Activity-Based Profiling of Papain-like Cysteine Proteases During Late-Stage Leaf Senescence in Barley
Abstract
1. Introduction
2. Results and Discussion
2.1. Isolation of PLCPs Using Ion-Exchange and Affinity Chromatography
2.2. Semiquantitative Tandem MS Analysis of Barley PLCPs

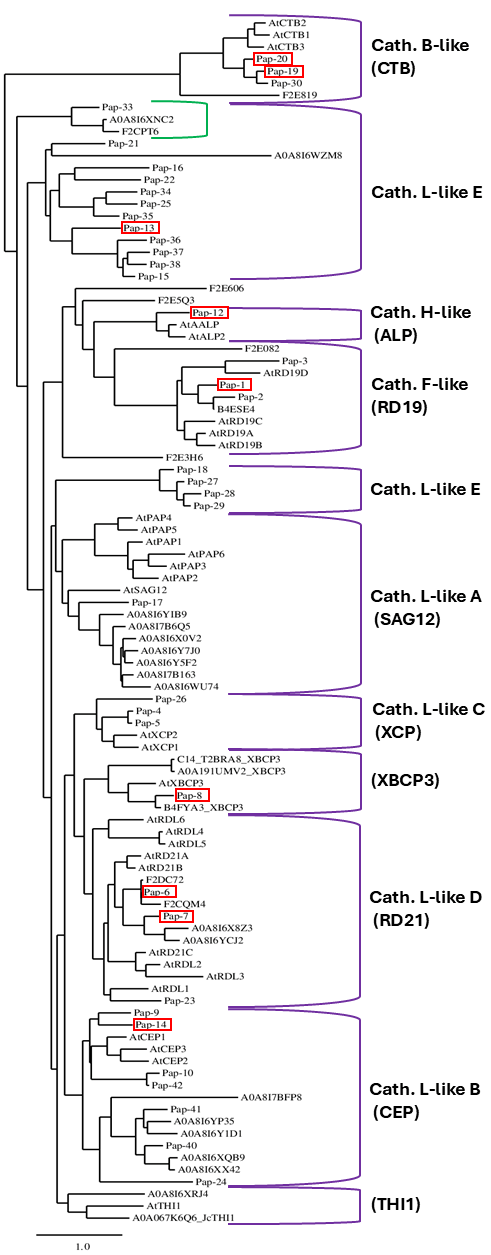
2.3. Phylogenetic Analysis of Barley PLCPs
3. Materials and Methods
3.1. Materials and Reagents
3.2. Plant Material
3.3. Ion-Exchange and Affinity Purification of PLCPs
3.4. Enzymatic Assays and Protein Determination
3.5. Gel-Based Tandem MS Analysis
3.6. Database Searching and Criteria for Protein Identification
3.7. Computational Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Newton, A.C.; Flavell, A.J.; George, T.S.; Leat, P.; Mullholland, B.; Ramsay, L.; Revoredo-Giha, C.; Russell, J.; Steffenson, B.J.; Swanston, J.S.; et al. Crops that feed the world 4. Barley: A resilient crop? Strengths and weaknesses in the context of food security. Food Secur. 2011, 3, 141–178. [Google Scholar] [CrossRef]
- Izydorczyk, M.S.; Edney, M. Wrigley, C., Batey, I., Miskelly, D., Eds.; Barley: Grain-Quality Characteristics and Management of Quality Requirements. In Cereal Grains: Assessing and Managing Quality, Woodhead Publishing Series in Food Science Technology and Nutrition, 2nd ed.; Woodhead Publishing: Sawston, UK, 2017; pp. 195–234. [Google Scholar]
- Quirino, B.F.; Noh, Y.S.; Himelblau, E.; Amasino, R.M. Molecular aspects of leaf senescence. Trends Plant Sci. 2000, 5, 278–282. [Google Scholar] [CrossRef]
- Lee, S.; Masclaux-Daubresse, C. Current understanding of leaf senescence in rice. Int. J. Mol. Sci. 2021, 22, 4515. [Google Scholar] [CrossRef]
- Lei, P.; Yu, F.; Liu, X.Y. Recent advances in cellular degradation and nuclear control of leaf senescence. J. Exp. Bot. 2023, 74, 5472–5486. [Google Scholar] [CrossRef]
- Diaz-Mendoza, M.; Velasco-Arroyo, B.; Santamaria, M.E.; González-Melendi, P.; Martinez, M.; Diaz, I. Plant senescence and proteolysis: Two processes with one destiny. Genet. Mol. Biol. 2016, 39, 329–338. [Google Scholar] [CrossRef]
- Roberts, I.N.; Caputo, C.; Criado, M.V.; Funk, C. Senescence-associated proteases in plants. Physiol. Plant. 2012, 145, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Mendoza, M.; Velasco-Arroyo, B.; González-Melendi, P.; Martínez, M.; Díaz, I. C1A cysteine protease-cystatin interactions in leaf senescence. J. Exp. Bot. 2014, 65, 3825–3833. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J.; Bateman, A. MEROPS: The peptidase database. Nucleic Acids Res. 2010, 38, D227–D233. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef]
- Verma, S.; Dixit, R.; Pandey, K.C. Cysteine proteases: Modes of activation and future prospects as pharmacological targets. Front. Pharmacol. 2016, 7, 107. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Shi, C.N.; Liu, S.; Lei, J.J.; Lu, Q.S.; Hu, H.C.; Ren, Y.; Zhang, N.; Sun, C.W.; Chen, L.; et al. A papain-like cysteine protease-released small signal peptide confers wheat resistance to wheat yellow mosaic virus. Nat. Commun. 2023, 14, 7773. [Google Scholar] [CrossRef]
- Richau, K.H.; Kaschani, F.; Verdoes, M.; Pansuriya, T.C.; Niessen, S.; Stüber, K.; Colby, T.; Overkleeft, H.S.; Bogyo, M.; Van der Hoorn, R.A.L. Subclassification and biochemical analysis of plant papain-like cysteine proteases displays subfamily-specific characteristics. Plant Physiol. 2012, 158, 1583–1599. [Google Scholar] [CrossRef] [PubMed]
- Martinez, M.; Diaz, I. The origin and evolution of plant cystatins and their target cysteine proteinases indicate a complex functional relationship. BMC Evol. Biol. 2008, 8, 198. [Google Scholar] [CrossRef]
- Misas-Villamil, J.C.; van der Hoorn, R.A.L.; Doehlemann, G. Papain-like cysteine proteases as hubs in plant immunity. New Phytol. 2016, 212, 902–907. [Google Scholar] [CrossRef]
- Masclaux-Daubresse, C.; Daniel-Vedele, F.; Dechorgnat, J.; Chardon, F.; Gaufichon, L.; Suzuki, A. Nitrogen uptake, assimilation and remobilization in plants: Challenges for sustainable and productive agriculture. Ann. Bot. 2010, 105, 1141–1157. [Google Scholar] [CrossRef]
- Hajibarat, Z.; Saidi, A. Senescence-associated proteins and nitrogen remobilization in grain filling under drought stress condition. J. Genet. Eng. Biotechnol. 2022, 20, 101. [Google Scholar] [CrossRef]
- Havé, M.; Marmagne, A.; Chardon, F.; Masclaux-Daubresse, C. Nitrogen remobilization during leaf senescence: Lessons from Arabidopsis to crops. J. Exp. Bot. 2017, 68, 2513–2529. [Google Scholar] [CrossRef] [PubMed]
- Vatov, E.; Ludewig, U.; Zentgraf, U. Disparate dynamics of gene body and cis-regulatory element evolution illustrated for the senescence-associated cysteine protease gene SAG12 of plants. Plants 2021, 10, 1380. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, C.; Leverentz, M.K.; Griffiths, G.; Thomas, B.; Chanasut, U.; Stead, A.D.; Rogers, H.J. Cysteine protease gene expression and proteolytic activity during senescence of Alstroemeria petals. J. Exp. Bot. 2002, 53, 233–240. [Google Scholar] [CrossRef]
- Parrott, D.L.; McInnerney, K.; Feller, U.; Fischer, A.M. Steam-girdling of barley (Hordeum vulgare) leaves leads to carbohydrate accumulation and accelerated leaf senescence, facilitating transcriptomic analysis of senescence-associated genes. New Phytol. 2007, 176, 56–69. [Google Scholar] [CrossRef]
- Jukanti, A.K.; Heidlebaugh, N.M.; Parrott, D.L.; Fischer, I.A.; McInnerney, K.; Fischer, A.M. Comparative transcriptome profiling of near-isogenic barley (Hordeum vulgare) lines differing in the allelic state of a major grain protein content locus identifies genes with possible roles in leaf senescence and nitrogen reallocation. New Phytol. 2008, 177, 333–349. [Google Scholar] [CrossRef]
- Cohen, M.; Hertweck, K.; Itkin, M.; Malitsky, S.; Dassa, B.; Fischer, A.M.; Fluhr, R. Enhanced proteostasis, lipid remodeling, and nitrogen remobilization define barley flag leaf senescence. J. Exp. Bot. 2022, 73, 6816–6837. [Google Scholar] [CrossRef]
- Hollmann, J.; Gregersen, P.L.; Krupinska, K. Identification of predominant genes involved in regulation and execution of senescence-associated nitrogen remobilization in flag leaves of field grown barley. J. Exp. Bot. 2014, 65, 3963–3973. [Google Scholar] [CrossRef]
- Hu, J.J.; Rampitsch, C.; Bykova, N.V. Advances in plant proteomics toward improvement of crop productivity and stress resistance. Front. Plant Sci. 2015, 6, 209. [Google Scholar] [CrossRef]
- Gregersen, P.L.; Holm, P.B.; Krupinska, K. Leaf senescence and nutrient remobilisation in barley and wheat. Plant Biol. 2008, 10, 37–49. [Google Scholar] [CrossRef]
- Distelfeld, A.; Avni, R.; Fischer, A.M. Senescence, nutrient remobilization, and yield in wheat and barley. J. Exp. Bot. 2014, 65, 3783–3798. [Google Scholar] [CrossRef] [PubMed]
- Nägler, D.K.; Ménard, R. Family C1 cysteine proteases:: Biological diversity or redundancy? Biol. Chem. 2003, 384, 837–843. [Google Scholar] [CrossRef] [PubMed]
- Kocks, C.; Maehr, R.; Overkleeft, H.S.; Wang, E.W.; Iyer, L.K.; Lennon-Duménil, A.M.; Ploegh, H.L.; Kessler, B.M. Functional proteomics of the active cysteine protease content in drosophila S2 cells. Mol. Cell. Proteom. 2003, 2, 1188–1197. [Google Scholar] [CrossRef]
- Bahmani, M.; O’Lone, C.E.; Juhász, A.; Nye-Wood, M.; Dunn, H.; Edwards, I.B.; Colgrave, M.L. Application of mass spectrometry-based proteomics to barley research. J. Agric. Food Chem. 2021, 69, 8591–8609. [Google Scholar] [CrossRef] [PubMed]
- Washburn, M.P.; Wolters, D.; Yates, J.R. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001, 19, 242–247. [Google Scholar] [CrossRef]
- Bantscheff, M.; Schirle, M.; Sweetman, G.; Rick, J.; Kuster, B. Quantitative mass spectrometry in proteomics: A critical review. Anal. Bioanal. Chem. 2007, 389, 1017–1031. [Google Scholar] [CrossRef] [PubMed]
- Gerster, S.; Kwon, T.; Ludwig, C.; Matondo, M.; Vogel, C.; Marcotte, E.M.; Aebersold, R.; Bühlmann, P. Statistical approach to protein quantification. Mol. Cell. Proteom. 2014, 13, 666–677. [Google Scholar] [CrossRef]
- Fonovic, M.; Bogyo, M. Activity-based probes as a tool for functional proteomic analysis of proteases. Expert Rev. Proteom. 2008, 5, 721–730. [Google Scholar] [CrossRef]
- Schepetkin, I.A.; Fischer, A.M. Cathepsin B- and L-like protease activities are induced during developmental barley leaf senescence. Plants 2024, 13, 3009. [Google Scholar] [CrossRef]
- Tsuji, A.; Tsukamoto, K.; Iwamoto, K.; Ito, Y.; Yuasa, K. Enzymatic characterization of germination-specific cysteine protease-1 expressed transiently in cotyledons during the early phase of germination. J. Biochem. 2013, 153, 73–83. [Google Scholar] [CrossRef]
- Hüynck, J.S.; Kaschani, F.; van der Linde, K.; Ziemann, S.; Müller, A.N.; Colby, T.; Kaiser, M.; Villamil, J.C.M.; Doehlemann, G. Proteases underground: Analysis of the maize root apoplast identifies organ specific papain-like cysteine protease activity. Front. Plant Sci. 2019, 10, 473. [Google Scholar] [CrossRef]
- Jinka, R.; Ramakrishna, V.; Rao, S.K.; Rao, R.P. Purification and characterization of cysteine protease from germinating cotyledons of horse gram. BMC Biochem. 2009, 10, 28. [Google Scholar] [CrossRef]
- Barrett, A.J.; Kembhavi, A.A.; Hanada, K. E-64 [L-trans-epoxysuccinyl-leucyl-amido(4-guanidino)butane] and related epoxides as inhibitors of cysteine proteinases. Acta Biol. Med. Ger. 1981, 40, 1513–1517. [Google Scholar] [PubMed]
- Holwerda, B.C.; Galvin, N.J.; Baranski, T.J.; Rogers, J.C. In vitro processing of aleurain, a barley vacuolar thiol protease. Plant Cell 1990, 2, 1091–1106. [Google Scholar] [CrossRef] [PubMed]
- Greenbaum, D.; Medzihradszky, K.F.; Burlingame, A.; Bogyo, M. Epoxide electrophiles as activity-dependent cysteine protease profiling and discovery tools. Chem. Biol. 2000, 7, 569–581. [Google Scholar] [CrossRef]
- Poret, M.; Chandrasekar, B.; van der Hoorn, R.A.L.; Coquet, L.; Jouenne, T.; Avice, J.C. Proteomic investigations of proteases involved in cotyledon senescence: A model to explore the genotypic variability of proteolysis machinery associated with nitrogen remobilization efficiency during the leaf senescence of oilseed rape. Proteomes 2017, 5, 29. [Google Scholar] [CrossRef]
- Havé, M.; Espinasse, C.; Cottyn-Boitte, B.; Puga-Freitas, R.; Bagard, M.; Balliau, T.; Zivy, M.; Ganeshan, S.; Chibbar, R.N.; Castell, J.F.; et al. Triticain alpha represents the major active papain-like cysteine protease in naturally senescing and ozone-treated leaves of wheat. Plant Physiol. Biochem. 2025, 219, 109380. [Google Scholar] [CrossRef]
- Cheah, J.S.; Yamada, S. A simple elution strategy for biotinylated proteins bound to streptavidin conjugated beads using excess biotin and heat. Biochem. Biophys. Res. Commun. 2017, 493, 1522–1527. [Google Scholar] [CrossRef]
- Frank, S.; Hollmann, J.; Mulisch, M.; Matros, A.; Carrión, C.C.; Mock, H.P.; Hensel, G.; Krupinska, K. Barley cysteine protease PAP14 plays a role in degradation of chloroplast proteins. J. Exp. Bot. 2019, 70, 6057–6069. [Google Scholar] [CrossRef]
- Sekhon, R.S.; Saski, C.; Kumar, R.; Flinn, B.S.; Luo, F.; Beissinger, T.M.; Ackerman, A.J.; Breitzman, M.W.; Bridges, W.C.; de Leon, N.; et al. Integrated genome-scale analysis identifies novel genes and networks underlying senescence in maize. Plant Cell 2019, 31, 1968–1989. [Google Scholar] [CrossRef] [PubMed]
- Pruzinská, A.; Shindo, T.; Niessen, S.; Kaschani, F.; Tóth, R.; Millar, A.H.; van der Hoorn, R.A.L. Major Cys protease activities are not essential for senescence in individually darkened Arabidopsis leaves. BMC Plant Biol. 2017, 17, 4. [Google Scholar] [CrossRef]
- Huang, J.; van der Hoorn, R.A.L. RD21-like proteases: Key effector hubs in plant-pathogen interactions. J. Exp. Bot. 2025, 76, 4212–4219. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Zhong, J.; Pandey, A. Common errors in mass spectrometry-based analysis of post-translational modifications. Proteomics 2016, 16, 700–714. [Google Scholar] [CrossRef]
- Schreiber, M.; Jayakodi, M.; Stein, N.; Mascher, M. Plant pangenomes for crop improvement, biodiversity and evolution. Nat. Rev. Genet. 2024, 25, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Gonzales, L.J.D.; Bowler-Barnett, E.H.; Rice, D.L.; Kim, M.; Wijerathne, S.; Luciani, A.; Kandasaamy, S.; Luo, J.; Watkins, X.; et al. The UniProt website API: Facilitating programmatic access to protein knowledge. Nucleic Acids Res. 2025, 53, W547–W553. [Google Scholar] [CrossRef]
- Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Adesina, A.; Ahmad, S.; Bowler-Barnett, E.H.; Bye-A-Jee, H.; Carpentier, D.; Denny, P.; et al. UniProt: The universal protein knowledgebase in 2025. Nucleic Acids Res. 2024, 52, D609–D617. [Google Scholar] [CrossRef]
- Jayakodi, M.; Lu, Q.X.; Pidon, H.; Rabanus-Wallace, M.T.; Bayer, M.; Lux, T.; Guo, Y.; Jaegle, B.; Badea, A.; Bekele, W.; et al. Structural variation in the pangenome of wild and domesticated barley. Nature 2024, 636, 654–662. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.J.; Hu, M.H.; Wang, Q.; Cheng, L.; Zhang, Z.B. Role of papain-like cysteine proteases in plant development. Front. Plant Sci. 2018, 9, 1717. [Google Scholar] [CrossRef]
- Szewinska, J.; Siminska, J.; Bielawski, W. The roles of cysteine proteases and phytocystatins in development and germination of cereal seeds. J. Plant Physiol. 2016, 207, 10–21. [Google Scholar] [CrossRef]
- Martinez, M.; Gómez-Cabellos, S.; Giménez, M.J.; Barro, F.; Diaz, I.; Diaz-Mendoza, M. Plant proteases: From key enzymes in germination to allies for fighting human gluten-related disorders. Front. Plant Sci. 2019, 10, 721. [Google Scholar] [CrossRef]
- Hu, G.S.; Evans, C.P.; Satterfield, K.; Ellberg, S.; Marshall, J.M.; Schroeder, K.L.; Obert, D.E. Registration of ‘GemCraft’ spring malting barley cultivar. J. Plant Regist. 2024, 18, 11–16. [Google Scholar] [CrossRef]
- Nickerson, J.L.; Doucette, A.A. Rapid and quantitative protein precipitation for proteome analysis by mass spectrometry. J. Proteome Res. 2020, 19, 2035–2042. [Google Scholar] [CrossRef] [PubMed]
- Wojtkiewicz, M.; Berg Luecke, L.; Kelly, M.I.; Gundry, R.L. Facile preparation of peptides for mass spectrometry analysis in bottom-up proteomics workflows. Curr. Protoc. 2021, 1, e85. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Nesvizhskii, A.I.; Kolker, E.; Aebersold, R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002, 74, 5383–5392. [Google Scholar] [CrossRef]
- Nesvizhskii, A.I.; Keller, A.; Kolker, E.; Aebersold, R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003, 75, 4646–4658. [Google Scholar] [CrossRef]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef] [PubMed]


| Fraction | Substrate | Effect/IC50 |
|---|---|---|
| E-64 | ||
| Fr. No 26–27 | R-AMC | No inhibition up to 25 µM |
| Fr. No 35–36 | Z-RR-AMC | 9.1 ± 2.2 nM |
| Z-FR-AMC | 15.3 ± 4.1 nM | |
| Bestatin | ||
| Fr. No 26–27 | R-AMC | 247 ± 38 µM |
| Fr. No 35–36 | Z-RR-AMC | No inhibition up to 1 mM |
| Z-FR-AMC | No inhibition up to 1 mM | |
| 1,10-Phenanthroline | ||
| Fr. No 26–27 | R-AMC | 68.5 ± 12.2 µM |
| Fr. No 35–36 | Z-RR-AMC | No inhibition up to 5 mM |
| Z-FR-AMC | No inhibition up to 5 mM | |
| Name | UniProt ID | Total Spectral Count (% of Coverage) * | |
|---|---|---|---|
| 43 kDa Band | 38 kDa Band | ||
| HvPap-1 | F2DDC9 | 5 (13) | 47 (24) |
| HvPap-6 | A0A8I6WYU4 | 270 (29) | 257 (25) |
| HvPap-7 | F2E6V2 | 8 (15) | 6 (17) |
| HvPap-8 | B4ESF0 | 5 (9.4) | 13 (22) |
| HvPap-12 | P05167 | 8 (14) | 83 (26) |
| HvPap-13 | A0A8I6XJP4 | 7 (16) | 40 (44) |
| HvPap-14 | B4ESF2 | 3 (9.5) | 73 (37) |
| HvPap-16 | B4ESF4 | 2 (4.9) | 10 (21) |
| HvPap-17 | A0A8I6Y6A5 | 6 (17) | 12 (21) |
| HvPap-19 | A0A8I7BBL8 | 7 (17) | 40 (33) |
| HvPap-20 | A0A8I7B6S7 | 5 (8.2) | 17 (18) |
| Name 1 | Subfamily 2 | Gene Identifier 3 | UniProt ID | nAA 4 | MW, Da 5 |
|---|---|---|---|---|---|
| HvPap-1 | F-like (RD19) | MOREX.r3.5HG0499760 | F2DDC9 | 377 | 41,016 |
| HvPap-2 | F-like (RD19) | MOREX.r3.2HG0159420 | A0A8I6WSN3 | 378 | 41,885 |
| HvPap-3 | F-like (RD19) | MOREX.r3.2HG0131510 | B4ESE5 | 368 | 38,969 |
| B4ESE4 | F-like (RD19) | Akashinriki.Proj.2HG00143120 | B4ESE4 | 381 | 41,931 |
| F2E082 | F-like (RD19) | N.F. | F2E082 | 341 | 37,057 |
| HvPap-12 | H-like (ALP) | MOREX.r3.5HG0480100 | P05167 | 362 | 39,122 |
| HvPap-19 | B-like (CTB) | MOREX.r3.4HG0339740 | A0A8I7BBL8 | 344 | 37,222 |
| HvPap-20 | B-like (CTB) | MOREX.r3.4HG0339730 | A0A8I7B6S7 | 353 | 38,423 |
| HvPap-30 | B-like (CTB) | MOREX.r3.4HG0339750 | A0A8I6X9J3 | 347 | 37,752 |
| F2E819 | B-like (CTB) | N.F. | F2E819 | 471 | 50,283 |
| HvPap-17 | L-like A (SAG12) | MOREX.r3.5HG0511140 | A0A8I6Y6A5 | 349 | 36,895 |
| A0A8I6YIB9 | L-like A (SAG12) | MOREX.r3.6HG0570590 | A0A8I6YIB9 | 343 | 36,737 |
| A0A8I7B6Q5 | L-like A (SAG12) | MOREX.r3.4HG0336140 | A0A8I7B6Q5 | 341 | 36,789 |
| A0A8I7B163 | L-like A (SAG12) | MOREX.r3.1HG0061980 | A0A8I7B163 | 339 | 36,752 |
| A0A8I6Y7J0 | L-like A (SAG12) | MOREX.r3.7HG0673530 | A0A8I6Y7J0 | 340 | 36,792 |
| A0A8I6Y5F2 | L-like A (SAG12) | MOREX.r3.6HG0623260 | A0A8I6Y5F2 | 339 | 37,020 |
| A0A8I6X0V2 | L-like A (SAG12) | MOREX.r3.2HG0103220 | A0A8I6X0V2 | 340 | 37,030 |
| A0A8I6WU74 | L-like A (SAG12) | MOREX.r3.1HG0061970 | A0A8I6WU74 | 340 | 37,333 |
| HvPap-9 | L-like B (CEP) | MOREX.r3.4HG0342040 | A0A8I6XX70 | 365 | 40,033 |
| HvPap-10 | L-like B (CEP) | MOREX.r3.3HG0308010 | A0A8I6XAH7 | 373 | 40,574 |
| HvPap-14 | L-like B (CEP) | MOREX.r3.3HG0304500 | B4ESF2 | 367 | 39,790 |
| HvPap-24 | L-like B (CEP) | MOREX.r3.2HG0211390 | A0A8I6WKU7 | 320 | 36,267 |
| HvPap-40 | L-like B (CEP) | MOREX.r3.6HG0548850 | A0A8I6XXP5 | 374 | 41,375 |
| HvPap-41 | L-like B (CEP) | MOREX.r3.6HG0543660 | A0A8I6XQ46 | 357 | 39,752 |
| HvPap-42 | L-like B (CEP) | MOREX.r3.3HG0308000 | A0A8I7BAT3 | 379 | 40,979 |
| A0A8I6YP35 | L-like B (CEP) | MOREX.r3.6HG0545100 | A0A8I6YP35 | 358 | 39,836 |
| A0A8I6XQB9 | L-like B (CEP) | MOREX.r3.6HG0545330 | A0A8I6XQB9 | 231 | 24,863 |
| A0A8I6XX42 | L-like B (CEP) | MOREX.r3.6HG0545210 | A0A8I6XX42 | 366 | 40,642 |
| A0A8I6Y1D1 | L-like B (CEP) | MOREX.r3.6HG0545130 | A0A8I6Y1D1 | 338 | 37,546 |
| A0A8I7BFP8 | L-like B (CEP) | MOREX.r3.7HG0747000 | A0A8I7BFP8 | 352 | 38,442 |
| HvPap-4 | L-like C (XCP) | MOREX.r3.5HG0462580 | B4ESE6 | 356 | 38,651 |
| HvPap-5 | L-like C (XCP) | MOREX.r3.1HG0004220 | B4ESE7 | 351 | 38,212 |
| HvPap-26 | L-like C (XCP) | MOREX.r3.6HG0609400 | A0A287UKW8 | 364 | 39,959 |
| HvPap-6 | L-like D (RD21) | MOREX.r3.2HG0204520 | A0A8I6WYU4 | 463 | 50,226 |
| HvPap-7 | L-like D (RD21) | MOREX.r3.2HG0212170 | F2E6V2 | 473 | 50,677 |
| HvPap-23 | L-like D (RD21) | N.F. | B4ESF8 | 190 | 21,595 |
| F2CQM4 | L-like D (RD21) | N.F. | F2CQM4 | 436 | 47,292 |
| A0A8I6X8Z3 | L-like D (RD21) | MOREX.r3.2HG0211940 | A0A8I6X8Z3 | 469 | 50,348 |
| F2DC72 | L-like D (RD21) | N.F. | F2DC72 | 289 | 31,994 |
| A0A8I6YCJ2 | L-like D (RD21) | MOREX.r3.7HG0685270 | A0A8I6YCJ2 | 477 | 51,115 |
| HvPap-13 | L-like E | MOREX.r3.5HG0506230 | A0A8I6XJP4 | 366 | 39,825 |
| HvPap-15 | L-like E | MOREX.r3.6HG0545500 | M0YYX8 | 406 | 44,371 |
| HvPap-16 | L-like E | MOREX.r3.7HG0749670 | B4ESF4 | 389 | 42,091 |
| HvPap-18 | L-like E | Akashinriki.Proj.4HG00376280 | F2CR43 | 365 | 39,599 |
| HvPap-21 | L-like E | MOREX.r3.2HG0205780 | A0A8I6WU79 | 355 | 38,776 |
| HvPap-22 | L-like E | MOREX.r3.1HG0004940 | A0A8I6WPU1 | 345 | 37,350 |
| HvPap-25 | L-like E | MOREX.r3.1HG0085150 | A0A8I7B430 | 355 | 38,295 |
| HvPap-27 | L-like E | MOREX.r3.3HG0245120 | A0A8I6WQD4 | 357 | 38,641 |
| HvPap-28 | L-like E | MOREX.r3.5HG0424540 | A0A8I6XSH6 | 396 | 43,119 |
| HvPap-29 | L-like E | MOREX.r3.3HG0245250 | A0A8I7B9F2 | 359 | 38,974 |
| HvPap-33 | L-like E | MOREX.r3.2HG0104490 | A0A8I6W9L7 | 355 | 37,217 |
| HvPap-34 | L-like E | MOREX.r3.2HG0106320 | F2EC73 | 360 | 39,066 |
| HvPap-35 | L-like E | MOREX.r3.2HG0212960 | A0A8I6WXF8 | 349 | 37,351 |
| HvPap-36 | L-like E | MOREX.r3.6HG0539010 | A0A8I6YPN0 | 383 | 41,925 |
| HvPap-37 | L-like E | MOREX.r3.4HG0418460 | F2DVR5 | 389 | 42,527 |
| HvPap-38 | L-like E | MOREX.r3.3HG0230090 | A0A8I6XUD2 | 390 | 42,342 |
| A0A8I6XNC2 | L-like E | MOREX.r3.3HG0271470 | A0A8I6XNC2 | 350 | 37,378 |
| F2CPT6 | L-like E | 10TJ18.Proj.3HG00173970 | F2CPT6 | 225 | 23,480 |
| HvPap-8 | (XBCP3) | MOREX.r3.1HG0076400 | B4ESF0 | 457 | 48,431 |
| A0A8I6XRJ4 | (THI1) | MOREX.r3.5HG0492390 | A0A8I6XRJ4 | 418 | 44,588 |
| HvPap-31 | N.S. | N.F. | F2E606 | 329 | 35,447 |
| HvPap-32 | N.S. | N.F. | F2E3H6 | 365 | 39,351 |
| HvPap-39 | N.S. | N.F. | F2E5Q3 | 333 | 37,013 |
| A0A8I6WZM8 | N.S. | MOREX.r3.2HG0208910 | A0A8I6WZM8 | 330 | 36,201 |
| Name | Species | UniProt ID | nAA | M.W., Da | Comparison with HvPap-6 | |
|---|---|---|---|---|---|---|
| Identity (%) | Similarity (%) | |||||
| HvPap-6 | Hordeum vulgare subsp. vulgare | A0A8I6WYU4 | 463 | 50,226 | - | - |
| HvPap-7 | Hordeum vulgare subsp. vulgare | F2E6V2 | 473 | 50,677 | 56.2 | 66.3 |
| Triticain α | Triticum aestivum | Q0WXG8 | 461 | 50,407 | 90.2 | 92.3 |
| Mir3 | Zea mays | O22500 | 480 | 51,787 | 71.5 | 77.8 |
| RD21A | Arabidopsis thaliana | P43297 | 462 | 50,966 | 62.2 | 73.6 |
| No. | Subfamily of PLCPs | A. thaliana by [13] | H. vulgare (Present Study) | Z. mays | ||
|---|---|---|---|---|---|---|
| By [13] | By [8] | By [37] | By [13] | |||
| 1 | RD21 | Cathepsin L-like D | 9 | 7 | 12 | 13 |
| 2 | CEP | Cathepsin L-like B | 3 | 12 | 7 | 3 |
| 3 | XCP | Cathepsin L-like C | 2 | 3 | 3 | 2 |
| 4 | XBCP3 | N.S. | 1 | 1 | 1 | 1 |
| 5 | THI1 | N.S. | 1 | 1 | 10 | 8 |
| 6 | SAG12 | Cathepsin L-like A | 6 | 8 | 11 | 5 |
| 7 | RD19 | Cathepsin F-like | 4 | 4 | 5 | 2 |
| 8 | ALP | Cathepsin H-like | 2 | 1 | 2 | 5 |
| 9 | CTB | Cathepsin B-like | 3 | 4 | 2 | 1 |
| N.S. | Cathepsin L-like E | N.F. | 18 | N.F. | N.F. | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schepetkin, I.A.; Fischer, A.M. Activity-Based Profiling of Papain-like Cysteine Proteases During Late-Stage Leaf Senescence in Barley. Plants 2025, 14, 3132. https://doi.org/10.3390/plants14203132
Schepetkin IA, Fischer AM. Activity-Based Profiling of Papain-like Cysteine Proteases During Late-Stage Leaf Senescence in Barley. Plants. 2025; 14(20):3132. https://doi.org/10.3390/plants14203132
Chicago/Turabian StyleSchepetkin, Igor A., and Andreas M. Fischer. 2025. "Activity-Based Profiling of Papain-like Cysteine Proteases During Late-Stage Leaf Senescence in Barley" Plants 14, no. 20: 3132. https://doi.org/10.3390/plants14203132
APA StyleSchepetkin, I. A., & Fischer, A. M. (2025). Activity-Based Profiling of Papain-like Cysteine Proteases During Late-Stage Leaf Senescence in Barley. Plants, 14(20), 3132. https://doi.org/10.3390/plants14203132







