Identification and Functional Analysis of Two UGT84 Glycosyltransferases in Flavonoid Biosynthesis of Carthamus tinctorius
Abstract
1. Introduction
2. Results
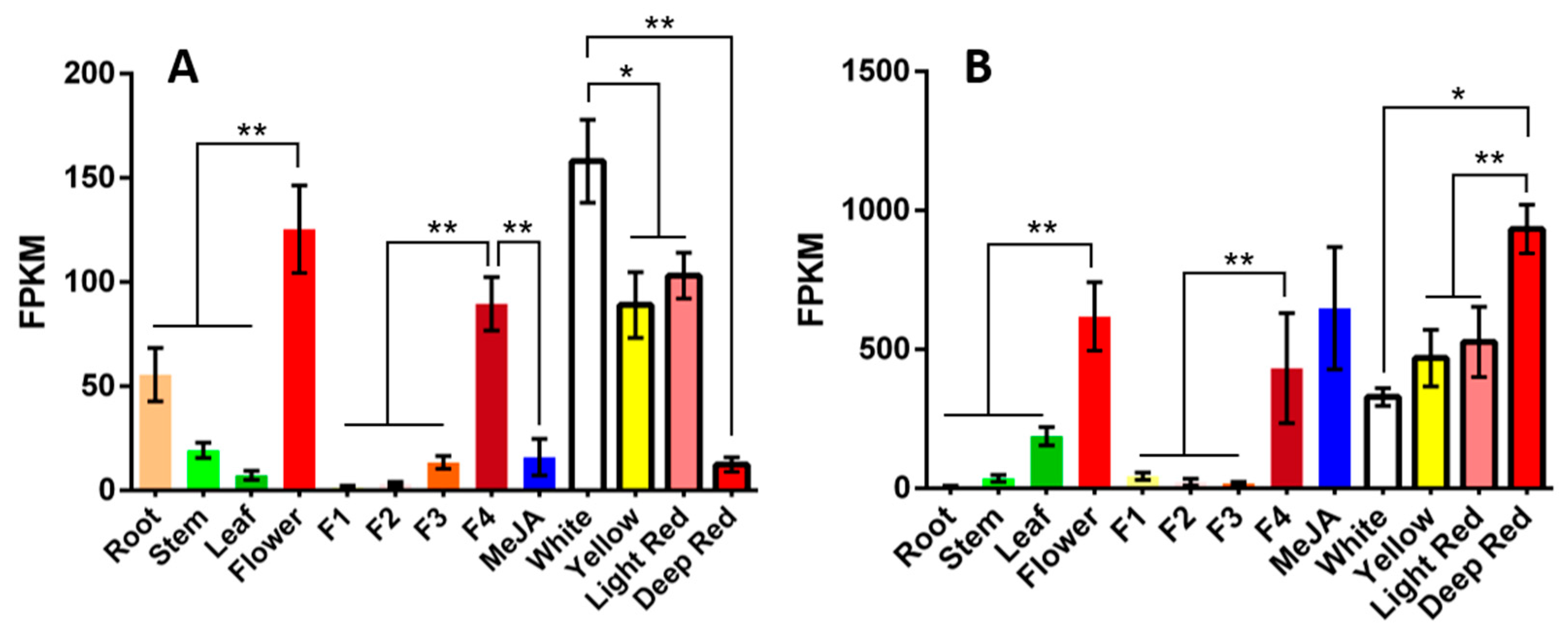
2.1. Mining and Bioinformatics Analysis of Candidate Genes
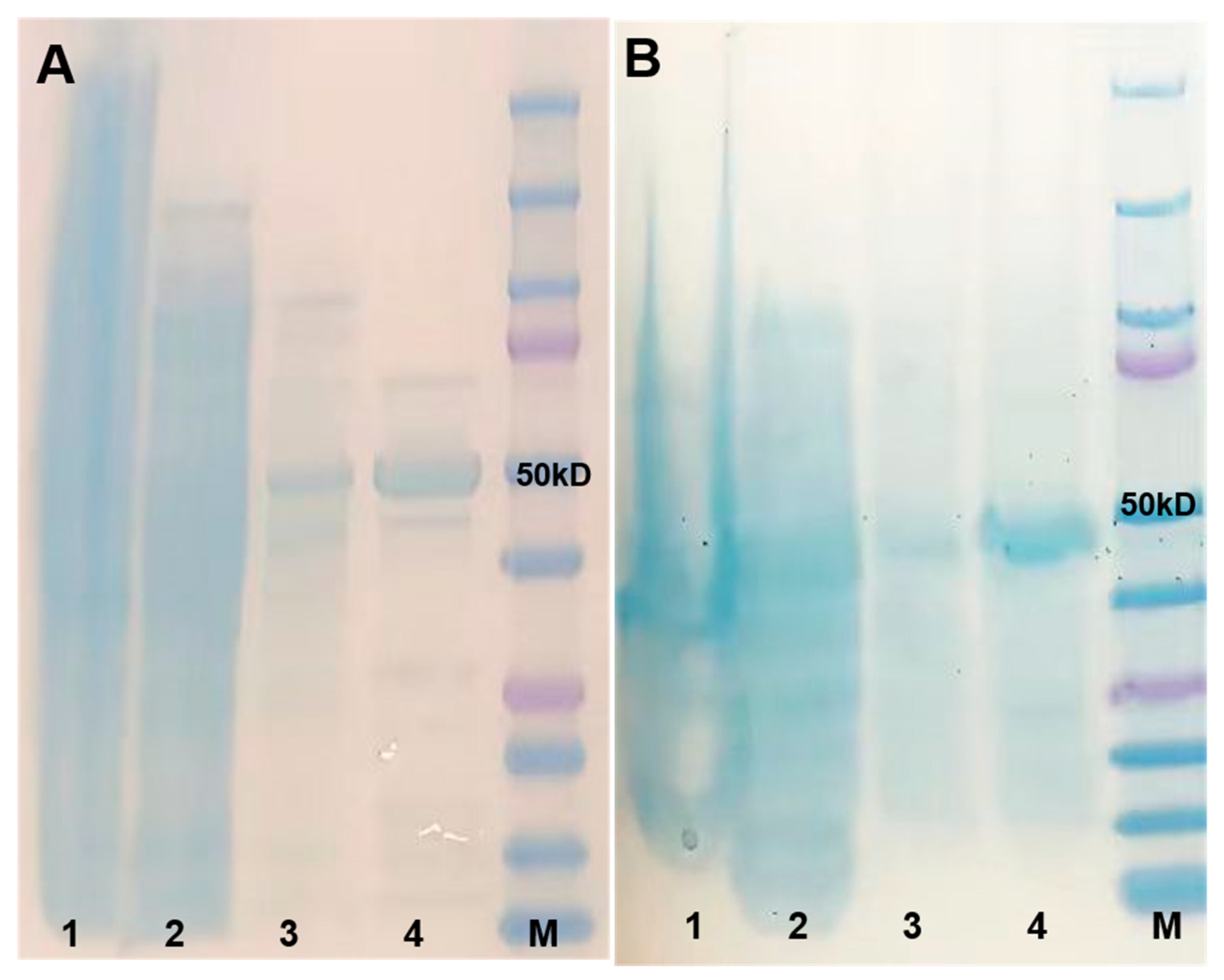
2.2. Molecular Cloning, Recombinant Protein Expression, and Purification of UGTs
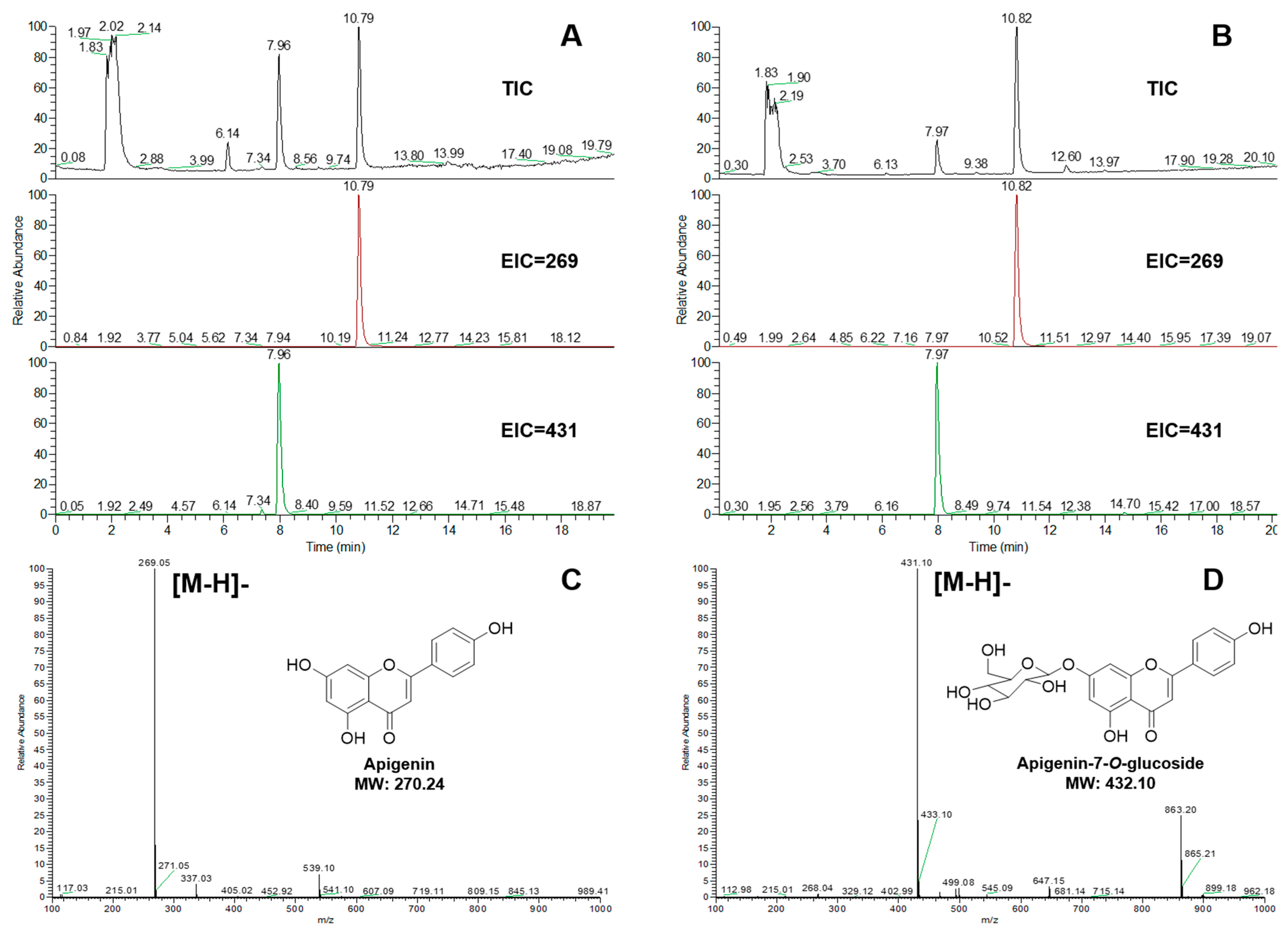
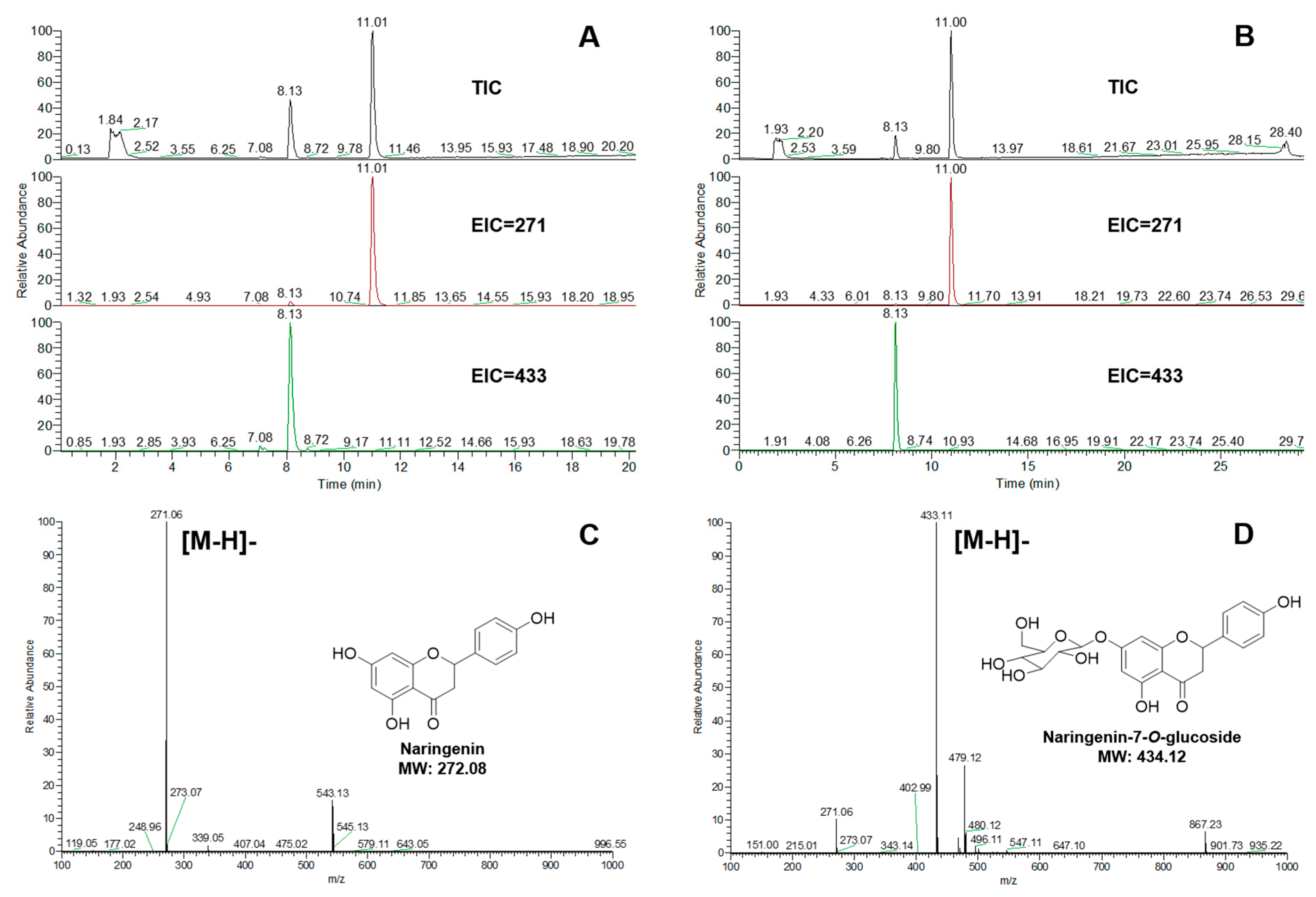
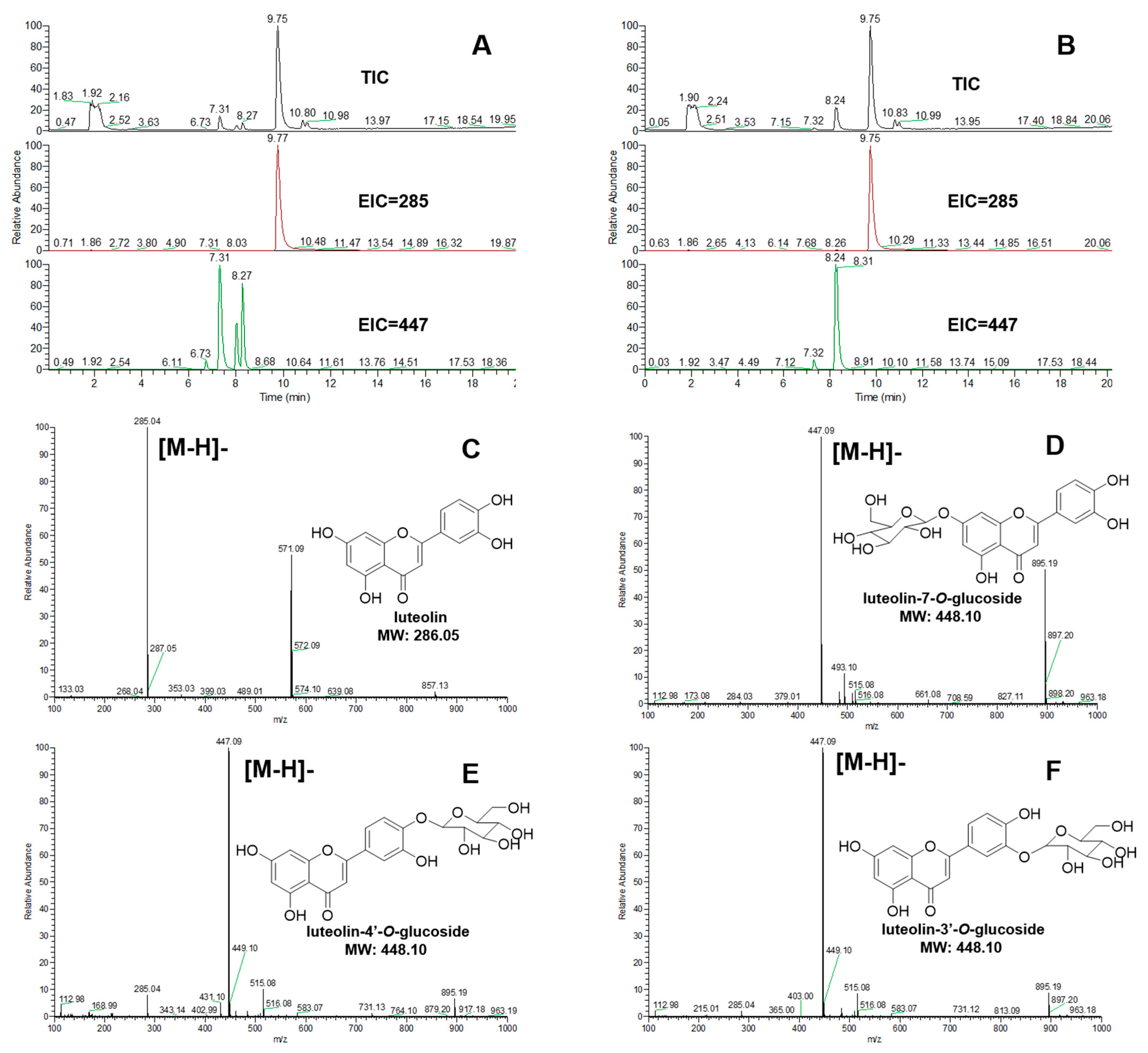
2.3. Validation of the Enzymatic Activity of UGTs In Vitro
2.4. Biochemical Properties of UGT84A28 and UGT84B3
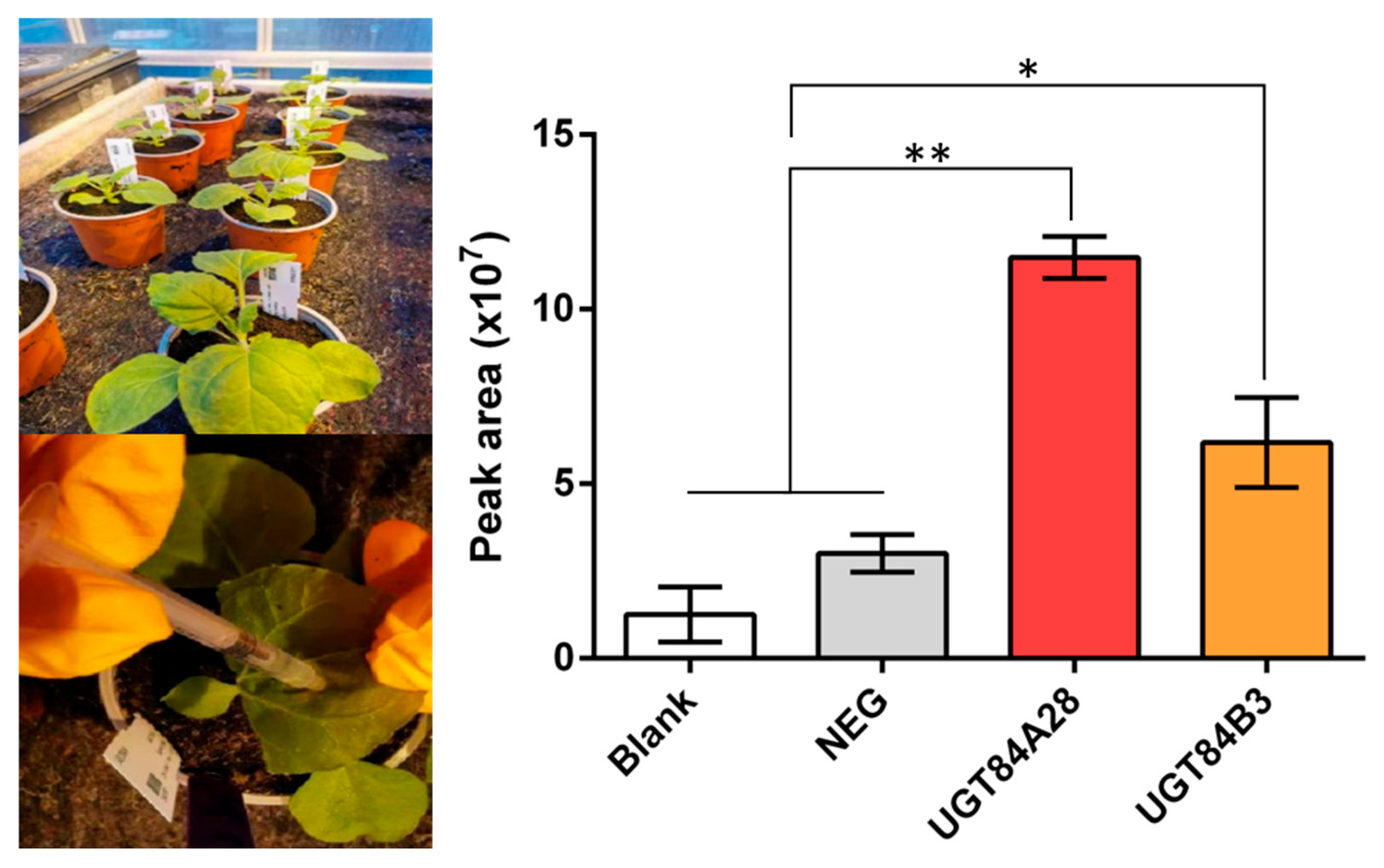
2.5. Validation of UGT84A28 and UGT84B3 Activity in Tobacco
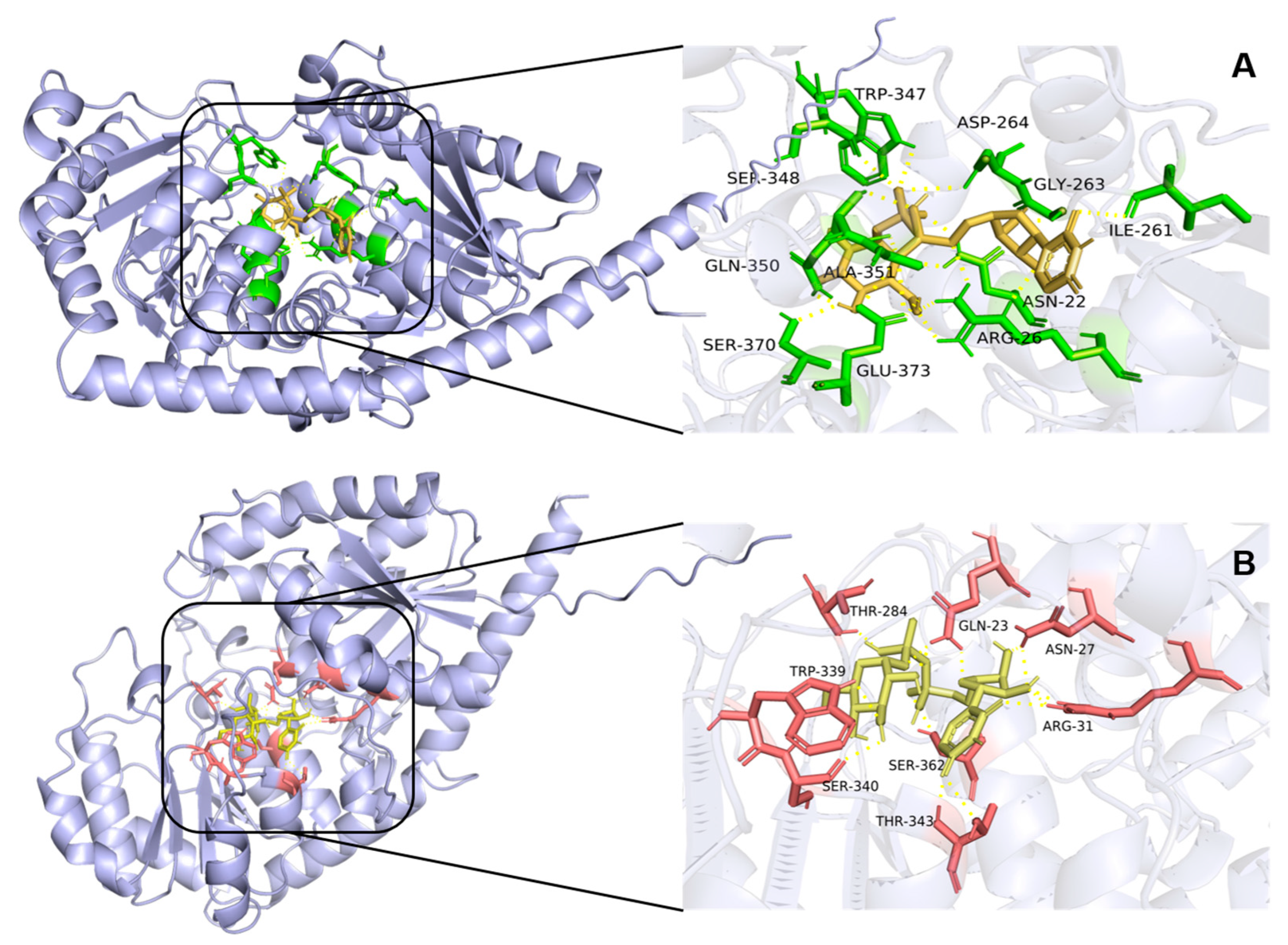
2.6. Molecular Docking of UGTs
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Screening and Bioinformatics Analysis of Candidate Genes
4.3. Molecular Cloning of Two Candidate UGT Genes and Expression of Recombination Proteins
4.4. Enzyme Activity Assays In Vitro
4.5. Determination of the Biochemical Properties
4.6. Enzyme Activity Validation in Nicotiana benthamiana
4.7. Molecular Docking
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, S.; Chen, Y.; Yang, Z.; Xiang, H.; Kang, P.; Li, J. Active substances and molecular mechanisms of the anti-myocardial ischemia effects of Carthami flos by network pharmacology and in vitro experiments. Heliyon 2023, 9, e13877. [Google Scholar] [CrossRef]
- Ju, S.; Liu, P.; Tan, L.; Tan, Y.; Li, X.; He, B.; Xia, Y.; Wang, M. Hydroxysafflor Yellow A Regulates Inflammation and Oxidative Stress by Suppressing the HIF-1α/JAK/STAT3 Signaling Pathway to Attenuate Osteoarthritis. Rev. Bras. Farmacogn. 2023, 33, 1022–1030. [Google Scholar] [CrossRef]
- Park, Y.G.; Cho, J.H.; Choi, J.; Ju, E.; Adam, G.; Hwang, D.; Lee, J.H.; An, S.Y.; Choi, H.K.; Park, C.B.; et al. Immunomodulatory effects of Curcuma longa L. and Carthamus tinctorius L. on RAW 264.7 macrophages and cyclophosphamide-induced immunosuppression C57BL/6 mouse models. J. Funct. Foods. 2022, 91, 105000. [Google Scholar] [CrossRef]
- Wang, R.; Ren, C.; Dong, S.; Chen, C.; Xian, B.; Wu, Q.; Wang, J.; Pei, J.; Chen, J. Integrated Metabolomics and Transcriptome Analysis of Flavonoid Biosynthesis in Safflower (Carthamus tinctorius L.) with Different Colors. Front. Plant Sci. 2021, 12, 712038. [Google Scholar] [CrossRef]
- Xian, B.; Wang, R.; Jiang, H.; Zhou, Y.; Yan, J.; Huang, X.; Chen, J.; Wu, Q.; Chen, C.; Xi, Z.; et al. Comprehensive review of two groups of flavonoids in Carthamus tinctorius L. Biomed. Pharmacother. 2022, 153, 113462. [Google Scholar] [CrossRef]
- Liu, W.; Feng, Y.; Yu, S.; Fan, Z.; Li, X.; Li, J.; Yin, H. The Flavonoid Biosynthesis Network in Plants. Int. J. Mol. Sci. 2021, 22, 12824. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, X.; Liu, Y.; Cao, B. Regulation of flavanone 3-hydroxylase gene involved in the flavonoid biosynthesis pathway in response to UV-B radiation and drought stress in the desert plant, Reaumuria soongorica. Plant Physiol. Biochem. 2013, 73, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Fu, H.; Guo, X.; Zhang, J.; Wang, P.; Mu, X.; Zhang, S.; Du, J. Genome-Wide Identification of MYB Family Genes in Cerasus humilis Reveals Candidate Genes Involved in Flavonoid Biosynthesis. Plant Mol. Biol. Rep. 2023, 41, 699–712. [Google Scholar] [CrossRef]
- Wang, Z.; Yu, Q.; Shen, W.; El Mohtar, C.A.; Zhao, X.; Gmitter, F.G. Functional study of CHS gene family members in citrus revealed a novel CHS gene affecting the production of flavonoids. BMC Plant Biol. 2018, 18, 189. [Google Scholar] [CrossRef]
- Chen, J.; Tang, X.; Ren, C.; Wei, B.; Wu, Y.; Wu, Q.; Pei, J. Full-length transcriptome sequences and the identification of putative genes for flavonoid biosynthesis in safflower. BMC Genom. 2018, 19, 548. [Google Scholar] [CrossRef]
- Choi, S.H.; Kim, H.S.; Yoon, Y.J.; Kim, D.; Lee, E.Y. Glycosyltransferase and its application to glycodiversification of natural products. J. Ind. Eng. Chem. 2012, 18, 1208–1212. [Google Scholar] [CrossRef]
- Rehman, H.M.; Nawaz, M.A.; Shah, Z.H.; Ludwig-Müller, J.; Chung, G.; Ahmad, M.Q.; Yang, S.H.; Lee, S.I. Author Correction: Comparative genomic and transcriptomic analyses of Family-1 UDP glycosyltransferase in three Brassica species and Arabidopsis indicates stress-responsive regulation. Sci. Rep. 2018, 8, 6237. [Google Scholar] [CrossRef]
- Yao, Y.; Gu, J.; Luo, Y.; Wang, Y.; Pang, Y.; Shen, G.; Guo, B. Genome-wide analysis of UGT gene family identified key gene for the biosynthesis of bioactive flavonol glycosides in Epimedium pubescens Maxim. Synth. Syst. Biotechnol. 2022, 7, 1095–1107. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Huang, X.X.; Zhao, S.M.; Xiao, D.W.; Xiao, L.T.; Tong, J.H.; Wang, W.S.; Li, Y.J.; Ding, Z.; Hou, B.K. IPyA glucosylation mediates light and temperature signaling to regulate auxin-dependent hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2020, 117, 6910–6917. [Google Scholar] [CrossRef]
- Sasaki, N.; Nishizaki, Y.; Yamada, E.; Tatsuzawa, F.; Nakatsuka, T.; Takahashi, H.; Nishihara, M. Identification of the glucosyltransferase that mediates direct flavone C-glucosylation in Gentiana triflora. FEBS Lett. 2015, 589, 182–187. [Google Scholar] [CrossRef]
- Li, T.; Borg, A.J.E.; Krammer, L.; Weber, H.; Breinbauer, R.; Nidetzky, B. Discovery, characterization, and comparative analysis of new UGT72 and UGT84 family glycosyltransferases. Commun. Chem. 2024, 7, 147. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Zhang, S.; Qiu, N.; Liu, L.; Wang, W.; Xie, Q.; Chang, Q.; Chen, Q. Identification and Characterization of Glucosyltransferase That Forms 1-Galloyl-β-d-Glucogallin in Canarium album L., a Functional Fruit Rich in Hydrolysable Tannins. Molecules 2021, 26, 4650. [Google Scholar] [CrossRef]
- Dembitsky, V.M. Chemistry and biodiversity of the biologically active natural glycosides. Chem. Biodivers. 2004, 1, 673–781. [Google Scholar] [CrossRef] [PubMed]
- Mateo-Bonmatí, E.; Casanova-Sáez, R.; Šimura, J.; Ljung, K. Broadening the roles of UDP-glycosyltransferases in auxin homeostasis and plant development. New Phytol. 2021, 232, 642–654. [Google Scholar] [CrossRef]
- Gao, J.; Chen, Y.; Gao, M.; Zhao, Y.; Wang, Y. WRKY14-DPB Module Enhances Drought Tolerance by Activating the Expression of UGT84B1 Involved in Hydrolyzable Tannin Biosynthesis. Plant Cell Environ. 2025. published online. [Google Scholar] [CrossRef]
- Xie, K.; Chen, R.; Li, J.; Wang, R.; Chen, D.; Dou, X.; Dai, J. Exploring the catalytic promiscuity of a new glycosyltransferase from Carthamus tinctorius. Org. Lett. 2014, 16, 4874–4877. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Chen, R.; Chen, D.; Li, J.; Wang, R.; Yang, L.; Dai, J. Enzymatic N-glycosylation of diverse arylamine aglycones by a promiscuous glycosyltransferase from Carthamus tinctorius. Adv. Synth. Catal. 2017, 359, 603–608. [Google Scholar] [CrossRef]
- Ren, C.; Xi, Z.; Xian, B.; Chen, C.; Huang, X.; Jiang, H.; Chen, J.; Peng, C.; Pei, J. Identification and Characterization of CtUGT3 as the Key Player of Astragalin Biosynthesis in Carthamus tinctorius L. J. Agric. Food Chem. 2023, 71, 16221–16232. [Google Scholar] [CrossRef] [PubMed]
- Xi, Z.; Li, Y.; Liu, S.; Wang, D.; Guo, J.; Xian, B.; Rao, K.; Chen, C.; Peng, Y.; Zhou, Y.; et al. Functional analysis and molecular characterization of UGT95A2, a specialized glycosyltransferase for flavonoid 3’-O-glycosylation in Carthamus tinctorius L. Plant J. 2025, 122, e70213. [Google Scholar] [CrossRef]
- Sui, S.Y.; Guo, R.M.; Xie, K.B.; Yang, L.; Dai, J.G. UGT88B2: A promiscuous O-glycosyltransferase from Carthamus tinctorius. Chin. Herb. Med. 2020, 12, 440–445. [Google Scholar] [CrossRef]
- Wang, Z.L.; Wang, H.T.; Chang, G.; Ye, G.; Zhang, M.; Chen, J.; Ye, M. Elucidation of the biosynthetic pathway of hydroxysafflor yellow A. Nat. Commun. 2025, 16, 4489. [Google Scholar] [CrossRef]
- Liu, N.; Zou, Y.; Jiang, Z.; Tu, L.; Wu, X.; Li, D.; Wang, J.; Huang, L.; Xu, C.; Gao, W. Multiomics driven identification of glycosyltransferases in flavonoid glycoside biosynthesis in safflower. Hortic. Plant J. 2024. [Google Scholar] [CrossRef]
- Wilson, A.E.; Feng, X.; Ono, N.N.; Holland, D.; Amir, R.; Tian, L. Characterization of a UGT84 Family Glycosyltransferase Provides New Insights into Substrate Binding and Reactivity of Galloylglucose Ester-Forming UGTs. Biochemistry 2017, 56, 6389–6400. [Google Scholar] [CrossRef]
- Xia, T.; Eiteman, M.A. Quercetin Glucoside Production by Engineered Escherichia coli. Appl. Biochem. Biotechnol. 2017, 182, 1358–1370. [Google Scholar] [CrossRef]
- Mashima, K.; Hatano, M.; Suzuki, H.; Shimosaka, M.; Taguchi, G. Identification and Characterization of Apigenin 6-C-Glucosyltransferase Involved in Biosynthesis of Isosaponarin in Wasabi (Eutrema japonicum). Plant Cell Physiol. 2019, 60, 2733–2743. [Google Scholar] [CrossRef]
- Jackson, R.G.; Kowalczyk, M.; Li, Y.; Higgins, G.; Ross, J.; Sandberg, G.; Bowles, D.J. Over-expression of an Arabidopsis gene encoding a glucosyltransferase of indole-3-acetic acid: Phenotypic characterisation of transgenic lines. Plant J. 2002, 32, 573–583. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New docking methods, expanded force field, and python bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Janson, G.; Paiardini, A. PyMod 3: A complete suite for structural bioinformatics in PyMOL. Bioinformatics 2021, 37, 1471–1472. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, C.; Guo, J.; Liu, S.; Xian, B.; Li, Y.; Yang, C.; Peng, C.; Pei, J.; Chen, J. Identification and Functional Analysis of Two UGT84 Glycosyltransferases in Flavonoid Biosynthesis of Carthamus tinctorius. Plants 2025, 14, 3112. https://doi.org/10.3390/plants14193112
Ren C, Guo J, Liu S, Xian B, Li Y, Yang C, Peng C, Pei J, Chen J. Identification and Functional Analysis of Two UGT84 Glycosyltransferases in Flavonoid Biosynthesis of Carthamus tinctorius. Plants. 2025; 14(19):3112. https://doi.org/10.3390/plants14193112
Chicago/Turabian StyleRen, Chaoxiang, Jinxin Guo, Siyu Liu, Bin Xian, Yuhang Li, Changyan Yang, Cheng Peng, Jin Pei, and Jiang Chen. 2025. "Identification and Functional Analysis of Two UGT84 Glycosyltransferases in Flavonoid Biosynthesis of Carthamus tinctorius" Plants 14, no. 19: 3112. https://doi.org/10.3390/plants14193112
APA StyleRen, C., Guo, J., Liu, S., Xian, B., Li, Y., Yang, C., Peng, C., Pei, J., & Chen, J. (2025). Identification and Functional Analysis of Two UGT84 Glycosyltransferases in Flavonoid Biosynthesis of Carthamus tinctorius. Plants, 14(19), 3112. https://doi.org/10.3390/plants14193112





