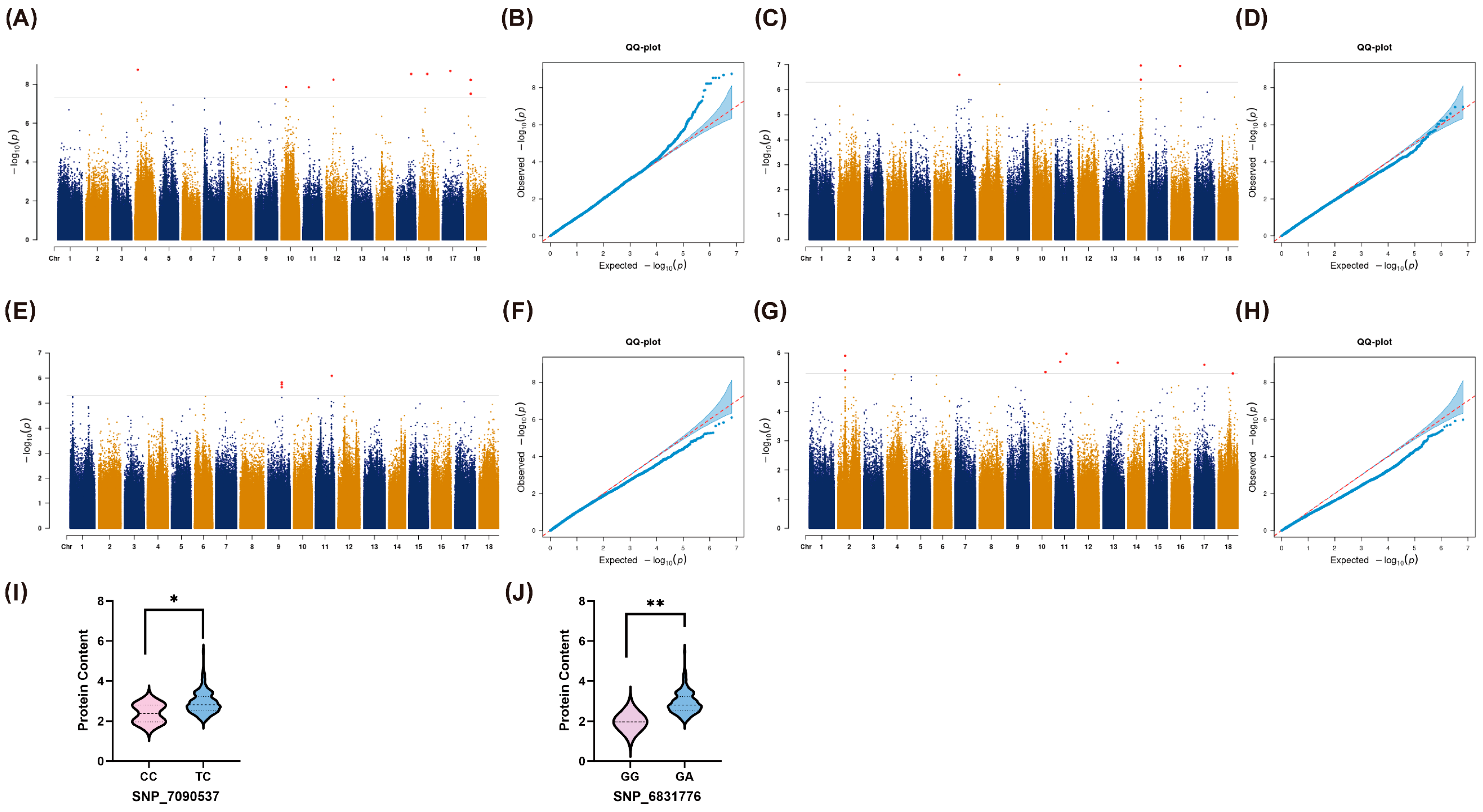
2.3. Genome-Wide Association Study of Storage Root Protein Content
2.4. Identification of Candidate Genes
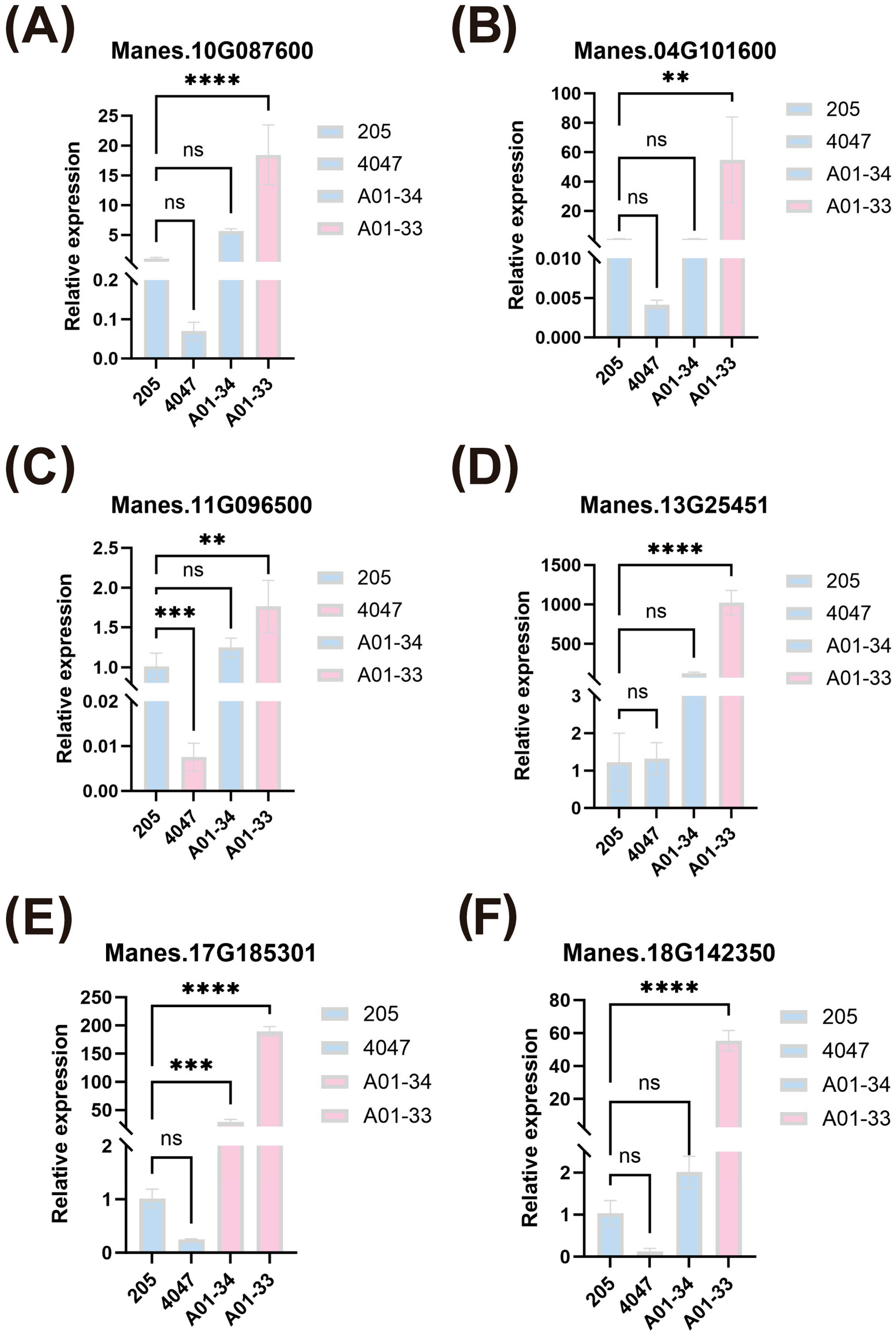
2.5. Quantitative Analysis of Candidate Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Field Trials
4.2. Phenotypic Trait Measurement
4.3. Genotyping and Polymorphism Analysis
4.4. Genome-Wide Association Study
4.5. Candidate Gene Identification and Functional Annotation
4.6. Candidate Gene Expression Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Otekunrin, O.A. Cassava (Manihot esculenta Crantz): A global scientific footprint—Production, trade, and bibliometric insights. Discov. Agric. 2024, 2, 94. [Google Scholar] [CrossRef]
- Burns, A.; Gleadow, R.; Cliff, J.; Zacarias, A.; Cavagnaro, T. Cassava: The Drought, War and Famine Crop in a Changing World. Sustainability 2010, 2, 3572–3607. [Google Scholar] [CrossRef]
- Fu, H.; Qu, Y.; Pan, Y. Efficiency of Cassava Production in China: Empirical Analysis of Field Surveys from Six Provinces. Appl. Sci. 2018, 8, 1356. [Google Scholar] [CrossRef]
- Chamorro, A.F.; Palencia, M.; Lerma, T.A. Physicochemical Characterization and Properties of Cassava Starch: A Review. Polymers 2025, 17, 1663. [Google Scholar] [CrossRef]
- Breuninger, W.F.; Piyachomkwan, K.; Sriroth, K. Tapioca/cassava starch: Production and use. In Starch; Elsevier: Amsterdam, The Netherlands, 2009; pp. 541–568. [Google Scholar]
- Latif, S.; Müller, J. Potential of cassava leaves in human nutrition: A review. Trends Food Sci. Technol. 2015, 44, 147–158. [Google Scholar] [CrossRef]
- Lisi, A.G.; Weatherall, J.O. A Geometric Theory of Everything. Sci. Am. 2010, 303, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chen, X.; Lu, C.; Ye, J.; Zou, M.; Lu, K.; Feng, S.; Pei, J.; Liu, C.; Zhou, X.; et al. Genome-Wide Association Studies of 11 Agronomic Traits in Cassava (Manihot esculenta Crantz). Front. Plant Sci. 2018, 9, 503. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Yang, S.; Zeng, Y.; Lu, W.; Ruan, M.; Liu, Y.; Zhai, L.; Li, X.; Wang, Y.; Xia, Z.; et al. Genome-wide association study identifies candidate genes for latex yield in rubber tree. Int. J. Mol. Sci. 2020, 21, 5799. [Google Scholar] [CrossRef]
- Huang, W.; Hong, Z. Genetic Mechanism of Cassava Disease Resistance: From Traditional Breeding to CRISPR/Cas Application. Genom. Appl. Biol. 2024, 15, 47–53. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, L.; Li, Y.; Wang, Y.; Wang, S.; Zhao, Y.; Zhang, D.; Li, H.; Li, Y.; Yang, X.; et al. Genome-wide association analysis of protein and starch content in maize. Crop J. 2016, 4, 343–353. [Google Scholar] [CrossRef]
- Li, Y.; Guo, Y.; Li, C.; Shi, Y.; Gong, J.; Zhang, M.; Zhang, Y.; Wang, Z.; Wang, Y.; Wang, X.; et al. Genetic dissection of seed protein architecture in soybean by genome-wide association study. Front. Plant Sci. 2020, 11, 583. [Google Scholar] [CrossRef]
- Chen, P.; Lou, G.; Wang, Y.; Chen, J.; Chen, W.; Fan, Z.; Liu, Q.; Sun, B.; Mao, X.; Yu, H.; et al. The genetic basis of grain protein content in rice by genome-wide association analysis. Mol. Breed. 2022, 43, 1. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Ji, C.; Liang, Z.; Ye, J.; Ou, W.; Ding, Z.; Zhou, G.; Tie, W.; Yan, Y.; Yang, J.; et al. Resequencing of 388 cassava accessions identifies valuable loci and selection for variation in heterozygosity. Genome Biol. 2021, 22, 316. [Google Scholar] [CrossRef]
- Chen, L.; Chen, R.; Atwa, E.M.; Mabrouk, M.; Jiang, H.; Mou, X.; Ma, X. Nutritional Quality Assessment of Miscellaneous Cassava Tubers Using Principal Component Analysis and Cluster Analysis. Foods 2024, 13, 1861. [Google Scholar] [CrossRef] [PubMed]
- Izawa, I.; Nishizawa, M.; Ohtakara, K.; Ohtsuka, K.; Inada, H.; Inagaki, M. Identification of Mrj, a DnaJ/Hsp40 Family Protein, as a Keratin 8/18 Filament Regulatory Protein. J. Biol. Chem. 2000, 275, 34521–34527. [Google Scholar] [CrossRef]
- Hankoua, B.B.; Diao, M.; Ligaba-Osena, A.; Garcia, R.A.; Harun, S.; Ahlawat, Y.K. Constitutive overexpression of Qui-Quine Starch gene simultaneously improves starch and protein content in bioengineered cassava (Manihot esculenta Crantz). Front. Plant Sci. 2024, 15, 1442324. [Google Scholar] [CrossRef]
- Sojikul, P.; Kongsawadworakul, P.; Viboonjun, U.; Thaiprasit, J.; Intawong, B.; Narangajavana, J.; Svasti, M.R.J. AFLP-based transcript profiling for cassava genome-wide expression analysis in the onset of storage root formation. Physiol. Plant. 2010, 140, 189–298. [Google Scholar] [CrossRef]
- Zhang, K.; Liu, S.; Li, W.; Liu, S.; Li, X.; Fang, Y.; Zhang, J.; Wang, Y.; Xu, S.; Zhang, J.; et al. Identification of QTNs Controlling Seed Protein Content in Soybean Using Multi-Locus Genome-Wide Association Studies. Front. Plant Sci. 2018, 9, 1690. [Google Scholar] [CrossRef]
- Yang, X.; Shaw, R.K.; Li, L.; Jiang, F.; Sun, J.; Fan, X. Discovery of candidate genes enhancing kernel protein content in tropical maize introgression lines. BMC Plant Biol. 2024, 24, 1110. [Google Scholar] [CrossRef]
- Yan, Y.; Liang, C.; Liu, X.; Tan, Y.; Lu, Y.; Zhang, Y.; Luo, H.; He, C.; Cao, J.; Tang, C.; et al. Genome-wide association study identifies candidate genes responsible for inorganic phosphorus and sucrose content in rubber tree latex. Trop. Plants 2023, 2, 24. [Google Scholar] [CrossRef]
- Rabbi, I.Y.; Kayondo, S.I.; Bauchet, G.; Yusuf, M.; Aghogho, C.I.; Ogunpaimo, K.; Uwugiaren, R.; Smith, I.A.; Peteti, P.; Agbona, A.; et al. Genome-wide association analysis reveals new insights into the genetic architecture of defensive, agro-morphological and quality-related traits in cassava. Plant Mol. Biol. 2022, 109, 195–213. [Google Scholar] [CrossRef]
- Lu, Y.; Guo, Z.; Ke, B.; Zheng, H.; Zeng, Z.; Cai, Z.; Zeng, H.; Liao, J.; Chen, M. Genome-Wide Association Study and Transcriptome Analysis Provide Candidate Genes for Agronomic Traits of Agaricus bisporus. Horticulturae 2024, 10, 691. [Google Scholar] [CrossRef]
- Li, C.; Jia, Y.; Zhou, R.; Liu, L.; Cao, M.; Zhou, Y.; Wang, Z.; Di, H. GWAS and RNA-seq analysis uncover candidate genes associated with alkaline stress tolerance in maize (Zea mays L.) seedlings. Front. Plant Sci. 2022, 13, 963874. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Chen, B.; Cheng, Y.; Su, Y.; Song, M.; Guo, R.; Wang, M.; Deng, K.; Lan, T.; Bao, S.; et al. Integration of GWAS and RNA-Seq Analysis to Identify SNPs and Candidate Genes Associated with Alkali Stress Tolerance at the Germination Stage in Mung Bean. Genes 2023, 14, 1294. [Google Scholar] [CrossRef]
- Mæhre, H.K.; Dalheim, L.; Edvinsen, G.K.; Elvevoll, E.O.; Jensen, I.-J. Protein Determination—Method Matters. Foods 2018, 7, 5. [Google Scholar] [CrossRef]
- Langyan, S.; Bhardwaj, R.; Radhamani, J.; Yadav, R.; Gautam, R.K.; Kalia, S.; Kumar, A. A Quick Analysis Method for Protein Quantification in Oilseed Crops: A Comparison with Standard Protocol. Front. Nutr. 2022, 9, 892695. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Graffelman, J.; Jain, D.; Weir, B. A genome-wide study of Hardy-Weinberg equilibrium with next generation sequence data. Hum. Genet. 2017, 136, 727–741. [Google Scholar] [CrossRef]
- Mas-Gomez, J.; Cantin, C.M.; Moreno, M.A.; Martinez-Garcia, P.J. Genetic Diversity and Genome-Wide Association Study of Morphological and Quality Traits in Peach Using Two Spanish Peach Germplasm Collections. Front. Plant Sci. 2022, 13, 854770. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Xu, G.; Cheng, Y.; Wang, X.; Dai, Z.; Kang, Z.; Ye, Z.; Pan, Y.; Zhou, L.; Xie, D.; Sun, J. Identification of Single Nucleotide Polymorphic Loci and Candidate Genes for Seed Germination Percentage in Okra under Salt and No-Salt Stresses by Genome-Wide Association Study. Plants 2024, 13, 588. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2022, 51, D587–D592. [Google Scholar] [CrossRef] [PubMed]
- Jozefczuk, J.; Adjaye, J. Chapter Six—Quantitative Real-Time PCR-Based Analysis of Gene Expression. In Methods in Enzymology; Jameson, D., Verma, M., Westerhoff, H.V., Eds.; Academic Press: New Work, NY, USA, 2011; Volume 500, pp. 99–109. [Google Scholar]
- Zhao, J.; Yang, J.; Wang, X.; Xiong, Y.; Xiong, Y.; Dong, Z.; Lei, X.; Yan, L.; Ma, X. Selection and Validation of Reference Genes for qRT-PCR Gene Expression Analysis in Kengyilia melanthera. Genes 2022, 13, 1445. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Yang, M.; Fang, Y.; Luo, Y.; Gao, S.; Xiao, X.; An, Z.; Zhou, B.; Zhang, B.; Tan, X. The rubber tree genome reveals new insights into rubber production and species adaptation. Nat. Plants 2016, 2, 16073. [Google Scholar] [CrossRef]



| Trait | Year | Range | Mean | SD | CV |
|---|---|---|---|---|---|
| Protein Content | 2021 | 2.25–6.07% | 3.40% | 0.39 | 0.12 |
| 2022 | 1.01–5.76% | 2.37% | 0.87 | 0.37 | |
| 2023 | 2.25–7.12% | 3.42% | 0.47 | 0.14 |
| Individuals | Protein Content | Individuals | Protein Content |
|---|---|---|---|
| A01-007 | 6.07% | A01-422 | 4.03% |
| A01-008 | 4.96% | A01-430 | 4.57% |
| A01-033 | 5.07% | A01-453 | 4.15% |
| A01-034 | 5.03% | A01-455 | 5.47% |
| A01-340 | 4.20% | A01-460 | 4.66% |
| A01-387 | 5.05% | A01-505 | 4.39% |
| A01-415 | 7.12% | A01-531 | 4.20% |
| A01-416 | 4.00% | A01-562 | 4.16% |
| A01-418 | 4.05% | A01-609 | 4.51% |
| A01-420 | 4.00% | A01-620 | 5.76% |
| A01-421 | 4.03% |
| Type | Count | Ratio |
|---|---|---|
| Downstream | 2,666,688 | 17.83% |
| Exon | 410,429 | 2.75% |
| Intergenic | 7,858,808 | 52.56% |
| Intron | 1,074,878 | 7.19% |
| Splice site acceptor | 2331 | 0.02% |
| Splice site donor | 2427 | 0.02% |
| Splice site region | 27,842 | 0.12% |
| Upstream | 2,780,654 | 18.60% |
| Missense variant | 242,359 | 1.62% |
| Genome total length | 645,399,631 | |
| Genome effective length | 645,399,631 | |
| Total SNPs | 9,492,337 | |
| Chromosome | Year | SNP | Position | p-Value | R2 | REF | ALT |
|---|---|---|---|---|---|---|---|
| 02 | 2023 | SNP_792192 | 13148897 | 1.23 × 10−6 | 0.15 | C | T |
| 04 | 2021 | SNP_1641946 | 4696409 | 1.80 × 10−9 | 0.16 | G | A |
| 07 | 2022 | SNP_2959024 | 7028219 | 2.54 × 10−7 | 0.19 | A | C |
| 09 | 2022 | SNP_4104234 | 24914148 | 1.77 × 10−6 | 0.15 | T | C |
| SNP_4104235 | 24914151 | 1.49 × 10−6 | 0.16 | C | G | ||
| 2023 | SNP_4104236 | 24914164 | 2.28 × 10−6 | 0.15 | A | G | |
| 10 | 2023 | SNP_4622659 | 22538541 | 4.41 × 10−6 | 0.12 | A | C |
| 11 | 2021 | SNP_4878878 | 9927394 | 1.44 × 10−8 | 0.18 | G | A |
| 2023 | SNP_5140201 | 28175312 | 8.09 × 10−7 | 0.16 | A | G | |
| 2021–2023 Average | SNP_4878249 | 9889485 | 1.97 × 10−6 | 0.13 | T | C | |
| SNP_5017565 | 20325110 | 1.05 × 10−6 | 0.14 | G | A | ||
| 12 | 2021 | SNP_5378426 | 13017554 | 5.96 × 10−9 | 0.17 | G | A |
| 13 | 2021–2023 Average | SNP_5090879 | 25501246 | 2.13 × 10−6 | 0.14 | C | A |
| 14 | 2022 | SNP_6427875 | 22931608 | 3.99 × 10−7 | 0.17 | T | G |
| SNP_6428019 | 22940512 | 1.08 × 10−7 | 0.20 | T | C | ||
| 15 | 2021 | SNP_6831776 | 25558507 | 2.97 × 10−9 | 0.35 | G | A |
| 16 | 2021 | SNP_7090537 | 13958220 | 2.97 × 10−9 | 0.35 | T | C |
| 2022 | SNP_7117572 | 16338879 | 1.12 × 10−7 | 0.17 | T | C | |
| 17 | 2021 | SNP_7485551 | 13939320 | 2.07 × 10−9 | 0.19 | C | T |
| 2021–2023 Average | SNP_7536143 | 18444849 | 2.53 × 10−6 | 0.15 | C | T | |
| 18 | 2021–2023 Average | SNP_8079404 | 25680373 | 4.94 × 10−6 | 0.12 | G | A |
| Chr | Pos | Gene | Description |
|---|---|---|---|
| 02 | 13148897 | Manes.02G165100 | Belongs to the peroxidase family |
| 10 | 22538541 | Manes.10G087600 | TIR domain |
| 11 | 9889485 | Manes.04G101600 | Chaperone protein dnaJ 8 |
| 11 | 20325110 | Manes.11G096500 | N-acetyltransferase-like |
| 13 | 25501246 | Manes.13G254510 | - |
| 17 | 18444849 | Manes.17G185301 | - |
| 18 | 25680373 | Manes.18G142350 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.; Liu, Q.; Xie, X.; Zhang, J.; Xiao, J.; Wang, W. Genomic-Wide Association Markers and Candidate Genes for the High-Protein Trait in Storage Roots of Cassava (Manihot esculenta). Plants 2025, 14, 3162. https://doi.org/10.3390/plants14203162
Wang D, Liu Q, Xie X, Zhang J, Xiao J, Wang W. Genomic-Wide Association Markers and Candidate Genes for the High-Protein Trait in Storage Roots of Cassava (Manihot esculenta). Plants. 2025; 14(20):3162. https://doi.org/10.3390/plants14203162
Chicago/Turabian StyleWang, Dantong, Qi Liu, Xianhai Xie, Junyu Zhang, Jin Xiao, and Wenquan Wang. 2025. "Genomic-Wide Association Markers and Candidate Genes for the High-Protein Trait in Storage Roots of Cassava (Manihot esculenta)" Plants 14, no. 20: 3162. https://doi.org/10.3390/plants14203162
APA StyleWang, D., Liu, Q., Xie, X., Zhang, J., Xiao, J., & Wang, W. (2025). Genomic-Wide Association Markers and Candidate Genes for the High-Protein Trait in Storage Roots of Cassava (Manihot esculenta). Plants, 14(20), 3162. https://doi.org/10.3390/plants14203162





