Genome-Wide Identification and Expression Analysis of the SRS Gene Family in Hylocereus undatus
Abstract
1. Background
2. Methods
2.1. Identification and Physicochemical Properties of HuSRS Gene Family Members
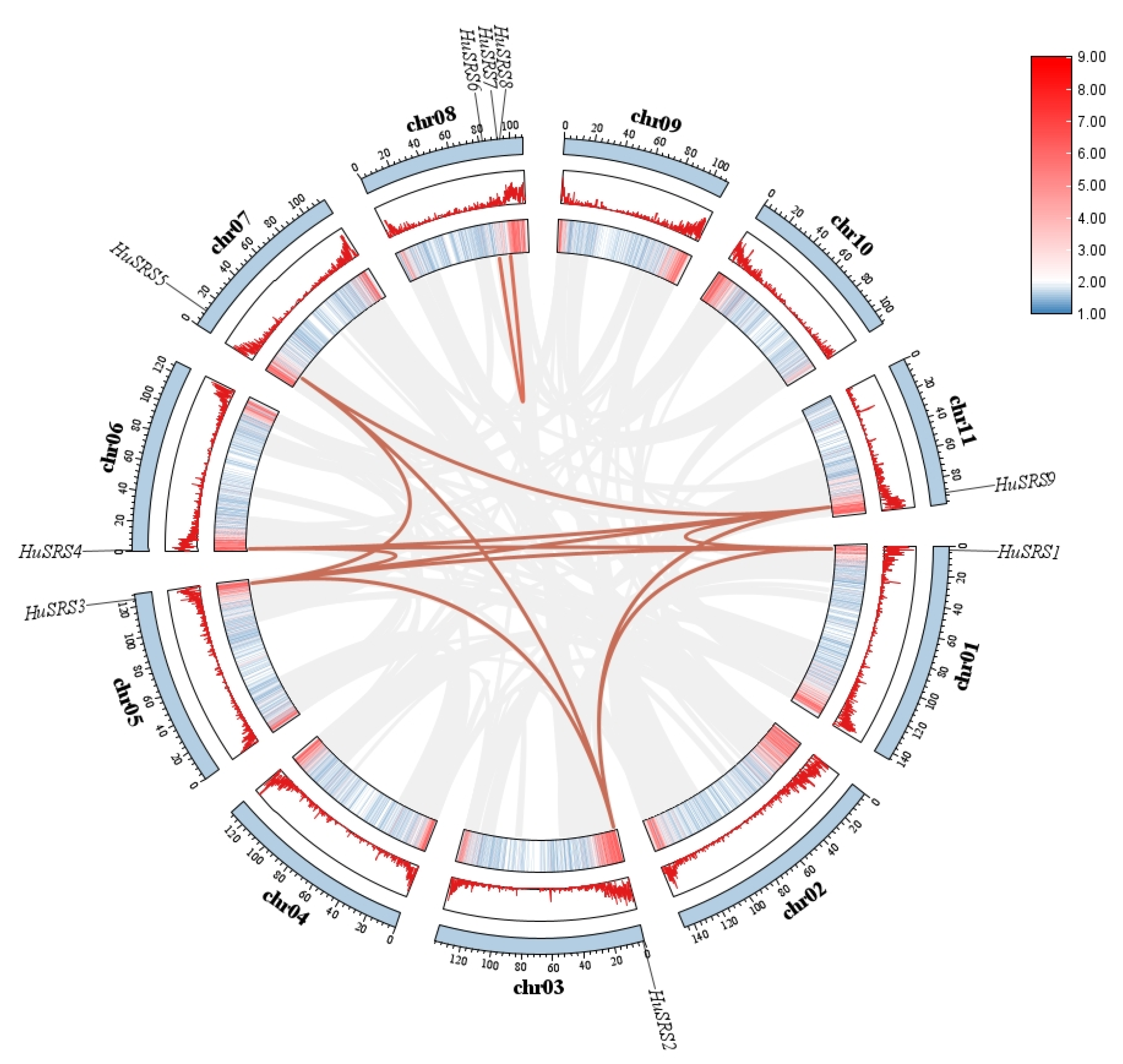
2.2. Chromosomal Localization and Collinearity Analysis of the HuSRS Gene Family
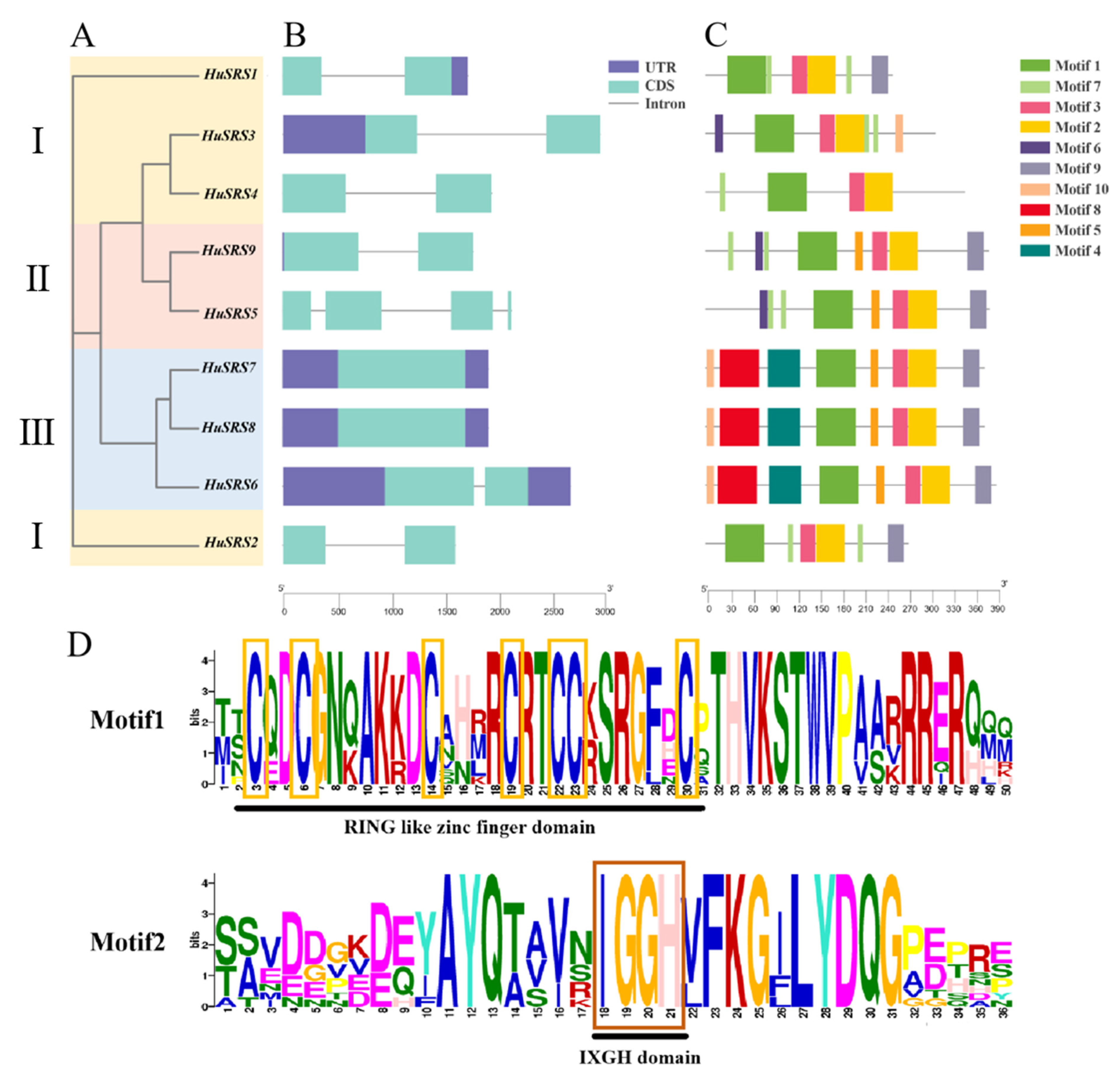
2.3. Conserved Motifs and Gene Structures of the HuSRS Gene Family
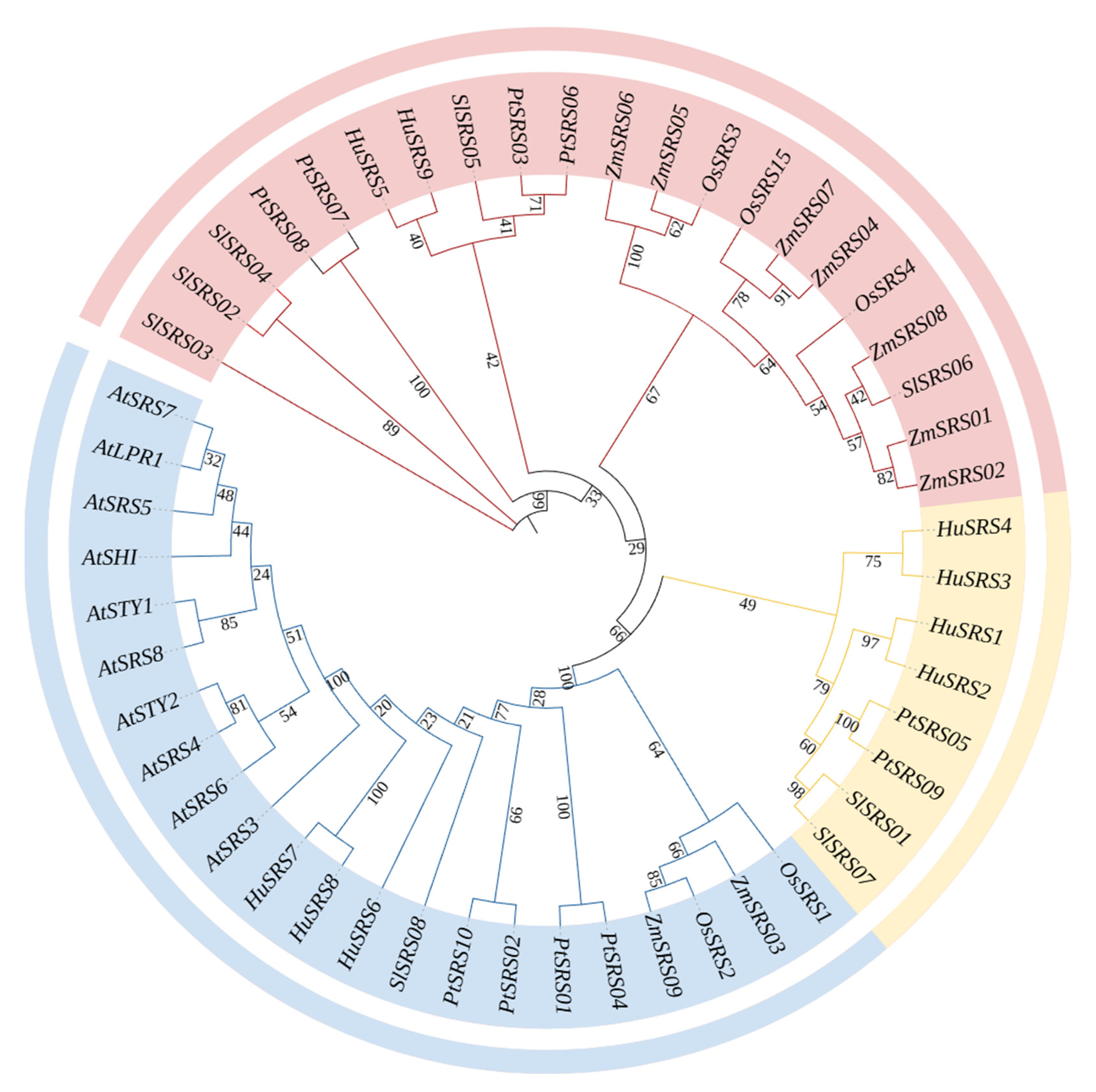
2.4. Phylogenetic Tree Construction of the HuSRS Gene Family
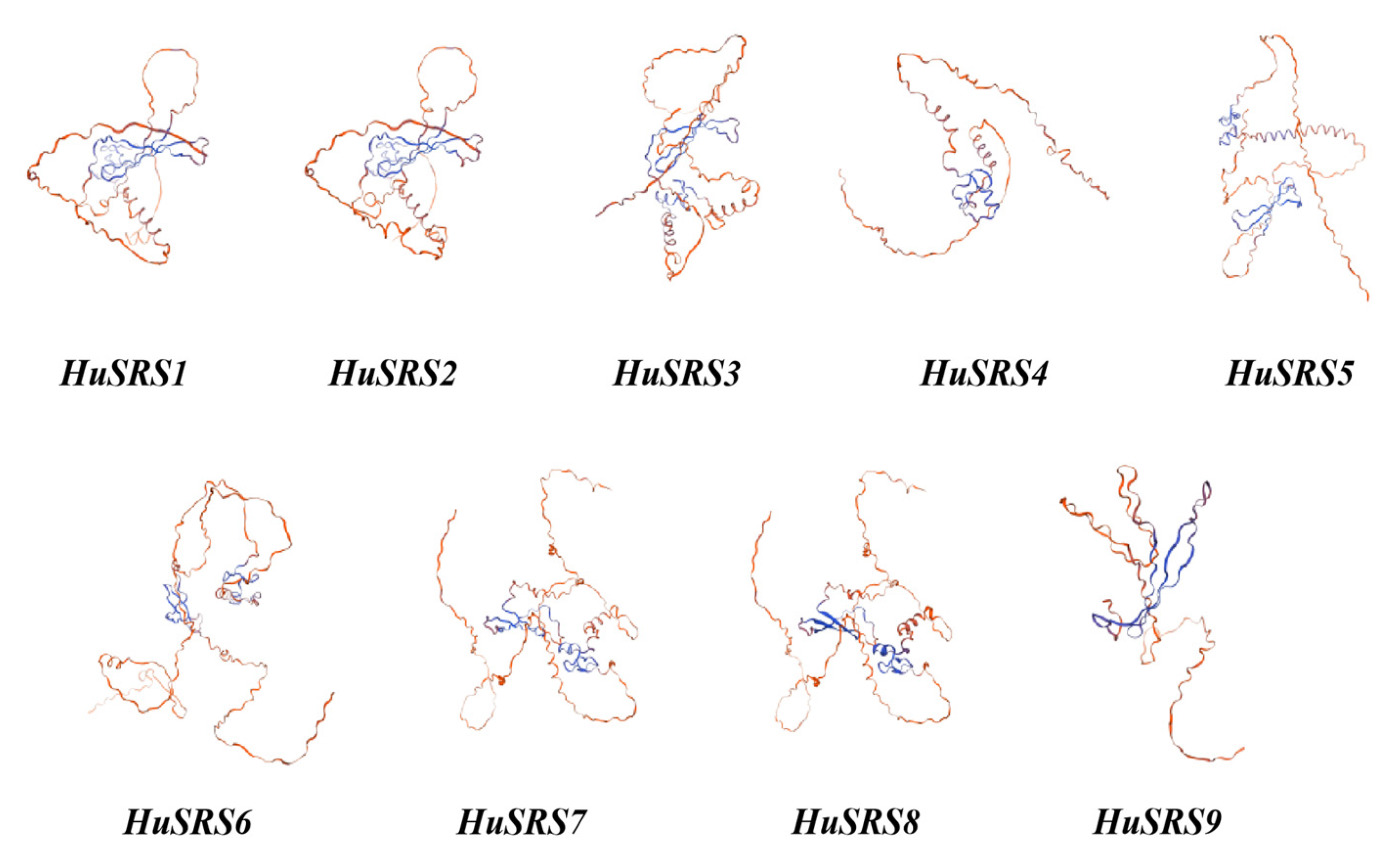
2.5. Secondary and Tertiary Structural Prediction and Protein–Protein Interaction Analysis of the HuSRS Gene Family
2.6. Temporal and Spatial Expression Patterns and Cis-Regulatory Element Analysis of the HuSRS Gene Family
2.7. Physiological Index Determination of Pitaya Callus and Spatial Expression Quantitative Analysis of HuSRS Gene Family
Plant Materials and Treatments
2.8. Coomassie Brilliant Blue G-250 Staining Method
2.9. Determination of Relative Electrolyte Conductivity (REC)
2.10. Expression of the HuSRS Genes in Pitaya Callus and Seedlings
3. Results
3.1. Identification of the HuSRS Gene Family Members and Analysis of Their Encoded Proteins’ Basic Physicochemical Properties
3.2. Chromosomal Distribution, Replication Events and Collinearity Analysis of the HuSRS Gene Family
3.3. Gene Structures and Conserved Motifs of the HuSRS Gene Family
3.4. Phylogenetic Tree Analysis of the HuSRS Gene Family
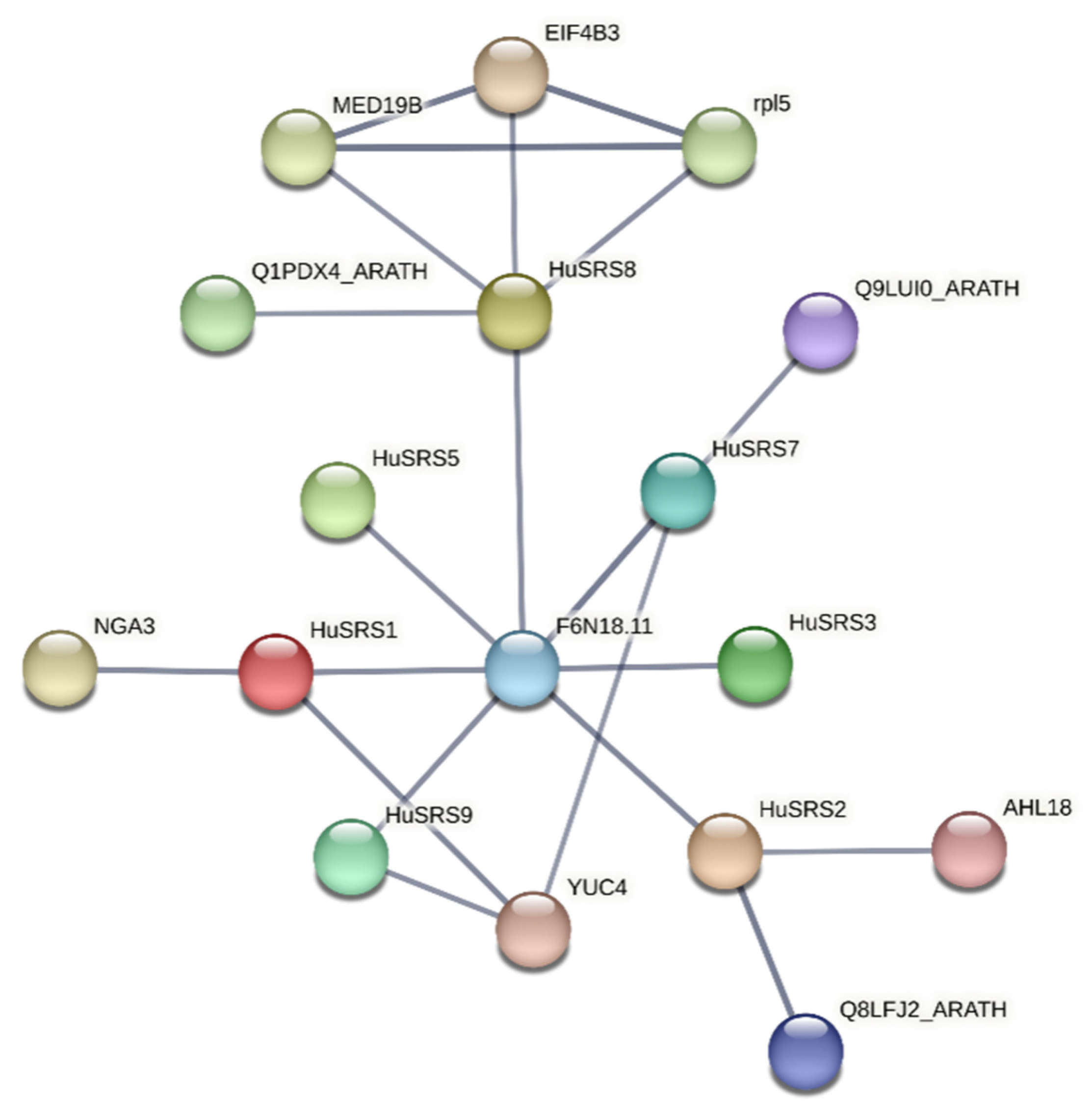
3.5. Protein Structure and Protein–Protein Interaction Analysis of the HuSRS Protein Family
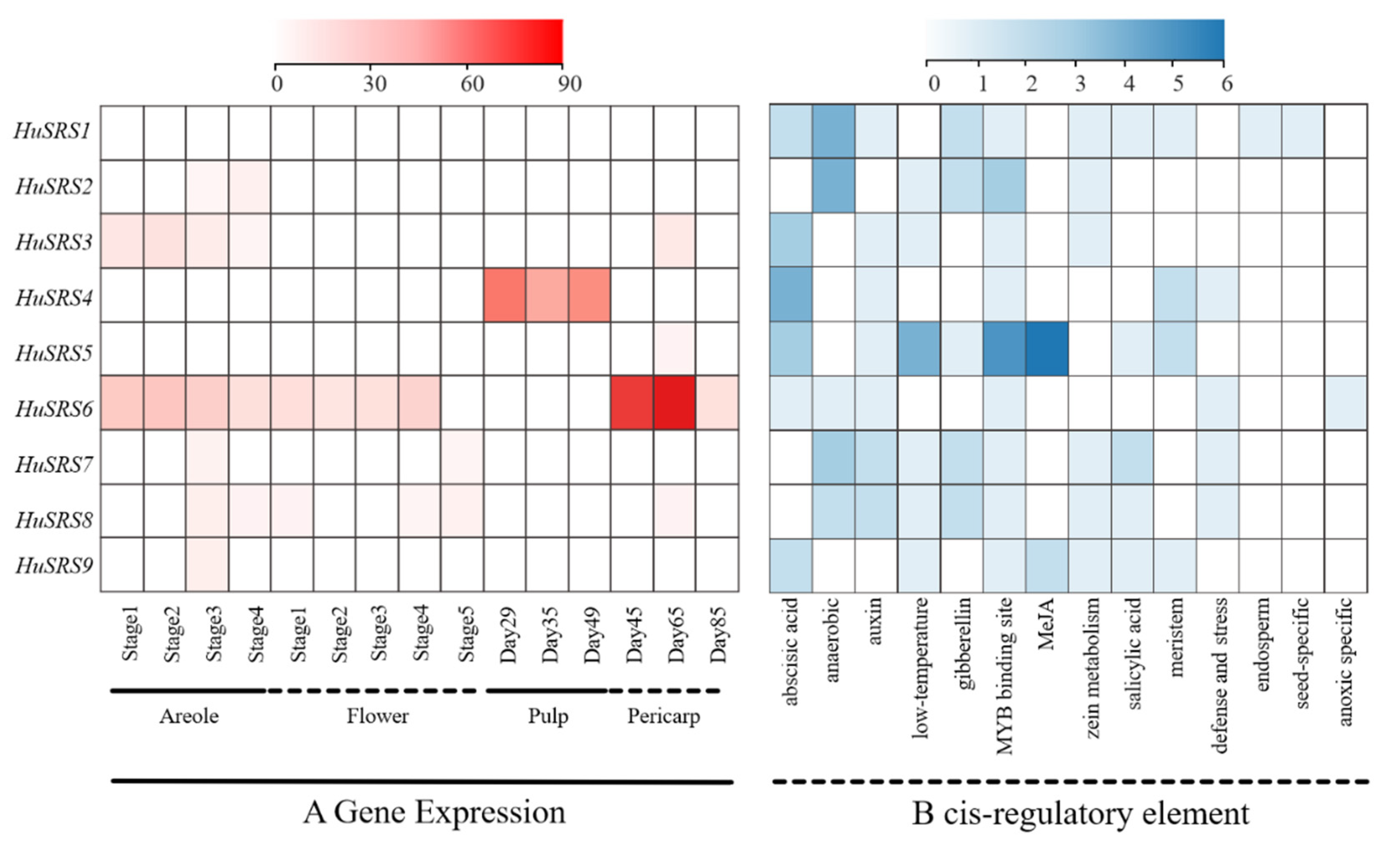
3.6. Temporal-Spatial Expression Transcriptome and Cis-Regulatory Element Analysis of HuSRS Genes
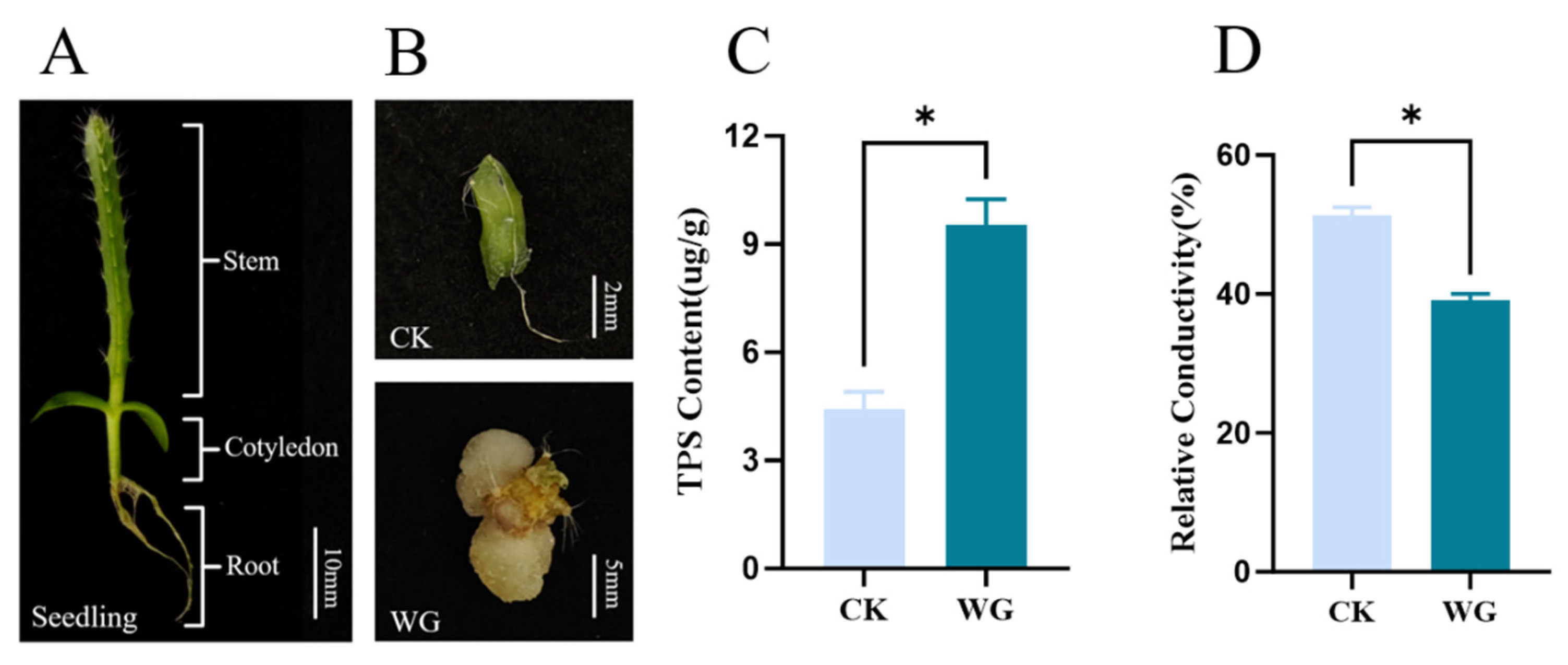
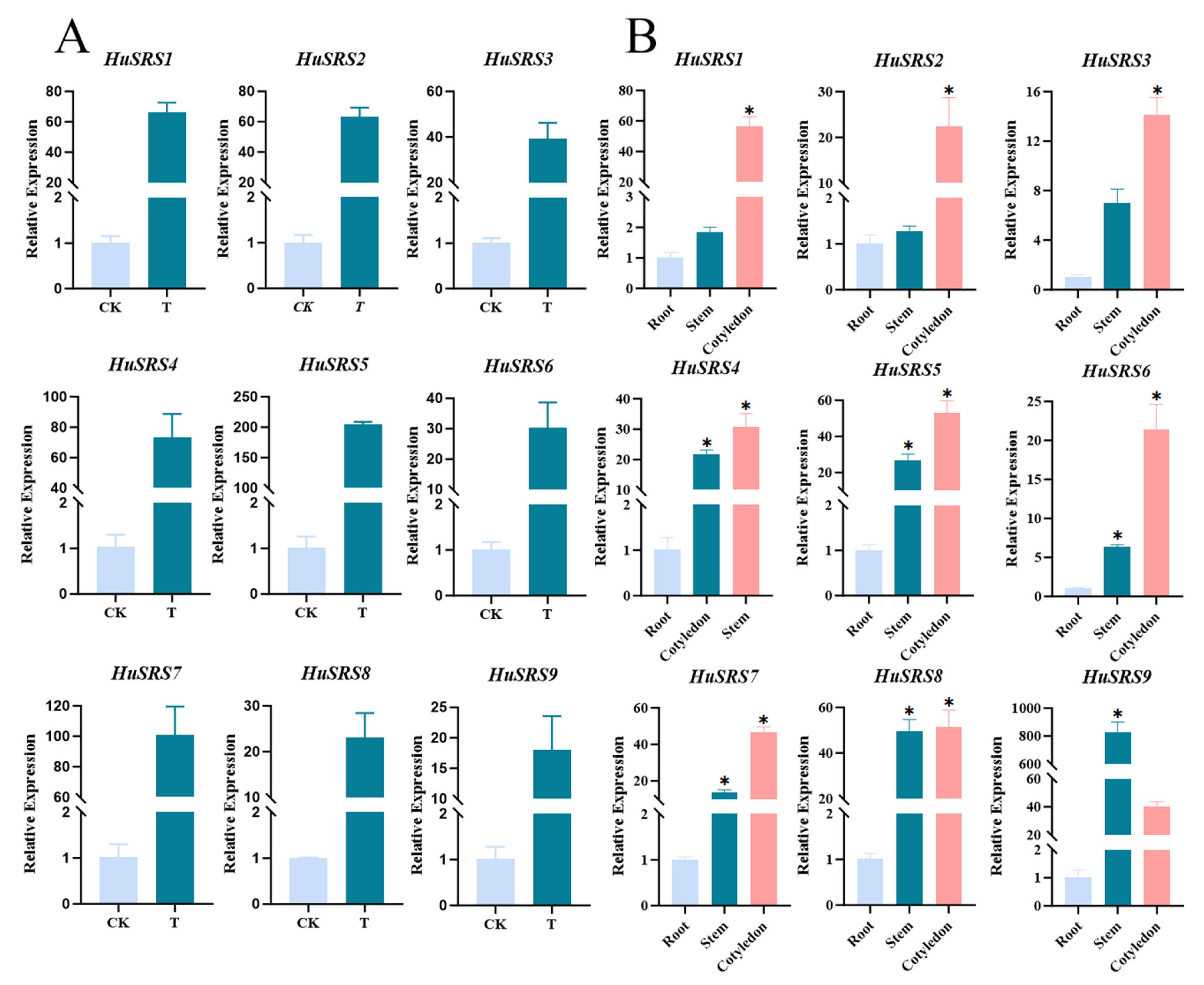
3.7. Physiological Characteristics of Pitaya Callus and Quantitative Analysis of Temporal-Spatial Expression of HuSRS Gene Family
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, J.; Sabir, I.A.; Qin, Y. From challenges to opportunities: Unveiling the secrets of pitaya through omics studies. Sci. Hortic. 2023, 321, 112357. [Google Scholar] [CrossRef]
- Tel-Zur, N. Breeding an underutilized fruit crop: A long-term program for Hylocereus. Hortic. Res. 2022, 9, uhac078. [Google Scholar] [CrossRef] [PubMed]
- Gomariz-Fernández, A.; Sánchez-Gerschon, V.; Fourquin, C.; Ferrándiz, C. The role of SHI/STY/SRS genes in organ growth and carpel development is conserved in the distant eudicot species Arabidopsis thaliana and Nicotiana benthamiana. Front. Plant Sci. 2017, 8, 814. [Google Scholar] [CrossRef] [PubMed]
- Baylis, T.; Cierlik, I.; Sundberg, E.; Mattsson, J. SHORT INTERNODES/STYLISH genes, regulators of auxin biosynthesis, are involved in leaf vein development in Arabidopsis thaliana. New Phytol. 2013, 197, 737–750. [Google Scholar] [CrossRef]
- Tian, Q.; Uhlir, N.J.; Reed, J.W. Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 2002, 14, 301–319. [Google Scholar] [CrossRef]
- Lütken, H.; Jensen, L.S.; Topp, S.H.; Mibus, H.; Müller, R.; Rasmussen, S.K. Production of compact plants by overexpression of AtSHI in the ornamental Kalanchoe. Plant Biotechnol. J. 2010, 8, 211–222. [Google Scholar] [CrossRef]
- Fridborg, I.; Kuusk, S.; Moritz, T.; Sundberg, E. The Arabidopsis dwarf mutant shi exhibits reduced gibberellin responses conferred by overexpression of a new putative zinc finger protein. Plant Cell 1999, 11, 1019–1032. [Google Scholar] [CrossRef]
- Hong, J.K.; Kim, J.A.; Kim, J.S.; Lee, S.I.; Koo, B.S.; Lee, Y.H. Overexpression of Brassica rapa SHI-RELATED SEQUENCE genes suppresses growth and development in Arabidopsis thaliana. Biotechnol. Lett. 2012, 34, 1561–1569. [Google Scholar] [CrossRef]
- Kuusk, S.; Sohlberg, J.J.; Magnus Eklund, D.; Sundberg, E. Functionally redundant SHI family genes regulate Arabidopsis gynoecium development in a dose--dependent manner. Plant J. 2006, 47, 99–111. [Google Scholar] [CrossRef]
- Smith, D.L.; Fedoroff, N.V. LRP1, a gene expressed in lateral and adventitious root primordia of arabidopsis. Plant Cell 1995, 7, 735–745. [Google Scholar]
- Kim, S.G.; Lee, S.; Kim, Y.S.; Yun, D.J.; Woo, J.C.; Park, C.M. Activation tagging of an Arabidopsis SHI-RELATED SEQUENCE gene produces abnormal anther dehiscence and floral development. Plant Mol. Biol. 2010, 74, 337–351. [Google Scholar] [CrossRef] [PubMed]
- Duan, E.; Wang, Y.; Li, X.; Lin, Q.; Zhang, T.; Wang, Y.; Zhou, C.; Zhang, H.; Jiang, L.; Wang, J.; et al. OsSHI1 Regulates Plant Architecture Through Modulating the Transcriptional Activity of IPA1 in Rice. Plant Cell 2019, 31, 1026–1042. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Shi, P.; Lv, Y.; Gao, Z.; Chen, G. Gene coexpression network analysis reveals the role of SRS genes in senescence leaf of maize (Zea mays L.). J. Genet. 2020, 99, 3. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Cui, B.; Zhu, Z.; Zhou, S. Genome-Wide identification, structural analysis, and expression patterns of tomato SRS gene family. J. Plant Growth Regul. 2024, 43, 60–75. [Google Scholar] [CrossRef]
- Zhao, S.P.; Song, X.Y.; Guo, L.L.; Zhang, X.Z.; Zheng, W.J. Genome-Wide Analysis of the Shi-Related Sequence Family and Functional Identification of GmSRS18 Involving in Drought and Salt Stresses in Soybean. Int. J. Mol. Sci. 2020, 21, 1810. [Google Scholar] [CrossRef]
- Yang, Y.; Qi, L.; Nian, L.; Zhu, X.; Yi, X.; Jiyu, Z.; Qiu, J. Genome-wide identification and expression analysis of the SRS gene family in Medicago sativa. DNA Cell Biol. 2021, 40, 1539–1553. [Google Scholar] [CrossRef]
- Sun, C.; Yu, L.; Zhang, S.; Gu, Q.; Wang, M. Genome-wide characterization of the SHORT INTER-NODES/STYLISH and Shi-Related Sequence family in Gossypium hirsutum and functional identification of GhSRS21 under salt stress. Front. Plant Sci. 2023, 13, 1078083. [Google Scholar] [CrossRef]
- Chen, C.; Li, F.; Xie, F.; Chen, J.; Hua, Q.; Chen, J.; Wu, Z.; Zhang, Z.; Zhang, R.; Zhao, J.; et al. Pitaya Genome and Multiomics Database (PGMD): A Comprehensive and Integrative Resource of Selenicereus undatus. Genes 2022, 13, 745. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Yang, M.; Derbyshire, M.K.; Yamashita, R.A.; Marchler--Bauer, A. NCBI’s Conserved Domain Database and Tools for Protein Domain Analysis. Curr. Protoc. Bioinform. 2020, 69, e90. [Google Scholar] [CrossRef]
- Chou, K.-C.; Shen, H.-B. Cell-PLoc 2.0: An improved package of web-servers for predicting subcellular localization of proteins in various organisms. Nat. Sci. 2010, 2, 1090–1103. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Hernandez, M.G.; et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Fang, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Snyder, J.C.; Desborough, S.L. Rapid estimation of potato tuber total protein content with coomassie brilliant blue G-250. TAG Theor. Appl. Genet. Theor. Und Angew. Genet. 1978, 52, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Knoche, M.; Peschel, S.; Hinz, M.; Bukovac, M.J. Studies on water transport through the sweet cherry fruit surface: II. Conductance of the cuticle in relation to fruit development. Planta 2001, 213, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, J.; Hua, Q.; Tel-Zur, N.; Xie, F.; Zhang, Z.; Chen, J.; Zhang, R.; Hu, G.; Zhao, J.; et al. Identification of reliable reference genes for quantitative real-time PCR normalization in pitaya. Plant Methods 2019, 15, 70. [Google Scholar] [CrossRef]
- Liu, J.-H.; Peng, T.; Dai, W. Critical cis-Acting Elements and Interacting Transcription Factors: Key Players Associated with Abiotic Stress Responses in Plants. Plant Mol. Biol. Report. 2014, 32, 303–317. [Google Scholar] [CrossRef]
- Buyuk, I.; Okay, A.; Aras, S. Identification and Characterization of SRS Genes in Phaseolus vulgaris Genome and Their Responses Under Salt Stress. Biochem. Genet. 2022, 60, 482–503. [Google Scholar] [CrossRef]
- Sohlberg, J.J.; Myrenås, M.; Kuusk, S.; Lagercrantz, U.; Kowalczyk, M.; Sandberg, G.; Sundberg, E. STY1 regulates auxin homeostasis and affects apical-basal patterning of the Arabidopsis gynoecium. Plant J. 2006, 47, 112–123. [Google Scholar] [CrossRef]
- Yin, Z.; Li, H.; Li, J.; Guo, C.; Li, Z.; Zhang, H.; Wang, H.; Siqin, T.; Sun, P.; Wang, Y.; et al. Genome-Wide Identification of the SRS Gene Family in Poplar and Expression Analysis Under Drought Stress and Salt Stress. Forests 2025, 16, 302. [Google Scholar] [CrossRef]
- Yang, J.; Xu, P.; Yu, D. Genome-wide identification and characterization of the SHI-related sequence gene family in rice. Evol. Bioinform. 2020, 16, 1176934320941495. [Google Scholar] [CrossRef]
- Ma, B.; Nian, L.; Ain, N.U.; Liu, X.; Yang, Y.; Zhu, X.; Haider, F.U.; Lv, Y.; Bai, P.; Zhang, X.; et al. Genome-wide identification and expression profiling of the SRS gene family in Melilotus albus reveals functions in various stress conditions. Plants 2022, 11, 3101. [Google Scholar] [CrossRef]
- Stone, S.L.; Hauksdóttir, H.; Troy, A.; Herschleb, J.; Kraft, E.; Callis, J. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 2005, 137, 13–30. [Google Scholar] [CrossRef]
- Susmi, F.A.; Simi, T.I.; Hasan, M.N.; Rahim, M.A. Genome-wide identification, characterization and functional prediction of the SRS gene family in sesame (Sesamum indicum L.). Oil Crop Sci. 2024, 9, 69–80. [Google Scholar] [CrossRef]
- Chen, S.; Kaeppler, S.M.; Vogel, K.P.; Casler, M.D. Selection Signatures in Four Lignin Genes from Switchgrass Populations Divergently Selected for In Vitro Dry Matter Digestibility. PLoS ONE 2016, 11, e0167005. [Google Scholar] [CrossRef]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF Transcription Factor Regulatory Networks in Hormone and Abiotic Stress Responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef] [PubMed]
- Fuertes-Aguilar, J.; Matilla, A.J. Transcriptional Control of Seed Life: New Insights into the Role of the NAC Family. Int. J. Mol. Sci. 2024, 25, 5369. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wang, B.; Wang, X.; Wei, X. Genome-wide identification, structural analysis and expression profiles of short internodes related sequence gene family in quinoa. Front. Genet. 2022, 13, 961925. [Google Scholar] [CrossRef] [PubMed]
- Eklund, D.M.; Cierlik, I.; Ståldal, V.; Claes, A.R.; Vestman, D.; Chandler, J.; Sundberg, E. Expression of Arabidopsis SHORT INTERNODES/STYLISH family genes in auxin biosynthesis zones of aerial organs is dependent on a GCC box-like regulatory element. Plant Physiol. 2011, 157, 2069–2080. [Google Scholar] [CrossRef]
- Huang, H.; Song, J.; Feng, Y.; Zheng, L.; Chen, Y.; Luo, K. Genome-Wide Identification and Expression Analysis of the SHI-Related Sequence Family in Cassava. Genes 2023, 14, 870. [Google Scholar] [CrossRef]
- Hu, M.; Xie, M.; Cui, X.; Huang, J.; Cheng, X.; Liu, L.; Yan, S.; Liu, S.; Tong, C. Characterization and Potential Function Analysis of the SRS Gene Family in Brassica napus. Genes 2023, 14, 1421. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, X.; Yuan, D.; Jin, F.; Zhang, Y.; Xu, J. Transcript profiling reveals complex auxin signaling pathway and transcription regulation involved in dedifferentiation and redifferentiation during somatic embryogenesis in cotton. BMC Plant Biol. 2012, 12, 10. [Google Scholar] [CrossRef]
- Luo, X.; Luo, Y.; Xu, A.; Kong, X.; Wang, X.; Deng, Q.; Zhang, H.; Lin, L.; Jia, Y. Enhancing antioxidant defense systems and regulating fatty acid unsaturation: Salicylic acid-mediated alleviation of low-temperature stress in Pitaya seedlings. BMC Plant Biol. 2025, 25, 1028. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef]
| Member Name | ID | Number of Amino Acids | Molecular Weight/Da | pI | Hydrophobicity Coefficient | Subcellular Localization |
|---|---|---|---|---|---|---|
| HuSRS1 | HU01G00264 | 235 | 24883.69 | 9.13 | −0.528 | Nucleus |
| HuSRS2 | HU03G00149 | 255 | 27036.96 | 8.98 | −0.509 | Nucleus |
| HuSRS3 | HU05G02071 | 289 | 31034.41 | 6.14 | −0.62 | Chloroplast |
| HuSRS4 | HU06G00092 | 326 | 34471.12 | 8.06 | −0.637 | Chloroplast |
| HuSRS5 | HU07G00744 | 357 | 38872.62 | 6.92 | −0.807 | Nucleus |
| HuSRS6 | HU08G00953 | 365 | 36488.21 | 8.68 | −0.336 | Cell membrane |
| HuSRS7 | HU08G01189 | 351 | 36500.44 | 9.04 | −0.565 | Nucleus |
| HuSRS8 | HU08G01241 | 351 | 36540.51 | 9.04 | −0.569 | Nucleus |
| HuSRS9 | HU11G01269 | 356 | 38240.04 | 7.18 | −0.802 | Nucleus |
| Protein | Alpha Helix | Extended Strand | Random Coil |
|---|---|---|---|
| HuSRS1 | 8.51% | 8.09% | 83.4% |
| HuSRS2 | 10.20% | 8.24% | 81.57% |
| HuSRS3 | 8.65% | 8.30% | 83.04% |
| HuSRS4 | 11.35% | 10.74% | 77.91% |
| HuSRS5 | 6.72% | 9.24% | 84.03% |
| HuSRS6 | 6.85% | 7.12% | 86.03% |
| HuSRS7 | 8.83% | 6.27% | 84.90% |
| HuSRS8 | 10.54% | 9.97% | 79.49% |
| HuSRS9 | 7.02% | 6.18% | 86.80% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, F.; Zhou, L.; Liu, S.; Huang, R.; Xu, G.; Yang, Z. Genome-Wide Identification and Expression Analysis of the SRS Gene Family in Hylocereus undatus. Plants 2025, 14, 3139. https://doi.org/10.3390/plants14203139
Peng F, Zhou L, Liu S, Huang R, Xu G, Yang Z. Genome-Wide Identification and Expression Analysis of the SRS Gene Family in Hylocereus undatus. Plants. 2025; 14(20):3139. https://doi.org/10.3390/plants14203139
Chicago/Turabian StylePeng, Fanjin, Lirong Zhou, Shuzhang Liu, Renzhi Huang, Guangzhao Xu, and Zhuanying Yang. 2025. "Genome-Wide Identification and Expression Analysis of the SRS Gene Family in Hylocereus undatus" Plants 14, no. 20: 3139. https://doi.org/10.3390/plants14203139
APA StylePeng, F., Zhou, L., Liu, S., Huang, R., Xu, G., & Yang, Z. (2025). Genome-Wide Identification and Expression Analysis of the SRS Gene Family in Hylocereus undatus. Plants, 14(20), 3139. https://doi.org/10.3390/plants14203139






