Fine-Mapping of a Red-Skinned Taproot Gene in Radish (Raphanus sativus L.)
Abstract
1. Introduction
2. Results
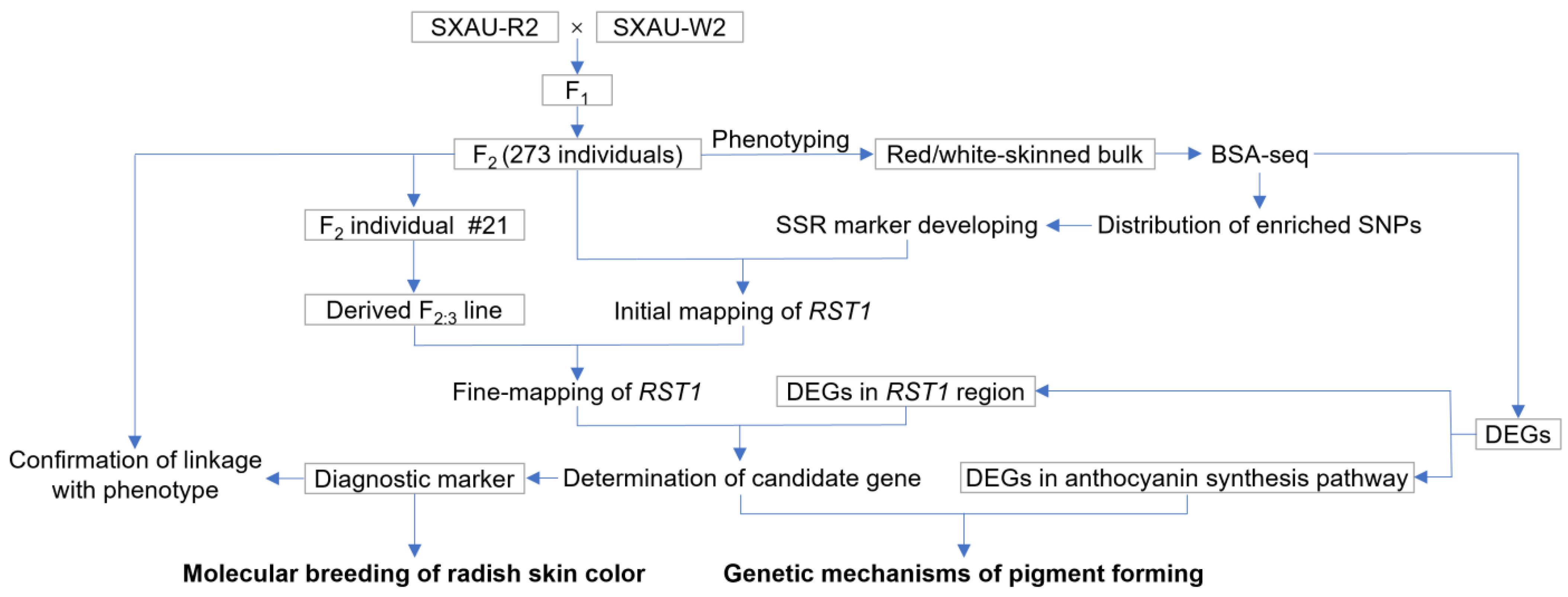
2.1. Genetic Dissection of Radish Skin Color
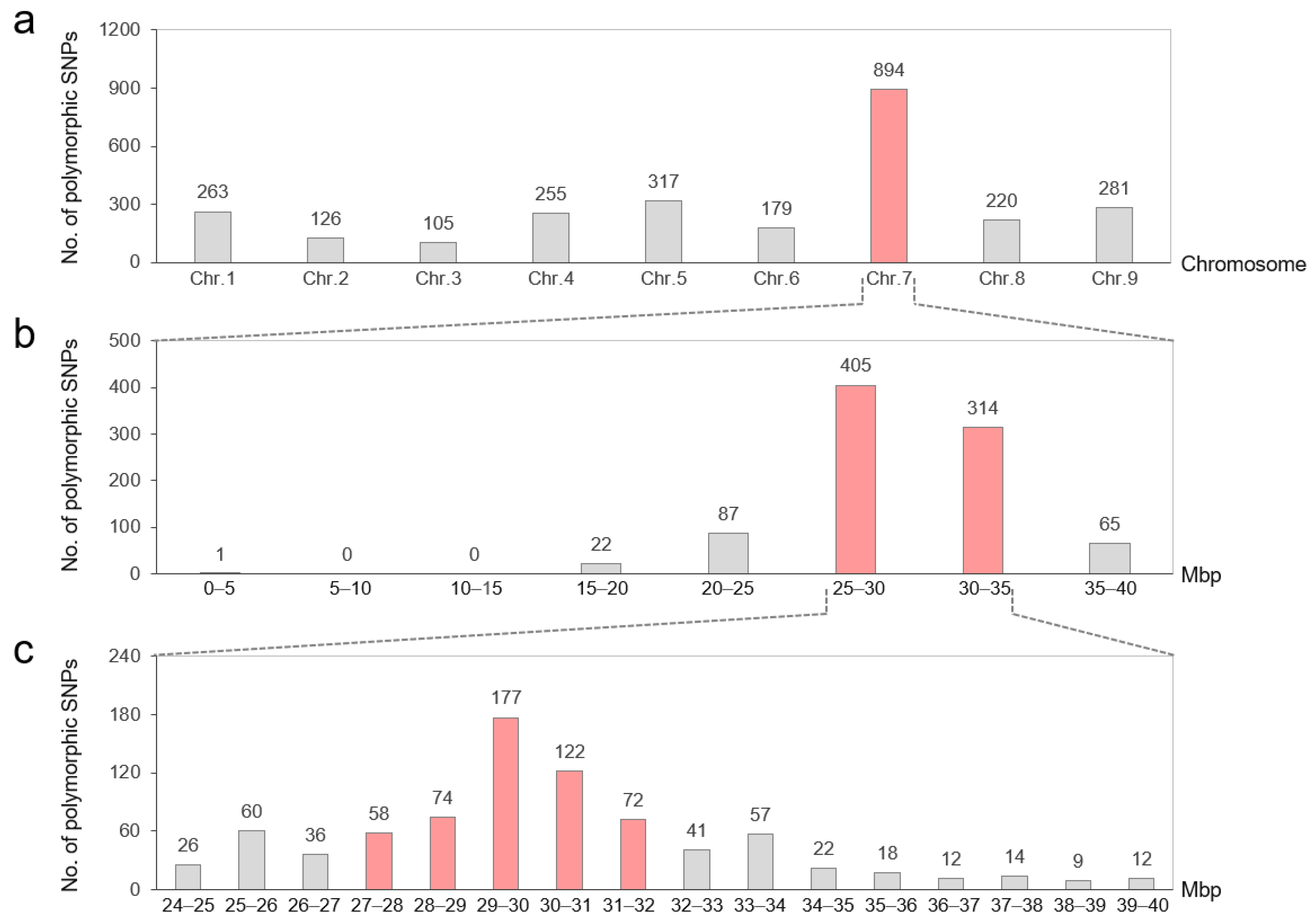
2.2. Distribution of Bulk-Polymorphic SNPs
2.3. Initial Mapping of RST1
2.4. Fine-Mapping of RST1
2.5. Determination of Candidate Gene
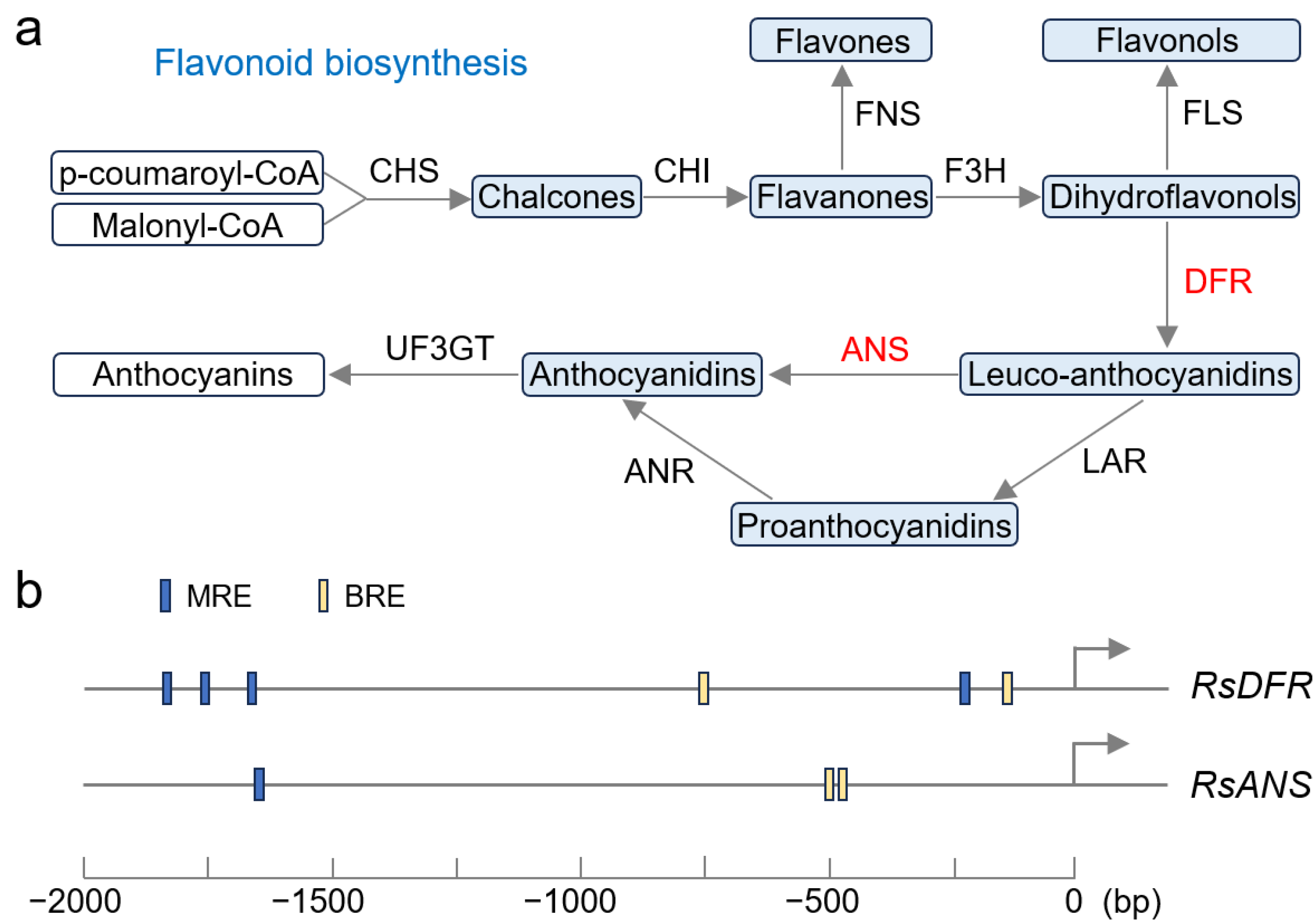
2.6. Expression Patterns of Genes Involving Anthocyanin Synthesis
3. Discussion
3.1. RST1 Can Be Used for the Radish Breeding of Skin Color
3.2. RST1 Participates in Regulating Anthocyanin Synthesis
3.3. Relationship Between RST1 and Anthocyanin-Related MYBs
4. Material and Methods
4.1. Plant Materials
4.2. Experimental Conditions and Phenotyping
4.3. BSA-Seq
4.4. Development of PCR-Based Markers
4.5. Mapping and Fine Mapping
4.6. Transcription Levels of Annotated Genes
4.7. RT-qPCR
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Singh, B.K. Radish (Raphanus sativus L.): Breeding for Higher Yield, Better Quality and Wider Adaptability. In Advances in Plant Breeding Strategies: Vegetable Crops; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer: Cham, Switzerland, 2021; pp. 275–304. [Google Scholar]
- Yamagishi, H.; Terachi, T. Multiple origins of cultivated radishes as evidenced by a comparison of the structural variations in mitochondrial DNA of Raphanus. Genome 2003, 46, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Shirasawa, K.; Fukino, N.; Hirakawa, H.; Akanuma, T.; Kitashiba, H. Identification of genome-wide SNPs among geographically diverse radish accessions. DNA Res. 2020, 27, dsaa001. [Google Scholar] [CrossRef]
- Zhang, J.; Qiu, X.; Tan, Q.; Xiao, Q.; Mei, S. A Comparative Metabolomics Study of Flavonoids in Radish with Different Skin and Flesh Colors (Raphanus sativus L.). J. Agric. Food Chem. 2020, 68, 14463–14470. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, Y.; Zhang, X.; Sun, Y.; Wang, H.; Song, J.; Li, X. Transcriptome analyses reveal key genes involved in skin color changes of ‘Xinlimei’ radish taproot. Plant Physiol. Biochem. 2019, 139, 528–539. [Google Scholar] [CrossRef]
- Chen, F.B.; Xing, C.Y.; Huo, S.P.; Cao, C.L.; Yao, Q.L.; Fang, P. Red Pigment Content and Expression of Genes Related to Anthocyanin Biosynthesis in Radishes (Raphanus sativus L.) with Different Colored Flesh. J. Agric. Sci. 2016, 8, 126–135. [Google Scholar] [CrossRef]
- Yu, R.; Du, X.; Li, J.; Liu, L.; Hu, C.; Yan, X.; Xia, Y.; Xu, H. Identification and differential expression analysis of anthocyanin biosynthetic genes in root-skin color variants of radish (Raphanus sativus L.). Genes Genom. 2020, 42, 413–424. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, J.; Tan, Q.; Qiu, X.; Mei, S. Comparative transcriptome analysis reveals key genes associated with pigmentation in radish (Raphanus sativus L.) skin and flesh. Sci. Rep. 2021, 11, 11434. [Google Scholar] [CrossRef]
- Liu, Z.; Qiao, N.; Lei, Y.; Zhang, X.; Li, Z.; Wang, S.; Qiao, L. Genetic Mapping of a Novel Gene GST1 for Green-Skinned Taproot in Radish (Raphanus sativus L.). J. Nucl. Agric. Sci. 2024, 38, 2066–2073. [Google Scholar]
- Lim, S.H.; Song, J.H.; Kim, D.H.; Kim, J.K.; Lee, J.Y.; Kim, Y.M.; Ha, S.H. Activation of Anthocyanin Biosynthesis by Expression of the Radish R2R3-MYB Transcription Factor Gene RsMYB1. Plant Cell Rep. 2016, 35, 641–653. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Wang, J.; Wu, C.; Zhang, Y.; Zhang, X.; Li, X.; Wang, H.; Song, J.; Li, X. Combined QTL-Seq and Traditional Linkage Analysis to Identify Candidate Genes for Purple Skin of Radish Fleshy Taproots. Front. Genet. 2019, 10, 808. [Google Scholar] [CrossRef]
- Fan, L.; Wang, Y.; Xu, L.; Tang, M.; Zhang, X.; Ying, J.; Li, C.; Dong, J.; Liu, L. A Genome-wide Association Study Uncovers a Critical Role of the RsPAP2 Gene in Red-Skinned Raphanus sativus L. Hortic. Res. 2020, 7, 164. [Google Scholar]
- Kim, J.; Jang, H.; Huh, S.M.; Cho, A.; Yim, B.; Jeong, S.H.; Kim, H.; Yu, H.J.; Mun, J.H. Effect of Structural Variation in the Promoter Region of RsMYB1.1 on the Skin Color of Radish Taproot. Front. Plant Sci. 2024, 14, 1327009. [Google Scholar] [CrossRef]
- Yi, G.; Kim, J.S.; Park, J.E.; Shin, H.; Yu, S.H.; Park, S.; Huh, J.H. MYB1 Transcription Factor Is a Candidate Responsible for Red Root Skin in Radish (Raphanus sativus L.). PLoS ONE 2018, 13, e0204241. [Google Scholar] [CrossRef]
- Luo, X.; Xu, L.; Wang, Y.; Dong, J.; Chen, Y.; Tang, M.; Fan, L.; Zhu, Y.; Liu, L. An Ultra-high-density Genetic Map Provides Insights into Genome Synteny, Recombination Landscape and Taproot Skin Colour in Radish (Raphanus sativus L.). Plant Biotechnol. J. 2020, 18, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Li, S.; Wang, Q.; Yuan, Y.; Ma, J.; Xu, M.; Yang, Y.; Zhang, C.; Chen, L.; Sun, Y. Construction of a High-Density Genetic Map Based on Specific-Locus Amplified Fragment Sequencing and Identification of Loci Controlling Anthocyanin Pigmentation in Yunnan Red Radish. Hortic. Res. 2022, 9, uhab031. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, Y.; Wu, X.; Shi, W.; Chen, N.; Pang, Y.; Zhang, L. Sequence and Epigenetic Variations of R2R3-MYB Transcription Factors Determine the Diversity of Taproot Skin and Flesh Colors in Different Cultivated Types of Radish (Raphanus sativus L.). Theor. Appl. Genet. 2024, 137, 33. [Google Scholar] [CrossRef] [PubMed]
- Karppinen, K.; Lafferty, D.J.; Albert, N.W.; Mikkola, N.; McGhie, T.; Allan, A.C.; Afzal, B.M.; Häggman, H.; Espley, R.V.; Jaakola, L. MYBA and MYBPA Transcription Factors Co-Regulate Anthocyanin Biosynthesis in Blue-Coloured Berries. New Phytol. 2021, 232, 1350–1367. [Google Scholar] [CrossRef] [PubMed]
- Park, N.I.; Xu, H.; Li, X.; Jang, I.H.; Park, S.; Ahn, G.H.; Lim, Y.P.; Kim, S.J.; Park, S.U. Anthocyanin Accumulation and Expression of Anthocyanin Biosynthetic Genes in Radish (Raphanus sativus). J. Agric. Food Chem. 2011, 59, 6034–6039. [Google Scholar] [CrossRef]
- Gonçalves, A.C.; Nunes, A.R.; Falcão, A.; Alves, G.; Silva, L.R. Dietary Effects of Anthocyanins in Human Health: A Comprehensive Review. Pharmaceuticals 2021, 14, 690. [Google Scholar] [CrossRef]
- Cappellini, F.; Marinelli, A.; Toccaceli, M.; Tonelli, C.; Petroni, K. Anthocyanins: From Mechanisms of Regulation in Plants to Health Benefits in Foods. Front. Plant Sci. 2021, 12, 748049. [Google Scholar] [CrossRef]
- Budahn, H.; Peterka, H.; Mousa, M.A.A.; Ding, Y.H.; Zhang, S.S.; Li, J.B. Molecular Mapping in Oil Radish (Raphanus sativus L.) and QTL Analysis of Resistance Against Beet Cyst Nematode (Heterodera schachtii). Theor. Appl. Genet. 2009, 118, 775–782. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Wang, L.J.; Gong, Y.Q.; Dai, W.H.; Wang, Y.; Zhu, X.W.; Wen, T.C.; Liu, L. Genetic Linkage Map Construction and QTL Mapping of Cadmium Accumulation in Radish (Raphanus sativus L.). Theor. Appl. Genet. 2012, 125, 659–670. [Google Scholar] [CrossRef]
- Yu, X.N.; Choi, S.R.; Dhandapani, V.; Rameneni, J.J.; Li, X.N.; Pang, W.X.; Lee, J.Y.; Lim, Y.P. Quantitative Trait Loci for Morphological Traits and Their Association with Functional Genes in Raphanus sativus. Front. Plant Sci. 2016, 7, 255. [Google Scholar] [CrossRef]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional Control of Flavonoid Biosynthesis by MYB–bHLH–WDR Complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef]
- Pan, J.; Zhou, X.; Ahmad, N.; Zhang, K.; Tang, R.; Zhao, H.; Jiang, J.; Tian, M.; Li, C.; Li, A.; et al. BSA-Seq and Genetic Mapping Identified Candidate Genes for Branching Habit in Peanut. Theor. Appl. Genet. 2022, 135, 4457–4468. [Google Scholar] [CrossRef]
- Ahmad, N.; Zhang, K.; Ma, J.; Yuan, M.; Zhao, S.; Wang, M.; Deng, L.; Ren, L.; Gangurde, S.S.; Pan, J.; et al. Transcriptional Networks Orchestrating Red and Pink Testa Color in Peanut. BMC Plant Biol. 2023, 23, 44. [Google Scholar] [CrossRef]
- LaFountain, A.M.; Yuan, Y.W. Repressors of Anthocyanin Biosynthesis. New Phytol. 2021, 231, 933–949. [Google Scholar] [CrossRef]
- Zhu, Z.; Wang, H.; Wang, Y.; Guan, S.; Wang, F.; Tang, J.; Zhang, R.; Xie, L.; Lu, Y. Characterization of the cis Elements in the proximal promoter regions of the Anthocyanin Pathway Genes Reveals a Common Regulatory Logic That Governs Pathway Regulation. J. Exp. Bot. 2015, 66, 3775–3789. [Google Scholar] [CrossRef]
- Zhang, X.H.; Liu, T.J.; Wang, J.L.; Wang, P.; Qiu, Y.; Zhao, W.; Pang, S.; Li, X.M.; Wang, H.P.; Song, J.P.; et al. Pan-genome of Raphanus Highlights Genetic Variation and Introgression Among Domesticated, Wild, and Weedy Radishes. Mol. Plant. 2021, 14, 2032–2055. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Wang, Y.; Dong, J.; Zhang, W.; Tang, M.; Zhang, W.; Wang, K.; Chen, Y.; Zhang, X.; He, Q.; et al. A Chromosome-Level Genome Assembly of Radish (Raphanus sativus L.) Reveals Insights into Genome Adaptation and Differential Bolting Regulation. Plant Biotechnol. J. 2023, 21, 990–1004. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Peng, S.; Yuan, S.; Ran, M.; Li, X.; Li, Y.; Liu, B.; Li, M.; Kong, C.; Yang, X.; et al. A Telomere-To-Telomere Genome Assembly of Radish (Raphanus sativus L.) Provides Insights into QTL Mapping of Bolting Traits. J. Genet. Genom. 2025, in press. [Google Scholar] [CrossRef]
- Lai, B.; Cheng, Y.; Liu, H.; Wang, Q.; Wang, Q.; Wang, C.; Su, R.; Chen, F.; Wang, H.; Du, L. Differential Anthocyanin Accumulation in Radish Taproot: Importance of RsMYB1 Gene Structure. Plant Cell Rep. 2020, 39, 217–226. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, Y.; Sun, H.; Sun, L.; Zhang, L. Transposon-Induced Methylation of the RsMYB1 Promoter Disturbs Anthocyanin Accumulation in Red-Fleshed Radish. J. Exp. Bot. 2020, 71, 2537–2550. [Google Scholar] [CrossRef]
- He, Q.; Wu, J.; Xue, Y.; Zhao, W.; Li, R.; Zhang, L. The Novel Gene BrMYB2, Located on Chromosome A07, With a Short Intron 1 Controls the Purple-Head Trait of Chinese Cabbage (Brassica rapa L). Hortic. Res. 2020, 7, 97. [Google Scholar] [CrossRef] [PubMed]
- Shoeva, O.Y.; Glagoleva, A.Y.; Khlestkina, E.K. The Factors Affecting the Evolution of the Anthocyanin Biosynthesis Pathway Genes in Monocot and Dicot Plant Species. BMC Plant Biol. 2017, 17, 256. [Google Scholar] [CrossRef]
- Doyle, J. Genomic Plant DNA Preparation from Fresh Tissue CTAB Method. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Chen, J.; Zhang, L.; Liu, Y.; Shen, X.; Guo, Y.; Ma, X.; Zhang, X.; Li, X.; Cheng, T.; Wen, H.; et al. RNA-Seq-Based WGCNA and Association Analysis Reveal the Key Regulatory Module and Genes Responding to Salt Stress in Wheat Roots. Plants 2024, 13, 274. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wei, A.; Chen, J.; Wu, L.; Li, T.; Qiao, L. Identification of Ethylene Response Factors in Wheat Reveals That TaERF16-B Contributes to Salt Tolerance. Plants 2025, 14, 621. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Li, Z.; Li, G.; Qiao, L. Fine-Mapping of a Red-Skinned Taproot Gene in Radish (Raphanus sativus L.). Plants 2025, 14, 3065. https://doi.org/10.3390/plants14193065
Liu Z, Li Z, Li G, Qiao L. Fine-Mapping of a Red-Skinned Taproot Gene in Radish (Raphanus sativus L.). Plants. 2025; 14(19):3065. https://doi.org/10.3390/plants14193065
Chicago/Turabian StyleLiu, Zhao, Zhenzhen Li, Gaizhen Li, and Linyi Qiao. 2025. "Fine-Mapping of a Red-Skinned Taproot Gene in Radish (Raphanus sativus L.)" Plants 14, no. 19: 3065. https://doi.org/10.3390/plants14193065
APA StyleLiu, Z., Li, Z., Li, G., & Qiao, L. (2025). Fine-Mapping of a Red-Skinned Taproot Gene in Radish (Raphanus sativus L.). Plants, 14(19), 3065. https://doi.org/10.3390/plants14193065




