Small Population Size and Low Levels of Genetic Diversity in an Endangered Species Endemic to the Western Tianshan Mountains
Abstract
1. Introduction
2. Results
2.1. SNPS from RAD-seq Analysis of A. nanus
2.2. Genetic Diversity of A. nanus
2.3. Genetic Structure of A. nanus
3. Discussion
3.1. Genetic Diversity of A. nanus
3.2. Genetic Structure of A. nanus
3.3. Conservation Implications
4. Materials and Methods
4.1. Plant Materials
4.2. DNA Extraction and RAD Sequencing
4.3. SNP Calling
4.4. Genetic Diversity and Structure
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ellwanger, C.; Steger, L.; Pollack, C.; Wells, R.; Benjamin Fant, J. Anthropogenic fragmentation increases risk of genetic decline in the threatened orchid Platanthera leucophaea. Ecol. Evol. 2022, 12, e8578. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, R.; Cristóbal-Pérez, E.J.; Balvino-Olvera, F.J.; de Jesús Aguilar-Aguilar, M.; Aguirre-Acosta, N.; Ashworth, L.; Lobo, J.A.; Martén-Rodríguez, S.; Fuchs, E.J.; Sanchez-Montoya, G.; et al. Habitat fragmentation reduces plant progeny quality: A global synthesis. Ecol. Lett. 2019, 22, 1163–1173. [Google Scholar] [CrossRef]
- Soons, M.B.; Messelink, J.H.; Jongejans, E.; Heil, G.W. Habitat fragmentation reduces grassland connectivity for both short-distance and long-distance wind-dispersed forbs. J. Ecol. 2005, 93, 1214–1225. [Google Scholar] [CrossRef]
- Schlaepfer, D.R.; Braschler, B.; Rusterholz, H.-P.; Baur, B. Genetic effects of anthropogenic habitat fragmentation on remnant animal and plant populations: A meta-analysis. Ecosphere 2018, 9, e02488. [Google Scholar] [CrossRef]
- Chen, X.Y. Effects of habitat fragmentation on genetic structure of plant populations and implications for the biodiversity conservation. Acta Ecol. Sin. 2000, 20, 9. [Google Scholar] [CrossRef]
- Honnay, O.; Jacquemyn, H. Susceptibility of common and rare plant species to the genetic consequences of habitat fragmentation. Conserv. Biol. 2007, 21, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Morden, C.W.; Loeffler, W. Fragmentation and genetic differentiation among subpopulations of the endangered Hawaiian mint Haplostachys haplostachya (Lamiaceae). Mol. Ecol. 1999, 8, 617–625. [Google Scholar] [CrossRef]
- Evans, M.J.; Banks, S.C.; Driscoll, D.A.; Hicks, A.J.; Melbourne, B.A.; Davies, K.F. Short- and long-term effects of habitat fragmentation differ but are predicted by response to the matrix. Ecology 2017, 98, 807–819. [Google Scholar] [CrossRef]
- Cui, H.B. Fabaceae (5). In Flora of China; Wu, Z.Y., Raven, P.H., Eds.; Science Press: Beijing, China, 1998; Volume 42, pp. 388–397. Available online: https://www.iplant.cn/info/Ammopiptanthus%20nanus?t=z (accessed on 12 October 2024).
- Cao, S.; Wang, Y.; Li, X.; Gao, F.; Feng, J.; Zhou, Y. Characterization of the AP2/ERF transcription factor family and expression profiling of DREB subfamily under cold and osmotic stresses in Ammopiptanthus nanus. Plants 2020, 9, 455. [Google Scholar] [CrossRef]
- Shen, G.M. Fabaceae-Apiaceae. In Flora Xinjiangensis; Shen, G.M., Mao, Z.M., Eds.; Xinjiang Science & Technology Press: Xinjiang, China, 2011; Volume 3, pp. 11–12. [Google Scholar]
- Fu, G.L. Rare and Endangered Plants in China; Science Press: Bejing, China, 1991; Volume 1, pp. 370–371. [Google Scholar]
- Brooks, E.; Slender, A.L.; Cu, S.; Breed, M.F.; Stangoulis, J.C. A range-wide analysis of population structure and genomic variation within the critically endangered spiny daisy (Acanthocladium dockeri). Conserv. Genet. 2022, 23, 1027–1037. [Google Scholar] [CrossRef]
- Chen, G.-Q.; Huang, H.-W.; Crawford, D.J.; Pan, B.-R.; Ge, X.-J. Mating system and genetic diversity of a rare desert legume Ammopiptanthus nanus (Leguminosae). J. Syst. Evol. 2009, 47, 57–66. [Google Scholar] [CrossRef]
- Su, Z.; Richardson, B.A.; Zhuo, L.; Jiang, X.; Li, W.; Kang, X. Genetic diversity and structure of an endangered desert shrub and the implications for conservation. AoB Plants 2017, 9, plx016. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, X.; Lu, T.; Wang, J.; Shi, W. Identification of the efficacy of ex situ conservation of Ammopiptanthus nanus based on its ETS-SSR markers. Plants 2023, 12, 2670. [Google Scholar] [CrossRef]
- Ge, X.-J.; Yu, Y.; Yuan, Y.-M.; Huang, H.-W.; Yan, C. Genetic diversity and geographic differentiation in endangered Ammopiptanthus (Leguminosae) populations in desert regions of northwest China as revealed by ISSR analysis. Ann. Bot. 2005, 95, 843–851. [Google Scholar] [CrossRef]
- Basak, M.; Uzun, B.; Yol, E. Genetic diversity and population structure of the Mediterranean sesame core collection with use of genome-wide SNPs developed by double digest RAD-Seq. PLoS ONE 2019, 14, e0223757. [Google Scholar] [CrossRef] [PubMed]
- Peterson, B.K.; Weber, J.N.; Kay, E.H.; Fisher, H.S.; Hoekstra, H.E. Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 2012, 7, e37135. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evol. Int. J. Org. Evol. 1984, 38, 1358–1370. [Google Scholar] [CrossRef]
- Wright, S. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 1965, 19, 395–420. [Google Scholar] [CrossRef]
- Potts, J.; Michael, V.N.; Meru, G.; Wu, X.; Blair, M.W. Dissecting the genetic diversity of USDA cowpea germplasm collection using kompetitive allele specific PCR-single nucleotide polymorphism markers. Genes 2024, 15, 362. [Google Scholar] [CrossRef]
- Atasagun, B. Assessment of the genetic diversity of a critically endangered species Centaurea amaena (Asteraceae). Arch. Biol. Sci. 2022, 74, 325–332. [Google Scholar] [CrossRef]
- Du, Y.; Yu, X.M.; Wang, P.; Li, Q.; Wang, Y.X.; Zhang, B. Genetic diversity analysis of wild Medicago falcata L. in Xinjiang based on SNP molecular markers. Feed Res. 2024, 47, 68–73. [Google Scholar] [CrossRef]
- Shaibu, A.S.; Ibrahim, H.; Miko, Z.L.; Mohammed, I.B.; Mohammed, S.G.; Yusuf, H.L.; Kamara, A.Y.; Omoigui, L.O.; Karikari, B. Assessment of the genetic structure and diversity of Soybean (Glycine max L.) germplasm using diversity array technology and single nucleotide polymorphism markers. Plants 2021, 11, 68. [Google Scholar] [CrossRef] [PubMed]
- Frankham, R. Conservation genetics. Ann. Rev. Genet. 1995, 29, 305–327. [Google Scholar] [CrossRef]
- Young, A.; Boyle, T.; Brown, T. The population genetic consequences of habitat fragmentation for plants. Trends Ecol. Evol. 1996, 11, 413–418. [Google Scholar] [CrossRef]
- Li, S.; Gan, X.; Han, H.; Zhang, X.; Tian, Z. Low within-population genetic diversity and high genetic differentiation among populations of the endangered plant Tetracentron sinense Oliver revealed by inter-simple sequence repeat analysis. Ann. For. Sci. 2018, 75, 74. [Google Scholar] [CrossRef]
- Du, Z.; He, Y.; Wang, H.; Wang, C.; Duan, Y. Potential geographical distribution and habitat shift of the genus Ammopiptanthus in China under current and future climate change based on the MaxEnt model. J. Arid Environ. 2021, 184, 104328. [Google Scholar] [CrossRef]
- Anto, M.; Anjala, M.; Jothish, P.S.; Rameshkumar, K.B.; Padmesh, P.; Anilkumar, C. Population genetic structure of Garcinia imberti Bourd. an endangered endemic tree of southern Western Ghats, India. Plant Sci. Today 2020, 7, 424–431. [Google Scholar] [CrossRef]
- Li, A.; Ma, M.; Li, H.; He, S.; Wang, S. Genetic diversity and population differentiation of a Chinese endangered plant Ammopiptanthus nanus (M. Pop.) Cheng f. Genes 2023, 14, 1020. [Google Scholar] [CrossRef]
- Chávez-Cortázar, A.; Oyama, K.; Ochoa-Zavala, M.; Mata-Rosas, M.; Veltjen, E.; Samain, M.S.; Quesada, M. Conservation genetics of relict tropical species of Magnolia (section Macrophylla). Conserv. Genet. 2021, 22, 259–273. [Google Scholar] [CrossRef]
- Cai, C.; Xiao, J.; Ci, X.; Conran, J.G.; Li, J. Genetic diversity of Horsfieldia tetratepala (Myristicaceae), an endangered plant species with extremely small populations to China: Implications for its conservation. Plant Syst. Evol. 2021, 307, 50. [Google Scholar] [CrossRef]
- Shi, W.; Su, Z.H.; Liu, P.L.; Pan, B.R.; Zhao, Y.F.; Wang, J.C. Molecular, Karyotypic, and Morphological evidence for Ammopiptanthus (Fabaceae) taxonomy. Ann. Mo. Bot. Gard. 2017, 102, 559–573. [Google Scholar] [CrossRef] [PubMed]
- Cristóbal-Pérez, E.J.; Fuchs, E.J.; Martén-Rodríguez, S.; Quesada, M. Habitat fragmentation negatively affects effective gene flow via pollen, and male and female fitness in the dioecious tree, Spondias purpurea (Anacardiaceae). Biol. Conserv. 2021, 256, 109007. [Google Scholar] [CrossRef]
- Zhuo, L.; Su, Z.; Zhao, H.; Jiang, X.; Zhang, L. Genetic structure of two endangered shrubs in Central Asia and northwestern China and the implications for conservation. Plant Syst. Evol. 2024, 310, 1. [Google Scholar] [CrossRef]
- Kashimshetty, Y.; Pelikan, S.; Rogstad, S.H. Effective seed harvesting strategies for the ex situ genetic diversity conservation of rare tropical tree populations. Biodivers. Conserv. 2017, 26, 1311–1331. [Google Scholar] [CrossRef]
- Chen, G.; Sun, W. The role of botanical gardens in scientific research, conservation, and citizen science. Plant Divers. 2018, 40, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Blackmore, S.; Gibby, M.; Rae, D. Strengthening the scientific contribution of botanic gardens to the second phase of the Global Strategy for Plant Conservation. Bot. J. Linn. Soc. 2011, 166, 267–281. [Google Scholar] [CrossRef]
- Ottewell, K.M.; Bickerton, D.C.; Byrne, M.; Lowe, A.J. Bridging the gap: A genetic assessment framework for population-level threatened plant conservation prioritization and decision-making. Divers. Distrib. 2016, 22, 174–188. [Google Scholar] [CrossRef]
- Wei, H.; Wu, P.; Ge, X.; Liu, M.; Wei, X. Chemical constituents of the seeds of Ammopiptanthus (Leguminosae) and their systematic and ecological significance. Biochem. Syst. Ecol. 2007, 35, 274–280. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Michaels, S.D.; John, M.C.; Amasino, R.M. Removal of polysaccharides from plant DNA by ethanol precipitation. BioTechniques 1994, 17, 274–276. [Google Scholar]
- Catchen, J.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Marinček, P.; Pittet, L.; Wagner, N.D.; Hörandl, E. Evolution of a hybrid zone of two willow species (Salix L.) in the European Alps analyzed by RAD-seq and morphometrics. Ecol. Evol. 2023, 13, e9700. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Mastretta-Yanes, A.; Arrigo, N.; Alvarez, N.; Jorgensen, T.H.; Piñero, D.; Emerson, B.C. Restriction site-associated DNA sequencing, genotyping error estimation and de novo assembly optimization for population genetic inference. Mol. Ecol. Resour. 2015, 15, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Rochette, N.C.; Catchen, J.M. Deriving genotypes from RAD-seq short-read data using Stacks. Nat. Protoc. 2017, 12, 2640–2659. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2012. [Google Scholar]
- Oksanen, J.; Kindt, R.; Legendre, P.; O’Hara, B.; Stevens, M.H.H.; Oksanen, M.J.; Suggests, M.A.S.S. The vegan package. Community Ecol. Package 2009, 10, 719. [Google Scholar]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef]
- Chen, H.; Boutros, P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Posada, D.; Crandall, K.A.; Templeton, A.R. GeoDis: A program for the cladistic nested analysis of the geographical distribution of genetic haplotypes. Mol. Ecol. 2000, 9, 487–488. [Google Scholar] [CrossRef] [PubMed]
- Pickrell, J.K.; Pritchard, J.K. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012, 8, e1002967. [Google Scholar] [CrossRef] [PubMed]
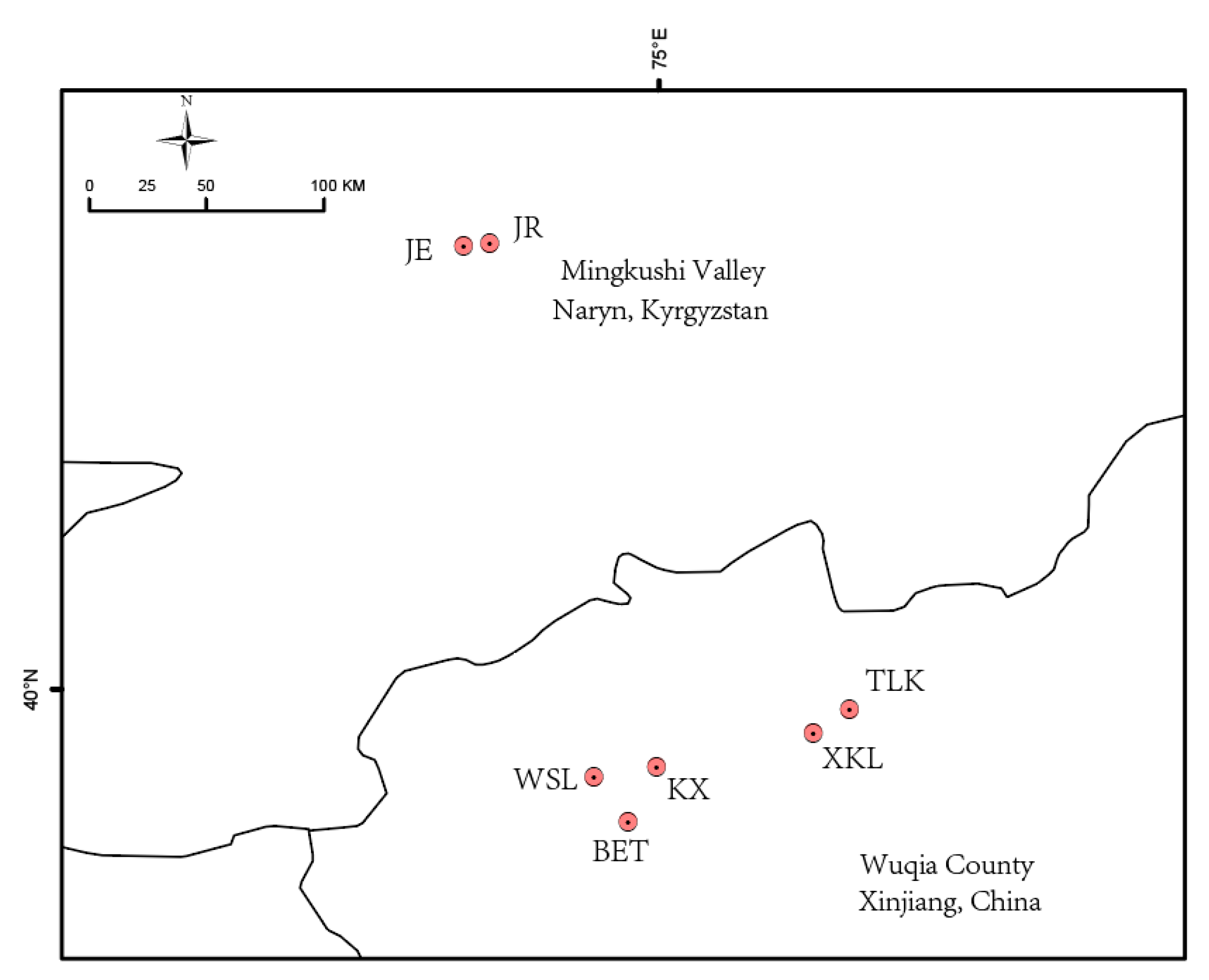
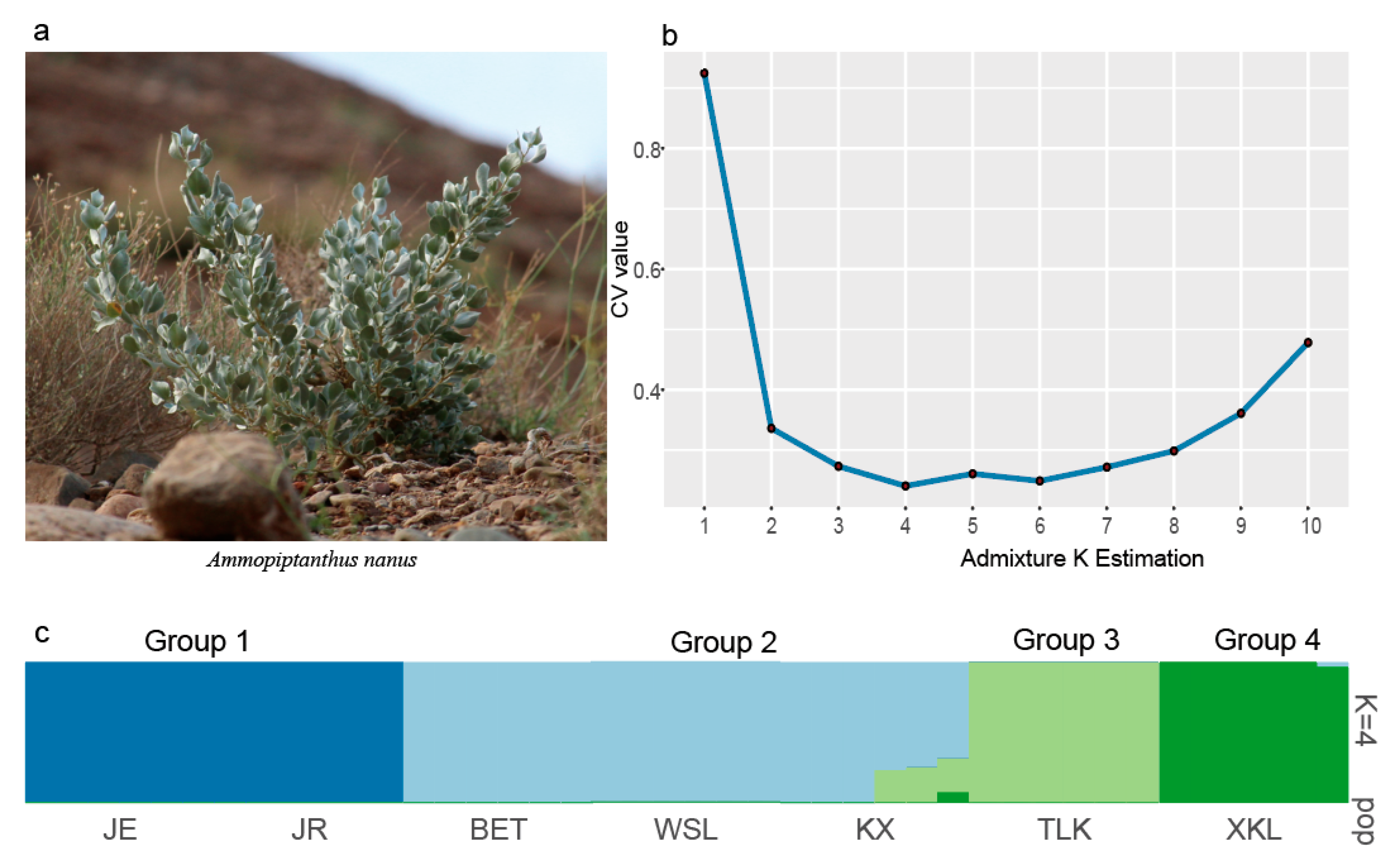





| Population ID | Latitude | Longitude | Altitude (m) | Sample | Ho | HE | π | FIS |
|---|---|---|---|---|---|---|---|---|
| JR | 74.35 | 41.71 | 1730 | 6 | 0.12 | 0.09 | 0.11 | −0.01 |
| JE | 74.25 | 41.70 | 1693 | 6 | 0.11 | 0.09 | 0.10 | −0.01 |
| WSL | 74.75 | 39.66 | 2290 | 6 | 0.09 | 0.08 | 0.09 | 0.00 |
| KX | 74.99 | 39.70 | 2167 | 6 | 0.10 | 0.11 | 0.13 | 0.05 |
| BET | 74.88 | 39.49 | 2512 | 6 | 0.11 | 0.10 | 0.11 | 0.01 |
| TLK | 75.73 | 39.92 | 2212 | 6 | 0.10 | 0.08 | 0.09 | −0.01 |
| XKL | 75.59 | 39.83 | 2109 | 6 | 0.10 | 0.09 | 0.10 | 0.01 |
| Population | JR | JE | WSL | KX | BET | TLK | XKL |
|---|---|---|---|---|---|---|---|
| JR | 0.1822 | 0.7002 | 0.6575 | 0.6687 | 0.7138 | 0.7024 | |
| JE | 0.7187 | 0.6810 | 0.6933 | 0.7339 | 0.7078 | ||
| WSL | 0.2550 | 0.2073 | 0.3966 | 0.3610 | |||
| KX | 0.1579 | 0.3177 | 0.3020 | ||||
| BET | 0.3469 | 0.3107 | |||||
| TLK | 0.3332 | ||||||
| XKL |
| Source of Variation | d.f. | Sum of Squares | Variance Components | Percentage of Variation |
|---|---|---|---|---|
| Among populations | 6 | 351,886.83 | 9671.65 | 93.99 * |
| Within populations | 35 | 21,627.00 | 617.91 | 6.01 * |
| Total | 41 | 373,513.84 | 10,289.56 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, Z.; Lin, Z.; Zhuo, L.; Jiang, X.; Jiang, Q. Small Population Size and Low Levels of Genetic Diversity in an Endangered Species Endemic to the Western Tianshan Mountains. Plants 2025, 14, 3105. https://doi.org/10.3390/plants14193105
Su Z, Lin Z, Zhuo L, Jiang X, Jiang Q. Small Population Size and Low Levels of Genetic Diversity in an Endangered Species Endemic to the Western Tianshan Mountains. Plants. 2025; 14(19):3105. https://doi.org/10.3390/plants14193105
Chicago/Turabian StyleSu, Zhihao, Zhiye Lin, Li Zhuo, Xiaolong Jiang, and Qichuan Jiang. 2025. "Small Population Size and Low Levels of Genetic Diversity in an Endangered Species Endemic to the Western Tianshan Mountains" Plants 14, no. 19: 3105. https://doi.org/10.3390/plants14193105
APA StyleSu, Z., Lin, Z., Zhuo, L., Jiang, X., & Jiang, Q. (2025). Small Population Size and Low Levels of Genetic Diversity in an Endangered Species Endemic to the Western Tianshan Mountains. Plants, 14(19), 3105. https://doi.org/10.3390/plants14193105







