Impact of Alien Chromosome Introgression from Thinopyrum ponticum on Wheat Grain Traits
Abstract
1. Introduction
2. Results
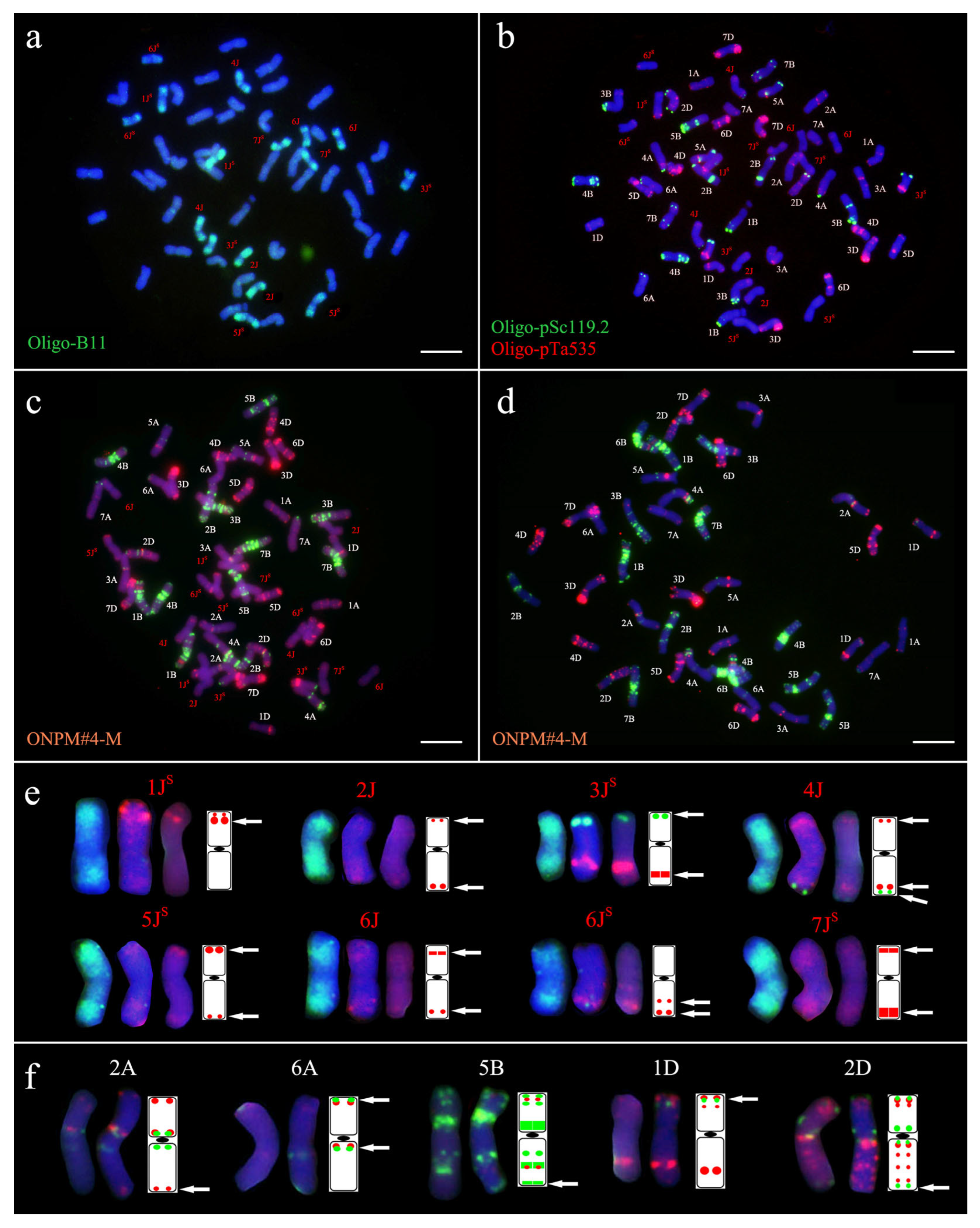
2.1. Chromosomal Composition of Xiaoyan 7430 and Yannong 1212
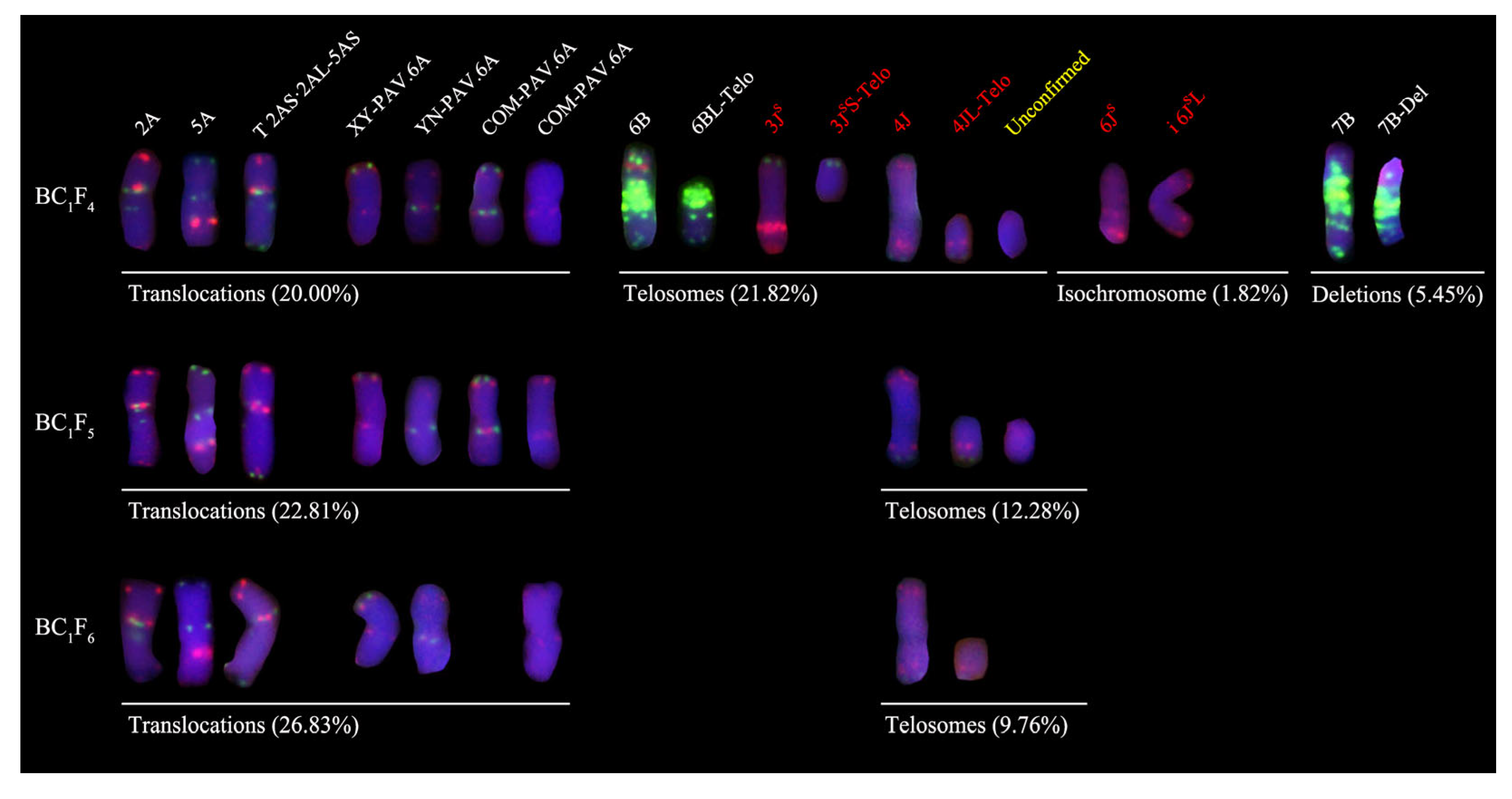
2.2. Analysis of Chromosomal Variations in BC1F4–BC1F6-Derived Lines
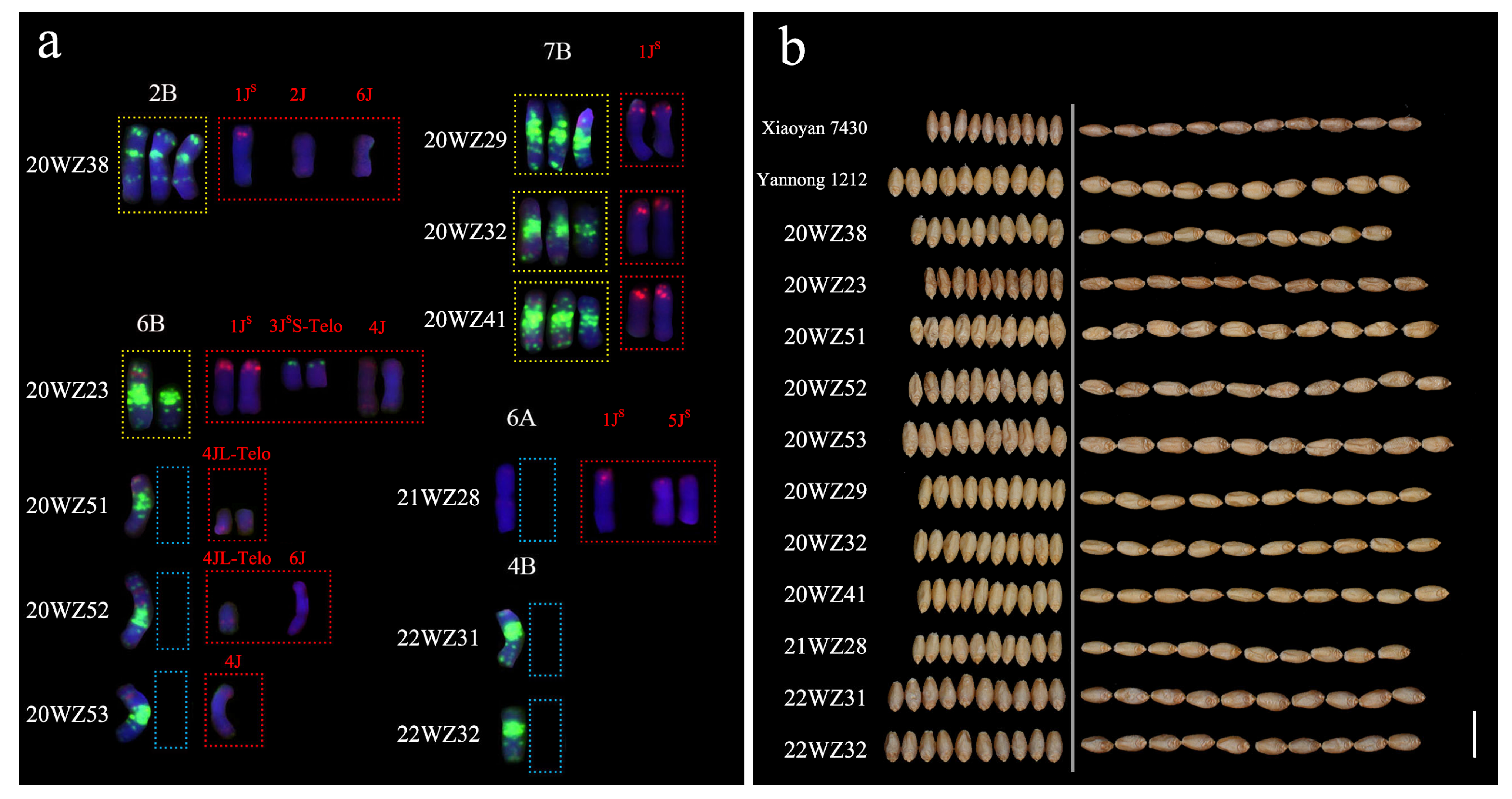
2.2.1. Karyotype Characterization of Derived Lines
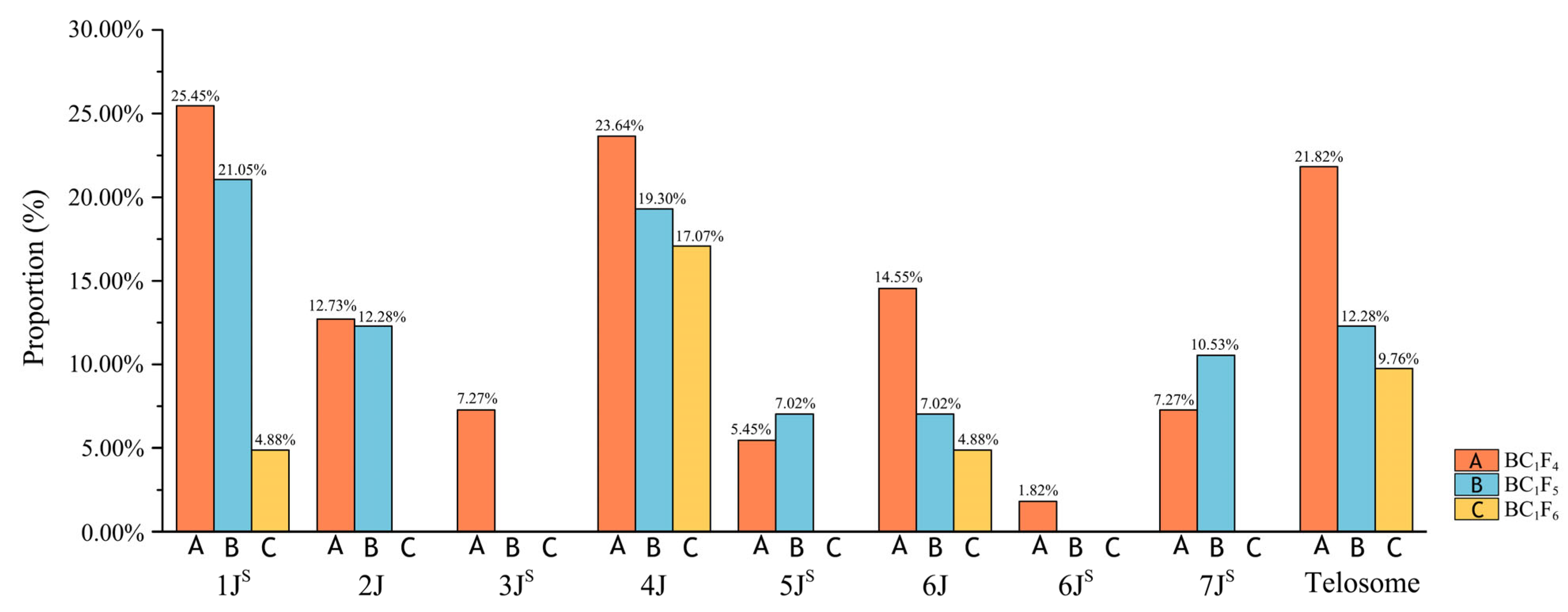
2.2.2. Analysis of Chromosomal Variations in Derived Progenies
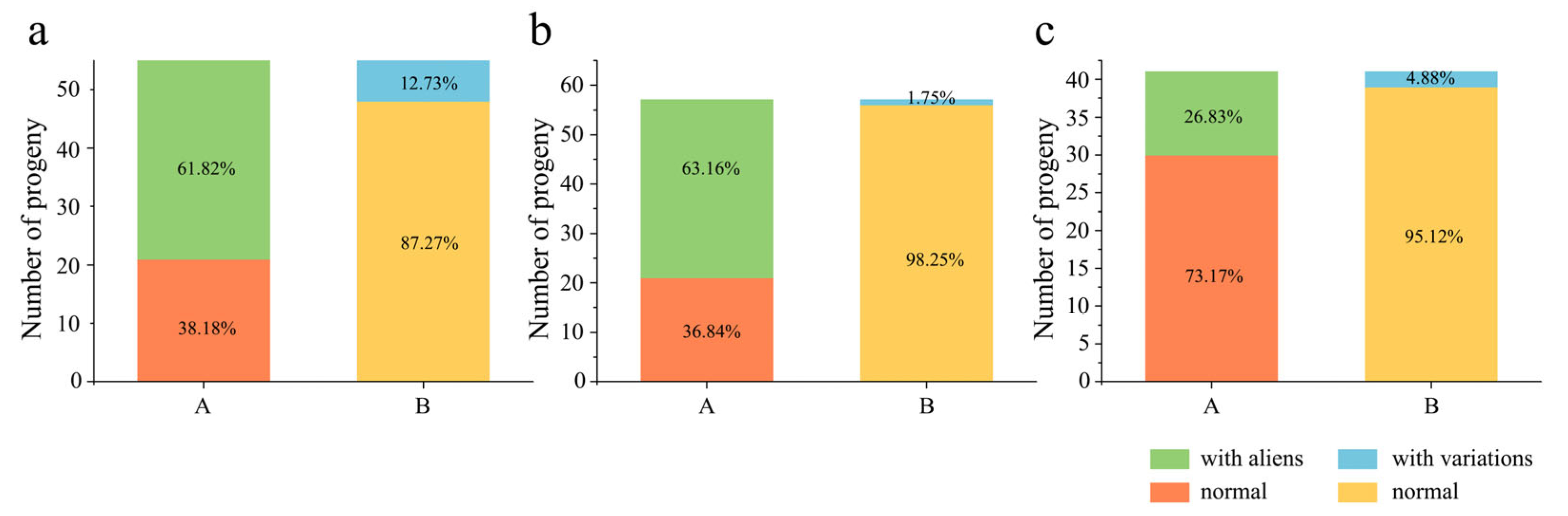
2.3. Alien Chromosome Transmission in Derived Progenies
2.4. Analysis of Genetic Effects of Chromosomal Variations on Grain Traits
2.4.1. Effects of Chromosomal Variations on Grain Traits
2.4.2. Analysis of Dose Effects of Alien Chromosomes on Grain Traits
2.4.3. Impact of Special Wheat Chromosomal Variations on Grain Traits
3. Discussion
3.1. Genetic Characteristics of Chromosome Variations in Wheat–Thinopyrum Ponticum-Derived Lines
3.2. Genetic Effects of Chromosomal Variations on Grain Traits
4. Materials and Methods
4.1. Materials
4.2. Plant Cultivation
4.3. Preparation of Root-Tip Mitotic Chromosomes
4.4. Fluorescence In Situ Hybridization (FISH)
4.5. Grain Trait Evaluation
4.6. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liu, H.; Li, H.F.; Hao, C.Y.; Wang, K.; Wang, Y.M.; Qin, L.; An, D.G.; Li, T.; Zhang, X.Y. TaDA1, a conserved negative regulator of kernel size, has an additive effect with TaGW2 in common wheat (Triticum aestivum L.). Plant Biotechnol. J. 2020, 18, 1330–1342. [Google Scholar] [CrossRef]
- Huang, Y.M.; Huang, W.; Meng, Z.; Braz, G.T.; Li, Y.F.; Wang, K.; Wang, H.; Lai, J.S.; Jiang, J.M.; Dong, Z.B.; et al. Megabase-scale presence-absence variation with Tripsacum origin was under selection during maize domestication and adaptation. Genome Biol. 2021, 22, 237. [Google Scholar] [CrossRef]
- Qin, P.; Lu, H.W.; Du, H.L.; Wang, H.; Chen, W.L.; Chen, Z.; He, Q.; Ou, S.J.; Zhang, H.Y.; Li, X.Z.; et al. Pan-genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations. Cell 2021, 184, 3542–3558.e3516. [Google Scholar] [CrossRef]
- Alkan, C.; Coe, B.P.; Eichler, E.E. Genome structural variation discovery and genotyping. Nat. Rev. Genet. 2011, 12, 363–376. [Google Scholar] [CrossRef]
- Gabur, I.; Chawla, H.S.; Snowdon, R.J.; Parkin, I.A.P. Connecting genome structural variation with complex traits in crop plants. Theor. Appl. Genet. 2019, 132, 733–750. [Google Scholar] [CrossRef]
- Lei, L.; Goltsman, E.; Goodstein, D.; Wu, G.A.; Rokhsar, D.S.; Vogel, J.P. Plant Pan-Genomics Comes of Age. Annu. Rev. Plant Biol. 2021, 72, 411–435. [Google Scholar] [CrossRef]
- Chalhoub, B.; Denoeud, F.; Liu, S.Y.; Parkin, I.A.P.; Tang, H.B.; Wang, X.Y.; Chiquet, J.; Belcram, H.; Tong, C.B.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Hurgobin, B.; Golicz, A.A.; Bayer, P.E.; Chan, C.K.K.; Tirnaz, S.; Dolatabadian, A.; Schiessl, S.V.; Samans, B.; Montenegro, J.D.; Parkin, I.A.P.; et al. Homoeologous exchange is a major cause of gene presence/absence variation in the amphidiploid Brassica napus. Plant Biotechnol. J. 2018, 16, 1265–1274. [Google Scholar] [CrossRef]
- Xu, X.; Liu, X.; Ge, S.; Jensen, J.D.; Hu, F.Y.; Li, X.; Dong, Y.; Gutenkunst, R.N.; Fang, L.; Huang, L.; et al. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 2012, 30, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Wang, C.H.; Xu, Q.; Feng, Y.; Yuan, X.P.; Yu, H.Y.; Wang, Y.P.; Tang, S.X.; Wei, X.H. Genome-wide copy number variations in Oryza sativa L. BMC Genom. 2013, 14, 649. [Google Scholar] [CrossRef] [PubMed]
- Schiessl, S.; Huettel, B.; Kuehn, D.; Reinhardt, R.; Snowdon, R.J. Targeted deep sequencing of flowering regulators in Brassica napus reveals extensive copy number variation. Sci. Data 2017, 4, 170013. [Google Scholar] [CrossRef] [PubMed]
- Gabur, I.; Chawla, H.S.; Liu, X.W.; Kumar, V.; Faure, S.; Tiedemann, A.; Jestin, C.; Dryzska, E.; Volkmann, S.; Breuer, F.; et al. Finding invisible quantitative trait loci with missing data. Plant Biotechnol. J. 2018, 16, 2102–2112. [Google Scholar] [CrossRef] [PubMed]
- Würschum, T.; Boeven, P.H.G.; Langer, S.M.; Longin, C.F.H.; Leiser, W.L. Multiply to conquer: Copy number variations at Ppd-B1 and Vrn-A1 facilitate global adaptation in wheat. BMC Genet. 2015, 16, 96. [Google Scholar] [CrossRef]
- Qian, L.W.; Voss-Fels, K.; Cui, Y.X.; Jan, H.U.; Samans, B.; Obermeier, C.; Qian, W.; Snowdon, R.J. Deletion of a Stay-Green Gene Associates with Adaptive Selection in Brassica napus. Mol. Plant 2016, 9, 1559–1569. [Google Scholar] [CrossRef]
- Samans, B.; Chalhoub, B.; Snowdon, R.J. Surviving a Genome Collision: Genomic Signatures of Allopolyploidization in the Recent Crop Species Brassica napus. Plant Genome 2017, 10, 1–15. [Google Scholar] [CrossRef]
- Francia, E.; Morcia, C.; Pasquariello, M.; Mazzamurro, V.; Milc, J.A.; Rizza, F.; Terzi, V.; Pecchioni, N. Copy number variation at the HvCBF4-HvCBF2 genomic segment is a major component of frost resistance in barley. Plant mol. Biol. 2016, 92, 161–175. [Google Scholar] [CrossRef]
- Mahmoud, M.; Gracz-Bernaciak, J.; Żywicki, M.; Karłowski, W.; Twardowski, T.; Tyczewska, A. Identification of Structural Variants in Two Novel Genomes of Maize Inbred Lines Possibly Related to Glyphosate Tolerance. Plants 2020, 9, 523. [Google Scholar] [CrossRef]
- Feldman, M.; Levy, A.A. Genome Evolution Due to Allopolyploidization in Wheat. Genetics 2012, 192, 763–774. [Google Scholar] [CrossRef]
- Wang, Z.H.; Wang, W.X.; Xie, X.M.; Wang, Y.F.; Yang, Z.Z.; Peng, H.R.; Xin, M.M.; Yao, Y.Y.; Hu, Z.R.; Liu, J.; et al. Dispersed emergence and protracted domestication of polyploid wheat uncovered by mosaic ancestral haploblock inference. Nat. Commun. 2022, 13, 3891. [Google Scholar] [CrossRef] [PubMed]
- Walkowiak, S.; Gao, L.L.; Monat, C.; Haberer, G.; Kassa, M.T.; Brinton, J.; Ramirez-Gonzalez, R.H.; Kolodziej, M.C.; Delorean, E.; Thambugala, D.; et al. Multiple wheat genomes reveal global variation in modern breeding. Nature 2020, 588, 277–283. [Google Scholar] [CrossRef]
- Badaeva, E.D.; Dedkova, O.S.; Gay, G.; Pukhalskyi, V.A.; Zelenin, A.V.; Bernard, S.; Bernard, M. Chromosomal rearrangements in wheat: Their types and distribution. Genome 2007, 50, 907–926. [Google Scholar] [CrossRef]
- Taketa, S.; Kawahara, T. C-banding analysis on wild Emmer (Triticum dicoccoides Körn) strains with and without spontaneous reciprocal translocations. Theor. Appl. Genet. 1996, 92, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.Y.; Zhu, M.Q.; Zhuang, L.F.; Zhang, S.Y.; Wang, J.J.; Chen, X.J.; Wang, D.R.; Chen, J.Y.; Bao, Y.G.; Guo, J.; et al. Structural chromosome rearrangements and polymorphisms identified in Chinese wheat cultivars by high-resolution multiplex oligonucleotide FISH. Theor. Appl. Genet. 2018, 131, 1967–1986. [Google Scholar] [CrossRef] [PubMed]
- Cavanagh, C.R.; Chao, S.; Wang, S.C.; Huang, B.E.; Stephen, S.; Kiani, S.; Forrest, K.; Saintenac, C.; Brown-Guedira, G.L.; Akhunova, A.; et al. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc. Natl. Acad. Sci. USA 2013, 110, 8057–8062. [Google Scholar] [CrossRef]
- Wu, N.; Lei, Y.H.; Pei, D.; Wu, H.; Liu, X.; Fang, J.X.; Guo, J.T.; Wang, C.L.; Guo, J.; Zhang, J.L.; et al. Predominant wheat-alien chromosome translocations in newly developed wheat of China. Mol. Breeding 2021, 41, 30. [Google Scholar] [CrossRef]
- Luo, P.G.; Luo, H.Y.; Chang, Z.J.; Zhang, H.Y.; Zhang, M.; Ren, Z.L. Characterization and chromosomal location of Pm40 in common wheat: A new gene for resistance to powdery mildew derived from Elytrigia intermedium. Theor. Appl. Genet. 2009, 118, 1059–1064. [Google Scholar] [CrossRef]
- Li, J.B.; Bao, Y.G.; Han, R.; Wang, X.L.; Xu, W.J.; Li, G.R.; Yang, Z.J.; Zhang, X.J.; Li, X.; Liu, A.F.; et al. Molecular and Cytogenetic Identification of Stem Rust Resistant Wheat-Thinopyrum intermedium Introgression Lines. Plant Dis. 2022, 106, 2447–2454. [Google Scholar] [CrossRef]
- Li, J.B.; Ryan, M.; Dong, C.M.; Forrest, K.L.; Hayden, M.J.; Singh, S.; Wang, Y.Q.; Ahmed, N.; McIntosh, R.A.; Zhang, P. Pseudo-linkage or real-linkage of rust resistance genes in a wheat-Thinopyrum intermedium translocation line. Theor. Appl. Genet. 2024, 138, 15. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.H.; Peng, H.; Zhang, H.; Li, L.X.; Saqlain, M.; Wu, D.D.; Zhu, W.; Xu, L.L.; Cheng, Y.R.; Wang, Y.; et al. Wheat-Psathyrostachys huashanica 4Ns Additional Line Confers Resistance to Fusarium Head Blight. Plants 2025, 14, 1104. [Google Scholar] [CrossRef] [PubMed]
- Du, P.; Zhuang, L.F.; Wang, Y.Z.; Yuan, L.; Wang, Q.; Wang, D.R.; Dawadondup; Tan, L.J.; Shen, J.; Xu, H.B.; et al. Development of oligonucleotides and multiplex probes for quick and accurate identification of wheat and Thinopyrum bessarabicum chromosomes. Genome 2017, 60, 93–103. [Google Scholar] [CrossRef]
- Zhao, J.J.; Zheng, X.W.; Qiao, L.; Yang, C.K.; Wu, B.B.; He, Z.M.; Tang, Y.Q.; Li, G.R.; Yang, Z.J.; Zheng, J.; et al. Genome-wide association study reveals structural chromosome variations with phenotypic effects in wheat (Triticum aestivum L.). Plant J. 2022, 112, 1447–1461. [Google Scholar] [CrossRef]
- Zhang, S.W.; Zhao, J.J.; Zhang, H.Y.; Fu, D.D.; Qiao, L.; Wu, B.B.; Li, X.H.; Hao, Y.Q.; Zheng, X.W.; Liang, Z.; et al. Structural chromosome variations from Jinmai 47 and Jinmai 84 affected agronomic traits and drought tolerance of wheat. J. Integr. Agr. 2025; in press. [Google Scholar] [CrossRef]
- Tang, Z.X.; Li, M.; Chen, L.; Wang, Y.Y.; Ren, Z.L.; Fu, S.L. New Types of Wheat Chromosomal Structural Variations in Derivatives of Wheat-Rye Hybrids. PLoS ONE 2014, 9, e110282. [Google Scholar] [CrossRef]
- Li, J.C.; Zhao, L.; Cheng, X.N.; Bai, G.H.; Li, M.; Wu, J.; Yang, Q.H.; Chen, X.H.; Yang, Z.J.; Zhao, J.X. Molecular cytogenetic characterization of a novel wheat-Psathyrostachys huashanica Keng T3DS-5NsL•5NsS and T5DL-3DS•3DL dual translocation line with powdery mildew resistance. BMC Plant Biol. 2020, 20, 163. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Xing, P.Y.; Qi, X.L.; Bao, Y.G.; Wang, H.G.; Wang, R.R.C.; Li, X.F. Characterization of chromosome constitution in three wheat-Thinopyrum intermedium amphiploids revealed frequent rearrangement of alien and wheat chromosomes. BMC Plant Biol. 2021, 21, 129. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.J.; Li, G.R.; Zeng, Z.X.; Chang, Z.J.; Liu, C.; Yang, Z.J. Molecular characterization of a wheat -Thinopyrum ponticum partial amphiploid and its derived substitution line for resistance to stripe rust. J. Appl. Genet. 2011, 52, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.X.; Huang, S.H.; Han, J.; Hou, C.C.; Zheng, D.S.; Zhang, Z.M.; Wu, J. Development and Molecular Cytogenetic Identification of a New Wheat-Psathyrostachys huashanica Keng Translocation Line Resistant to Powdery Mildew. Front. Plant Sci. 2021, 12, 689502. [Google Scholar] [CrossRef]
- Wang, T.T.; Li, G.R.; Jiang, C.Z.; Zhou, Y.W.; Yang, E.N.; Li, J.B.; Zhang, P.; Dundas, I.; Yang, Z.J. Development of a Set of Wheat-Rye Derivative Lines from Hexaploid Triticale with Complex Chromosomal Rearrangements to Improve Disease Resistance, Agronomic and Quality Traits of Wheat. Plants 2023, 12, 3885. [Google Scholar] [CrossRef]
- Jiang, C.Z.; Luo, Y.J.; Huang, D.D.; Chen, M.L.; Yang, E.N.; Li, G.R.; Yang, Z.J. Characterization of a New Stripe Rust Resistance Gene on Chromosome 2StS from Thinopyrum intermedium in Wheat. Plants 2025, 14, 1538. [Google Scholar] [CrossRef]
- Danilova, T.V.; Zhang, G.R.; Liu, W.X.; Friebe, B.; Gill, B.S. Homoeologous recombination-based transfer and molecular cytogenetic mapping of a wheat streak mosaic virus and Triticum mosaic virus resistance gene Wsm3 from Thinopyrum intermedium to wheat. Theor. Appl. Genet. 2017, 130, 549–556. [Google Scholar] [CrossRef]
- Lei, M.P.; Li, G.R.; Liu, C.; Yang, Z.J. Characterization of wheat: Secale africanum introgression lines reveals evolutionary aspects of chromosome 1R in rye. Genome 2012, 55, 765–774. [Google Scholar] [CrossRef]
- Liu, C.; Gong, W.P.; Han, R.; Guo, J.; Li, G.R.; Li, H.S.; Song, J.M.; Liu, A.F.; Cao, X.Y.; Zhai, S.N. Characterization, identification and evaluation of a set of wheat-Aegilops comosa chromosome lines. Sci. Rep. 2019, 9, 4773. [Google Scholar] [CrossRef]
- Song, L.Q.; Lu, Y.Q.; Zhang, J.P.; Pan, C.L.; Yang, X.M.; Li, X.Q.; Liu, W.H.; Li, L.H. Cytological and molecular analysis of wheat-Agropyron cristatum translocation lines with 6P chromosome fragments conferring superior agronomic traits in common wheat. Genome 2016, 59, 840–850. [Google Scholar] [CrossRef]
- Jiang, C.Z.; Luo, Y.J.; Qi, Y.L.; Li, L.; Jiang, T.T.; Yang, E.N.; Li, G.R.; Yang, Z.J. Molecular characterization of a new wheat-Thinopyrum ponticum translocation line with resistance to stripe rust. Theor. Appl. Genet. 2025, 138, 226. [Google Scholar] [CrossRef] [PubMed]
- Blanco, A.; Simeone, R.; Resta, P. The addition of Dasypyrum villosum (L.) Candargy chromosomes to durum wheat (Triticum durum Desf.). Theor. Appl. Genet. 1987, 74, 328–333. [Google Scholar] [CrossRef] [PubMed]
- He, L.M.; Dooner, H.K. Haplotype structure strongly affects recombination in a maize genetic interval polymorphic for Helitron and retrotransposon insertions. Proc. Natl. Acad. Sci. USA 2009, 106, 8410–8416. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.J.; Li, X.H.; Qiao, L.; Zheng, X.W.; Wu, B.B.; Guo, M.J.; Feng, M.C.; Qi, Z.J.; Yang, W.D.; Zheng, J. Identification of structural variations related to drought tolerance in wheat (Triticum aestivum L.). Theor. Appl. Genet. 2023, 136, 37. [Google Scholar] [CrossRef]
- Zou, Y.; Wan, L.R.; Luo, J.; Tang, Z.X.; Fu, S.L. FISH landmarks reflecting meiotic recombination and structural alterations of chromosomes in wheat (Triticum aestivum L.). BMC Plant Biol. 2021, 21, 167. [Google Scholar] [CrossRef]
- Mizuta, Y.; Harushima, Y.; Kurata, N. Rice pollen hybrid incompatibility caused by reciprocal gene loss of duplicated genes. Proc. Natl. Acad. Sci. USA 2010, 107, 20417–20422. [Google Scholar] [CrossRef]
- Kianian, S.F.; Quiros, C.F. Generation of a Brassica oleracea composite RFLP map: Linkage arrangements among various populations and evolutionary implications. Theor. Appl. Genet. 1992, 84, 544–554. [Google Scholar] [CrossRef]
- Luo, D.P.; Xu, H.; Liu, Z.L.; Guo, J.X.; Li, H.Y.; Chen, L.T.; Fang, C.; Zhang, Q.Y.; Bai, M.; Yao, N.; et al. A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nat. Genet. 2013, 45, 573–577. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, L.; Xiao, J.; Huang, N.; McCouch, S.R. Chromosomal regions associated with segregation distortion of molecular markers in F2, backcross, doubled haploid, and recombinant inbred populations in rice (Oryza sativa L.). Mol. Gen. Genet. 1997, 253, 535–545. [Google Scholar] [CrossRef]
- Borde, V. The multiple roles of the Mre11 complex for meiotic recombination. Chromosome Res. 2007, 15, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Saintenac, C.; Faure, S.; Remay, A.; Choulet, F.; Ravel, C.; Paux, E.; Balfourier, F.; Feuillet, C.; Sourdille, P. Variation in crossover rates across a 3-Mb contig of bread wheat (Triticum aestivum) reveals the presence of a meiotic recombination hotspot. Chromosoma. 2011, 120, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Klepikova, A.V.; Kasianov, A.S.; Gerasimov, E.S.; Logacheva, M.D.; Penin, A.A. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 2016, 88, 1058–1070. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, W.G.; Wang, H.W.; Dong, H.B.; Qi, X.L.; Zhao, M.Z.; Fang, Y.H.; Gao, C.; Hu, L. Progress in genetic improvement of grain yield and related physiological traits of Chinese wheat in Henan Province. Field Crop. Res. 2016, 199, 117–128. [Google Scholar] [CrossRef]
- Ramya, P.; Chaubal, A.; Kulkarni, K.; Gupta, L.; Kadoo, N.; Dhaliwal, H.S.; Chhuneja, P.; Lagu, M.; Gupta, V. QTL mapping of 1000-kernel weight, kernel length, and kernel width in bread wheat (Triticum aestivum L.). J. Appl. Genet. 2010, 51, 421–429. [Google Scholar] [CrossRef]
- Simmonds, J.; Scott, P.; Brinton, J.; Mestre, T.C.; Bush, M.; Del Blanco, A.; Dubcovsky, J.; Uauy, C. A splice acceptor site mutation in TaGW2-A1 increases thousand grain weight in tetraploid and hexaploid wheat through wider and longer grains. Theor. Appl. Genet. 2016, 129, 1099–1112. [Google Scholar] [CrossRef]
- Ma, L.; Li, T.; Hao, C.Y.; Wang, Y.Q.; Chen, X.H.; Zhang, X.Y. TaGS5-3A, a grain size gene selected during wheat improvement for larger kernel and yield. Plant Biotechnol. J. 2016, 14, 1269–1280. [Google Scholar] [CrossRef]
- Hou, J.; Jiang, Q.Y.; Hao, C.Y.; Wang, Y.Q.; Zhang, H.N.; Zhang, X.Y. Global selection on sucrose synthase haplotypes during a century of wheat breeding. Plant Physiol. 2014, 164, 1918–1929. [Google Scholar] [CrossRef]
- Wang, Y.H.; Wang, H.G. Characterization of Three Novel Wheat-Thinopyrum intermedium Addition Lines with Novel Storage Protein Subunits and Resistance to Both Powdery Mildew and Stripe Rust. J. Genet. Genom. 2016, 43, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.P.; Liu, W.H.; Yang, X.M.; Gao, A.N.; Li, X.Q.; Wu, X.Y.; Li, L.H. Isolation and characterization of two putative cytokinin oxidase genes related to grain number per spike phenotype in wheat. Mol. Biol. Rep. 2011, 38, 2337–2347. [Google Scholar] [CrossRef]
- Bednarek, J.; Boulaflous, A.; Girousse, C.; Ravel, C.; Tassy, C.; Barret, P.; Bouzidi, M.F.; Mouzeyar, S. Down-regulation of the TaGW2 gene by RNA interference results in decreased grain size and weight in wheat. J. Exp. Bot. 2012, 63, 5945–5955. [Google Scholar] [CrossRef]
- Mir, R.R.; Kumar, N.; Jaiswal, V.; Girdharwal, N.; Prasad, M.; Balyan, H.S.; Gupta, P.K. Genetic dissection of grain weight in bread wheat through quantitative trait locus interval and association mapping. Mol. Breeding. 2012, 29, 963–972. [Google Scholar] [CrossRef]
- Narasimhamoorthy, B.; Gill, B.S.; Fritz, A.K.; Nelson, J.C.; Brown-Guedira, G.L. Advanced backcross QTL analysis of a hard winter wheat × synthetic wheat population. Theor. Appl. Genet. 2006, 112, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Peleg, Z.; Fahima, T.; Korol, A.B.; Abbo, S.; Saranga, Y. Genetic analysis of wheat domestication and evolution under domestication. J. Exp. Bot. 2011, 62, 5051–5061. [Google Scholar] [CrossRef] [PubMed]
- Gegas, V.C.; Nazari, A.; Griffiths, S.; Simmonds, J.; Fish, L.; Orford, S.; Sayers, L.; Doonan, J.H.; Snape, J.W. A Genetic Framework for Grain Size and Shape Variation in Wheat(C)(W). Plant Cell 2010, 22, 1046–1056. [Google Scholar] [CrossRef]
- Gao, L.L.; Koo, D.H.; Juliana, P.; Rife, T.; Singh, D.; Lemes da Silva, C.; Lux, T.; Dorn, K.M.; Clinesmith, M.; Silva, P.; et al. The Aegilops ventricosa 2NvS segment in bread wheat: Cytology, genomics and breeding. Theor. Appl. Genet. 2021, 134, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, Y.L.; Su, H.D.; Guo, X.R.; Han, F.P. Centromere structure and function analysis in wheat-rye translocation lines. Plant J. 2017, 91, 199–207. [Google Scholar] [CrossRef]
- Monneveux, P.; Reynolds, M.P.; Aguilar, J.G.; Singh, R.P.; Weber, W.E. Effects of the 7DL.7Ag translocation from Lophopyrum elongatum on wheat yield and related morphophysiological traits under different environments. Plant Breeding 2003, 122, 379–384. [Google Scholar] [CrossRef]
- Sun, Y.Y.; Lyu, M.J.; Han, H.M.; Zhou, S.H.; Lu, Y.Q.; Liu, W.H.; Yang, X.M.; Li, X.Q.; Zhang, J.P.; Liu, X.; et al. Identification and fine mapping of alien fragments associated with enhanced grain weight from Agropyron cristatum chromosome 7P in common wheat backgrounds. Theor. Appl. Genet. 2021, 134, 3759–3772. [Google Scholar] [CrossRef]
- Boehm, J., Jr.; Cai, X.W. Enrichment and Diversification of the Wheat Genome via Alien Introgression. Plants 2024, 13, 339. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.H.; Wei, X.F.; Jiang, Y.R.; Hu, X.; Rong, J.K. Identification of Elite Agronomic Traits Using Chromosome Arm Substitution Lines of Triticum dicoccoides in the Background of Common Wheat. Agronomy 2025, 15, 752. [Google Scholar] [CrossRef]
- Lang, T.; La, S.X.; Li, B.; Yu, Z.H.; Chen, Q.H.; Li, J.B.; Yang, E.N.; Li, G.R.; Yang, Z.J. Precise identification of wheat-Thinopyrum intermedium translocation chromosomes carrying resistance to wheat stripe rust in line Z4 and its derived progenies. Genome 2018, 61, 177–185. [Google Scholar] [CrossRef]
- Fu, S.L.; Chen, L.; Wang, Y.Y.; Li, M.; Yang, Z.J.; Qiu, L.; Yan, B.J.; Ren, Z.L.; Tang, Z.X. Oligonucleotide Probes for ND-FISH Analysis to Identify Rye and Wheat Chromosomes. Sci. Rep. 2015, 5, 10552. [Google Scholar] [CrossRef] [PubMed]
- Xi, W.; Tang, Z.X.; Tang, S.Y.; Yang, Z.J.; Luo, J.; Fu, S.L. New ND-FISH-Positive Oligo Probes for Identifying Thinopyrum Chromosomes in Wheat Backgrounds. Int. J. Mol. Sci. 2019, 20, 2031. [Google Scholar] [CrossRef] [PubMed]

| Population | Chr. | Yannong 1212 Types | Xiaoyan 7430 Types | Translocation Type 3 | Translocation Type 4 | χ 2 | p |
|---|---|---|---|---|---|---|---|
| BC1F4 | 2A | 19 | 33 | 3 | 14.511 | 0.000 *** | |
| 6A | 41 | 6 | 3 | 5 | 27.128 | 0.000 *** | |
| 5B | 27 | 28 | 0.036 | 0.850 | |||
| 1D | 45 | 10 | 11.77 | 0.000 *** | |||
| 2D | 46 | 9 | 13.378 | 0.000 *** | |||
| BC1F5 | 2A | 31 | 24 | 2 | 17.223 | 0.000 *** | |
| 6A | 40 | 6 | 5 | 6 | 22.027 | 0.000 *** | |
| 5B | 30 | 27 | 0.035 | 0.852 | |||
| 1D | 53 | 4 | 25.024 | 0.000 *** | |||
| 2D | 48 | 9 | 14.445 | 0.000 *** | |||
| BC1F6 | 2A | 13 | 20 | 8 | 2.286 | 0.319 | |
| 6A | 30 | 3 | 8 | 14.126 | 0.000 *** | ||
| 5B | 21 | 20 | 0.049 | 0.825 | |||
| 1D | 34 | 7 | 9.332 | 0.002 ** | |||
| 2D | 24 | 17 | 0.443 | 0.506 |
| Trait | Chr. | Structural Variation/Alien | Yannong 1212 Type | Xiaoyan 7430 Type | Effect Value |
|---|---|---|---|---|---|
| TGW | 6A | YN-PAV.6A | 48.06 | 53.75 | −10.59% |
| 6A | XY-PAV.6A | 52.16 | 53.75 | −2.94% | |
| 6A | COM-PAV.6A | 52.09 | 53.75 | −3.09% | |
| 5B | YN-PAV.5B | 53.15 | 46.08 | 15.36% | |
| 2D | XY-PAV.2D | 49.92 | 51.50 | 3.16% | |
| 4J | 4J | 43.41 | 51.36 | −15.47% | |
| GL | 6A | YN-PAV.6A | 6.86 | 6.74 | 1.81% |
| 6A | XY-PAV.6A | 6.94 | 6.74 | 3.01% | |
| 6A | COM-PAV.6A | 6.68 | 6.74 | −0.91% | |
| 5B | YN-PAV.5B | 6.93 | 6.78 | 2.25% | |
| 1D | YN-PAV.1D | 6.83 | 7.05 | −3.14% | |
| 2D | XY-PAV.2D | 6.91 | 6.71 | −2.84% | |
| 1JS | 1JS | 6.82 | 6.82 | −0.09% | |
| 6J | 6J | 6.59 | 6.88 | −4.15% | |
| GW | 6A | YN-PAV.6A | 3.51 | 3.68 | −4.63% |
| 6A | XY-PAV.6A | 3.73 | 3.68 | 1.22% | |
| 6A | COM-PAV.6A | 3.64 | 3.68 | −1.18% | |
| 5B | YN-PAV.5B | 3.68 | 3.45 | 6.82% | |
| 4J | 4J | 3.31 | 3.63 | −8.71% | |
| LWR | 2A | YN-PAV.2A | 1.91 | 2.01 | −4.63% |
| 6A | YN-PAV.6A | 2.00 | 1.84 | 8.48% | |
| 6A | XY-PAV.6A | 1.89 | 1.84 | 2.58% | |
| 6A | COM-PAV.6A | 1.86 | 1.84 | 0.89% | |
| 1JS | 1JS | 2.00 | 1.93 | 3.62% | |
| 4J | 4J | 2.14 | 1.91 | 11.88% | |
| GD | 2A | YN-PAV.2A | 4.98 | 4.84 | 2.77% |
| 6A | YN-PAV.6A | 4.85 | 4.97 | −2.41% | |
| 6A | XY-PAV.6A | 5.04 | 4.97 | 1.38% | |
| 6A | COM-PAV.6A | 4.92 | 4.97 | −1.13% | |
| 5B | YN-PAV.5B | 5.01 | 4.78 | 4.90% | |
| 4J | 4J | 4.74 | 4.94 | −3.96% | |
| GC | 6A | YN-PAV.6A | 18.21 | 18.22 | −0.08% |
| 6A | XY-PAV.6A | 18.63 | 18.22 | 2.22% | |
| 6A | COM-PAV.6A | 18.13 | 18.22 | −0.51% | |
| 5B | YN-PAV.5B | 18.62 | 17.91 | 3.95% | |
| 1D | YN-PAV.1D | 18.19 | 18.78 | −3.11% | |
| 1JS | 1JS | 18.08 | 18.29 | −1.12% | |
| 6J | 6J | 17.83 | 18.33 | −2.75% | |
| GA | 6A | YN-PAV.6A | 18.73 | 19.64 | −4.61% |
| 6A | XY-PAV.6A | 20.17 | 19.64 | 2.74% | |
| 6A | COM-PAV.6A | 19.23 | 19.64 | −2.08% | |
| 5B | YN-PAV.5B | 19.95 | 18.15 | 9.87% | |
| 2D | XY-PAV.2D | 19.20 | 19.16 | −0.19% | |
| 4J | 4J | 17.91 | 19.38 | −7.59% |
| Probes | Modification Types | Probe Sequences |
|---|---|---|
| Oligo-B11 | 5′6-FAM | 5′TCCGCTCACCTTGATGACAACATCAGGTGGAATTCCGTTCGAGGG3′ |
| Oligo-pSc119.2 | 5′6-FAM | 5′CCGTTTTGTGGACTATTACTCACCGCTTTGGGGTCCCATAGCTAT3′ |
| Oligo-pTa535 | 5′TAMRA | 5′AAAAACTTGACGCACGTCACGTACAAATTGGACAAACTCTTTCGGAGTATCAGGGTTTC3′ |
| Oligo-(GAA)10 | 5′6-FAM | 5′GAAGAAGAAGAAGAAGAAGAAGAAGAAGAA 3′ |
| Oligo-pTa71 | 5′TAMRA | 5′GGGCAAAACCACGTACGTGGCACACGCCGCGTA3′ |
| Oligo-pAs1-1 | 5′TAMRA | 5′GGATGCACTTCGTGTACAAAACGGACAATCTCTTTCAAAGTATCAGGATTTCATCC3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Zhang, Y.; Hu, T.; Li, L.; Wang, Z.; Qiao, L.; Chang, L.; Li, X.; Chang, Z.; Zhang, P.; et al. Impact of Alien Chromosome Introgression from Thinopyrum ponticum on Wheat Grain Traits. Plants 2025, 14, 3072. https://doi.org/10.3390/plants14193072
Zhang S, Zhang Y, Hu T, Li L, Wang Z, Qiao L, Chang L, Li X, Chang Z, Zhang P, et al. Impact of Alien Chromosome Introgression from Thinopyrum ponticum on Wheat Grain Traits. Plants. 2025; 14(19):3072. https://doi.org/10.3390/plants14193072
Chicago/Turabian StyleZhang, Shuwei, Yu Zhang, Ting Hu, Linying Li, Zihao Wang, Linyi Qiao, Lifang Chang, Xin Li, Zhijian Chang, Peng Zhang, and et al. 2025. "Impact of Alien Chromosome Introgression from Thinopyrum ponticum on Wheat Grain Traits" Plants 14, no. 19: 3072. https://doi.org/10.3390/plants14193072
APA StyleZhang, S., Zhang, Y., Hu, T., Li, L., Wang, Z., Qiao, L., Chang, L., Li, X., Chang, Z., Zhang, P., & Zhang, X. (2025). Impact of Alien Chromosome Introgression from Thinopyrum ponticum on Wheat Grain Traits. Plants, 14(19), 3072. https://doi.org/10.3390/plants14193072





