Abstract
Drought severely threatens wheat production. Under drought conditions, root system architecture (DRSA)-related traits in common wheat significantly affect wheat production. In China, Zhoumai16 is a high-yield winter wheat variety in the Huang-Huai wheat region. It is suitable for high-fertilizer and high-water cultivation and has moderate drought tolerance. DK171 is a newly developed high-yield and stress-tolerant variety, with higher drought tolerance. Thus, identifying genetic loci associated with DRSA-related traits from DK171 and developing available molecular markers are of great importance for enhancing wheat stress tolerance breeding. In this study, DRSA-related traits, including the total root dry weight (DDRW), total root length (DTRL), total root area (DTRA), and the number of root tips (DNRT) under drought stress, were assessed using the hydroponic system in Zhoumai16/DK171 recombinant inbred lines (RIL) population. A total of five quantitative trait loci (QTL) for DRSA-related traits were identified, e.g., QDDRW.daas-1BL, QDTRS.daas-4AL, QDNRT.daas-4DS, QDTRL.daas-3AL, and QDDRW.daas-5D, and explained 6.1% to 18.9% of the phenotypic variances, respectively. Among these, QDTRS.daas-4AL and QDTRL.daas-3AL were consistent with previous reports, whereas the QDDRW.daas-1BL, QDNRT.daas-4DS, and QDDRW.daas-5D are novel. The favorable alleles of QDTRS.daas-4AL and QDNRT.daas-4DS were inherited from Zhoumai16, whereas the favorable alleles for QDDRW.daas-1BL, QDTRL.daas-3AL, and QDDRW.daas-5D were contributed by DK171. Furthermore, five kompetitive allele-specific PCR (KASP) markers, Kasp_1BL_DTRS (QDDRW.daas-1BL), Kasp_3AL_DTRS (QDTRL.daas-3AL), Kasp_4A_DTRS (QDTRA.daas-4A), Kasp_5D_DDRW (QDDRW.daas-5D), and Kasp_4D_DNRT (QDNRT.daas-4D), were developed and validated in a diverse panel with 108 wheat varieties mainly from China. Additionally, eight candidate genes related to plant hormone regulation, ABC transporters, and calcium-dependent lipid-binding domain proteins were identified. This study offers new loci, candidate genes, and available KASP markers for wheat drought tolerance breeding and facilitating progress in developing drought-tolerant wheat cultivars.
1. Introduction
Common wheat is one of the most crucial staple crops for human consumption and serves as the primary food crop in arid and semi-arid regions worldwide. Drought is the most significant abiotic stress affecting wheat production, severely constraining its sustainable cultivation [1]. Agricultural water inputs are required, resulting in considerable waste of labor and water resources. Therefore, breeding drought-tolerant and water-efficient varieties is the most effective, economical, and environmentally safe approach to reduce the losses caused by drought [2,3,4].
Wheat root system morphology and physiological regulation determine its drought tolerance. Drought tolerance of wheat seedlings is crucial for plant development and production. Root length, surface area, and number of root tips are key root system architecture under drought conditions (DRSA) traits [5,6,7,8]. These characteristics determine the root spatial distribution and primary composition, significantly affecting water and nutrient uptake. Maximum root length forms the basis for water uptake, while the root tip number, root diameter, root length, and root density are closely related to root dry weight, volume, and surface area. Together, these traits determine the plant anchoring strength in the soil profile and its capacity to absorb soil solutions [9,10]. However, the evaluation of these traits is limited by environmental conditions and measurement methods, making it time-consuming and labor-intensive [7,9,10]. Therefore, identifying stable and effective loci for wheat drought tolerance, developing molecular markers for breeding, and elucidating the genetic mechanisms of DRSA-related traits will accelerate the efficient improvement of drought tolerance.
Recently, advancements in high-throughput genotyping technologies, such as re-sequencing and SNP chips [11], effective analysis tools, such as linkage mapping [12,13,14,15,16], and association mapping [17,18] have become common methods for identifying loci of complex traits [6,8,12,13,14]. Over the past two decades, over 50 loci associated with DRSA-related traits have been reported and mainly distributed on chromosomes 1A, 2B, 3A, 3B, 5B, and 6D [6,8,19,20,21]. Although some genetic loci related to root traits in wheat seedlings under drought stress have been reported, many of these loci are linked to SSR markers, which are not efficient for practical use. Additionally, some loci are influenced by complex genetic backgrounds or are linked to non-desirable agronomic traits, making them unsuitable for breeding applications. Therefore, discovering new genetic loci and developing molecular markers that are applicable in breeding holds significant importance for optimizing wheat root systems and achieving high and stable yields. Zhoumai16 (Zhou8425B/Zhou 9) is a high-yield, disease-resistant wheat variety with a significant cultivation area in the Huang-Huai wheat region of China. It is suitable for high-fertilizer and high-water cultivation and has moderate drought tolerance. DK171 (Liangxing66/Shixin828) is a newly developed high-yield and stress-tolerant variety in recent years, with strong drought resistance inherited by Shixin828. In this study, five loci for seedling-stage DRSA-related traits were identified in the Zhoumai16/DK171 recombinant inbred line (RIL) population using the wheat 90K SNP array. The main goal of this study is to uncover the genetic basis of DRSA traits and develop breeding-friendly kompetitive allele-specific PCR (KASP) markers to enhance wheat DRSA-related trait improvement.
2. Results
2.1. Phenotypic Evaluation
Zhoumai16 is a high-yielding variety suitable for high-fertility, water-rich regions, while DK171 is a wheat variety that combines high yield with water conservation. For Zhoumai16, the means of total root length (DTRL), total root surface (DTRS), number of root tips (DNRT), and dry root weight (DDRW) were 35.9 cm, 8.0 cm2, 123.6, and 0.0312 g, respectively. For DK171, these values were 47.2 cm, 8.9 cm2, 168.9, and 0.0339 g. The DTRL, DTRS, DNRT, and DDRW of DK171 were significantly higher than Zhoumai16 (p < 0.05). All four DRSA-related traits exhibited continuous and significantly wide variation across the 262 RILs (Table A1 and Figure 1 and Figure A1). The means of DTRL, DTRS, DNRT, and DDRW were 40.2 cm (range: 24.1–63.8 cm), 7.8 cm2 (range: 5.4–10.7 cm2), 106.9 (range: 24.7–213.7), and 0.0282 g (range: 0.0162–0.0413 g). The standard deviation and coefficient of variation for DTRL, DTRS, DNRT, and DDRW were 6.58 cm (16.4%), 1.06 cm2 (13.6%), 35.7 (33.4%), and 0.0041 g (14.5%). Significant correlation was observed between DDRW, DTRL, DTRA, and DNRT, with a correlation coefficient of 0.603 (p < 0.05) between DTRL and DTRS (R2 = 0.56), DTRL and DNRT (R2 = 0.31), DTRL and DRW (R2 = 0.32), DTRS and DNRT (R2 = 0.33), and DNRT and DRW (R2 = 0.30) (p < 0.05).
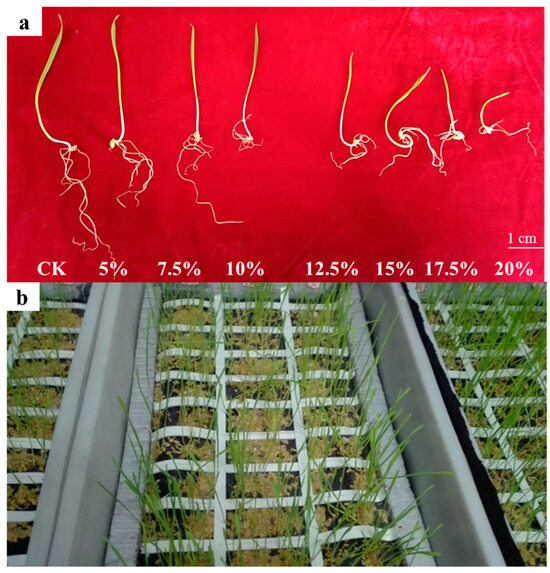
Figure 1.
The DRSA-related traits in the Zhoumai16/DK171 RIL population. (a) Root growth of wheat seedlings under different concentrations. Root development status under different concentrations of PEG6000, with 12.5% ultimately selected as the drought condition for root phenotypic characterization (5-day). PEG-6000 was used to simulate drought stress. At concentrations of 5–10%, there was no significant reduction in germination potential or germination rate, while concentrations of 12.5–20% significantly inhibited germination. Based on the comparative analysis of relative germination rates and germination potentials at different concentrations, a concentration of 12.5% was selected for the assessment of seedling-related phenotypes. (b) Bulk phenotyping of wheat seedling root traits. Bulk evaluation of root phenotypes under 12.5% PEG6000 conditions (8-day growth status).
2.2. QTL Identification
This genetic map includes all 21 chromosomes, with red representing QTL and black lines representing backbone SNP markers.
Two QTL for DDRW were identified on chromosomes 1BL and 5DL, referred to as QDDRW.daas-1BL (wsnp_Ex_rep_c67299_65845319-Excalibur_rep_c107035_354) and QDDRW.daas-5D (BobWhite_c5176_1164-RAC875_rep_c78046_324), respectively. These QTLs explained 18.9% (additive effect: −0.002 g) and 7.5% (additive effect: −0.0007 g) of the total phenotypic variances (PVEs) (Table 1; Figure 1). QDTRS.daas-4AL for DTRS was identified on 4AL chromosome (613.6–615.2 Mb, IAAV7132-wsnp_JD_c38619_27992279) and explained 9.6% of the PVEs with additive effect −0.275 cm2. QDNRT.daas-4DS for DNRT was identified on chromosome 4DS (1.2–3.6 Mb, RAC875_rep_c76650_164-Kukri_c15720_884) and explained 8.1% of the PVEs with additive effect -7.6. QDTRL.daas-3AL for DNRT was identified on chromosome 3AL (650.4–659.4 Mb, Kukri_rep_c69970_717-Kukri_rep_c103783_1380) and explained 6.1% (additive effect: −1.272 g) of the PVEs. The favorable allele of QDTRL.daas-3AL, QDDRW.daas-1BL and QDDRW.daas-5D were contributed by DK171, whereas the favorable allele of QDTRS.daas-4AL and QDNRT.daas-4DS were contributed by Zhoumai16 (Table 1; Figure 2).

Table 1.
QTL for DRSA-related traits in Zhoumai16/DK171 RIL population.
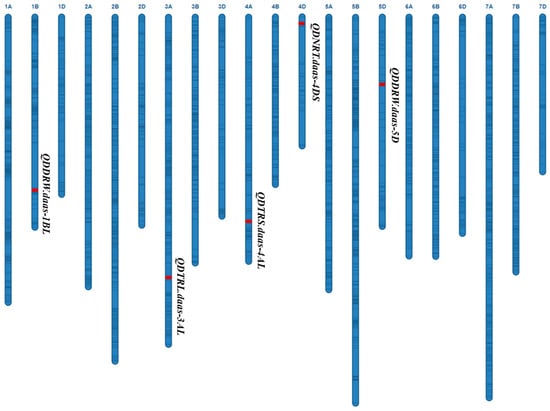
Figure 2.
QTL for the DRSA-related traits in the Zhoumai16/DK171 RIL population.
2.3. Candidate Genes Identification
Eight candidate genes were identified and involved in the biological metabolism, including the plant hormones, ABC transporter and calcium-dependent lipid-binding domain protein. Two candidate genes for QDDRW.daas-1BL were identified, e.g., TraesCS1B01G356600 and TraesCS1B01G373900, encoded the auxin-responsive protein and the ABC transporter family protein, respectively. Both TraesCS3A01G406200 and TraesCS3A01G416300 were candidate genes for QDTRL.daas-3AL and encoded the gibberellin 20 oxidase and the auxin transport protein, respectively. For QDTRS.daas-4AL, TraesCS4A01G326400 were selected as the candidate gene and encoded the ethylene-responsive transcription factor. TraesCS4D01G001900 (QDNRT.daas-4DS) encoded the calcium-dependent lipid-binding domain protein. TraesCS4D01G002400 of QDDRW.daas-5D encoded the ethylene-responsive transcription factor. TraesCS5D01G285900 identified at the genetic interval of QDDRW.daas-5D and encoded the auxin-induced in root cultures protein (Table 2 and Table A2, Figure 3). The expressions of the seven candidate genes in Zhoumai16 and DK171 were detected using the qRT-PCR. Of these, TraesCS1B01G356600, TraesCS4D01G002400, and TraesCS5D01G285900 showed no significant differences between the parents, whereas TraesCS4D01G002400 and TraesCS5D01G285900 showed more than 2.3–3.5 folds higher expression in Zhoumai16 compared to DK171; TraesCS3A01G416300, TraesCS4A01G326400, and TraesCS4D01G001900 showed more than 3.1–4.3 folds higher expression in DK171 compared to Zhoumai16 (Figure 4).

Table 2.
The candidate genes for DRSA-related traits identified in the Zhoumai16/DK171 RIL population.
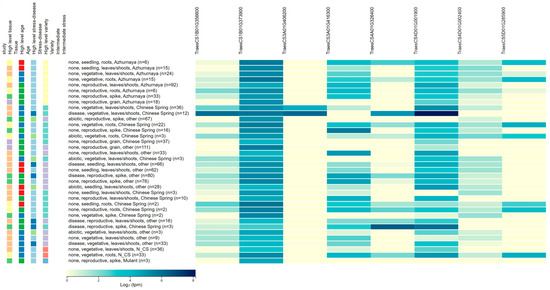
Figure 3.
The expression patterns for the eight candidate genes associated with DRSA-related traits. The left side of the figure represents the expression patterns of candidate genes in different tissues and at various developmental stages. The data originates from wheat expression data. The specific transcriptome data can be downloaded from this website (http://wheat-expression.com/, accessed on 24 June 2025).
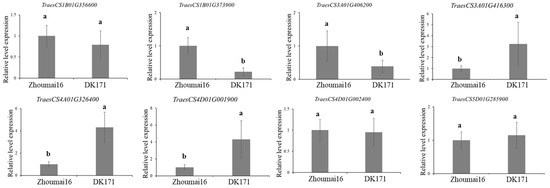
Figure 4.
The qRT-PCR results for the candidate genes identified in this study. Transcriptional analysis was conducted in the RIL population of Zhoumai16 and DK171, where different letters indicate significant differences at the p < 0.05 level.
2.4. QTL Validation
All five QTLs were employed in the development of KASP markers. A total of 5 KASP markers, Kasp-1BL-DDRW (QDDRW.daas-1BL, wsnp_Ex_rep_c67299_65845319, 586.3 Mb) and Kasp-3AL-DTRL (QDTRL.daas-3AL, Kukri_rep_c69970_717, 650.4 Mb), Kasp_DTRS_4A (QDTRL.daas-4A, wsnp_Ex_c7280_12498193, 725.6 Mb), Kasp_DDRW_5D (QDDRW.daas-5D, IACX2960, 347.5 Mb) and Kasp_DNRT_4D (QDNRT.daas-4D, Kukri_rep_c68594_530, 12.7 Mb), were successfully developed. To validate the efficacy of the 5 KASP markers, a diverse panel of 108 cultivars was employed. For Kasp-1BL-DDRW, the allele (AA) account for 60.2% (mean DDRW: 0.0262 g) exhibited lower DDRW compared to the allele (GG), which account for 38.9% with mean DDRW 0.0230 g (p < 0.05). For Kasp-3AL-DTRL, the allele (AA) account for 66.7% (mean DTRL: 63.52 cm) exhibited higher DTRL compared to the allele (GG), which account for 32.4% with mean DTRL of 56.60 cm2 (p < 0.05). For Kasp_DTRS_4A, the allele (AA) account for 38.9% (mean DTRL: 5.968 cm2) exhibited lower DTRS compared to the allele (GG), which account for 55.6% with mean DTRS 6.993 cm2 (p < 0.05). For Kasp_DDRW_5D, the favorable allele (CC 10.2%, a mean DRW of 0.0297 g) showed higher DDRW than unfavorable allele (TT, 88.9%, mean DDRW of 0.0244 g) at p = 0.05 level. For Kasp_DNRT_4D, the favorable allele (CC 65.7%, mean DNRT of 97.8) showed higher DNRT than the unfavorable allele (TT, 30.6%, mean DNRT of 83.0) (p = 0.05) (Table 3, Table 4 and Table A3).

Table 3.
Effects of Kasp_4AL_DTRS, Kasp_4DS_DNRT, and Kasp_5D_DDRW on DRSA-related traits in the natural population.

Table 4.
The primers of the KASP markers identified in this study.
To assess the accuracy of markers in detecting root phenotypes in natural populations, we selected the median value as the threshold for each trait. Phenotype values below the median were categorized as non-superior phenotypes, while those above were classified as superior phenotypes. We calculated the consistency rate between genotypes and phenotypes to provide a reference for the detection and evaluation of wheat root systems under drought conditions. The accuracy rates of Kasp_1BL_DTRS, Kasp_3AL_DTRS, Kasp_DTRS_4A, Kasp_DDRW_5D, and Kasp_DNRT_4D in detecting superior and non-superior phenotypes for DTRS, DDRW, and DNRT were 66.0% and 75.5%; 90.6% and 92.5%; 60.4% and 73.6%; 58.5% and 79.2%; 52.8% and 66.0%, respectively.
3. Discussion
Roots are the main parts of plants that take up water and nutrients. They also provide support and stability. Wheat has a fibrous root system. As it grows after germination, it develops adventitious roots. These roots are vital for anchoring the plant and absorbing water and nutrients [2,13]. They form the genetic foundation for desirable traits like drought tolerance, salt tolerance, and resistance to falling over (lodging). However, studying the root system of mature plants is difficult. Soil conditions and farming practices easily affect it. Traditional methods for taking root samples are often destructive and complicated, making it hard to measure root traits efficiently [5,14]. Research shows that the root systems of young seedlings are different. They are strongly inherited and less affected by the environment. These young roots can indicate the shape and spread of the mature root system and are closely linked to the plant’s ability to handle stress. A good root structure is the basis for having enough root volume and surface area to function well.
To adapt to diverse environments, wheat varieties have accumulated numerous genetic variations. Genetic enhancement of crop roots has seldom been analyzed [14,15]. The identification of QTL provides an effective strategy to develop molecular markers and identify candidate genes [16]. A thorough understanding of the genetic foundation of drought root system architecture traits would aid in optimizing root systems under conditions of nutrient deficiency [2,3,5,6,7,8,12,13,14]. Several QTLs associated with root system architecture-related traits have been uncovered in wheat [17,18]. Root system architecture is a highly adaptable trait across various environments [22]; QTLs governing root system architecture under drought conditions are crucial for enhancing drought tolerance. Over 10 loci influencing the number of root tips under drought stress have been mapped on chromosomes 4A, 4B, 4D, 5B, 1D, 6D, and 7D [23]. The locus on chromosome 4D (360.3–396.8 Mb) is different from the loci identified in this study (QDNRT.daas-4DS 1.2–3.6 Mb). Thus, QDNRT.daas-4DS is a new loci. Over 10 loci for root surface area under drought stress were identified on chromosomes 1D, 2A, 2B, 2D, 3B, 4A, 5B, 5D, 7A, and 7D in common wheat [23]. Of these, the locus on 4A (565.6–598.2 Mb) is nearly adjacent to the QDTRS.daas-4AL (613.6–615.2 Mb).
In total, 12 loci for total root length under drought stress were identified on chromosomes 1A, 1D, 2A, 2D, 3A, 3B, 3D, 4B, 5B, 5D, 7A, and 7D [23,24,25]. In this study, we have identified a locus for total root length under drought stress, QDTRL.daas-3AL (650.4–659.4 Mb), which is near the loci on 3AL (632.8–646.3 Mb). Until now, eight loci for DDRW were identified on chromosomes 1B, 2A, 2D, 4A, 4B, 5A, 7A, and 7D [1,26,27,28,29]. We identified two loci for DDRW, QDDRW.daas-1BL (586.3–609.0 Mb) and QDDRW.daas-5D (378.9–393.5 Mb), which differ from the loci located on chromosome 1B (120.2–159.6 Mb) and 5D (489.6–540.3 Mb) mentioned above. Thus, both QDDRW.daas-1BL and QDDRW.daas-5D are novel.
The genes associated with plant height and vernalization may also have significant effects on root system architecture-related traits [30]. Over the past 80 years, several genetic loci associated with plant height and vernalization have been identified in common wheat, and a number of functional genes have been cloned on chromosome 1B (Rht2/Rht10 at 19.18 Mb), 4D (SVP3-4D/BM1-4D at 469.46 Mb, Vrn2-4D/ZCCT1-4D at 509.43 Mb), and 5D (TaDEP1-5D at 329.11 Mb, Rht23 at 524.96 Mb, and Vrn1-5D at 470.00 Mb) [31], and 6A (Rht24, 411.93–414.88 Mb) [32]. Based on physical positions, the loci QDTRS.daas-4AL (613.6–615.2 Mb) and QDNRT.daas-4DS (1.2–3.6 Mb), QDTRL.daas-3AL (650.4–659.4 Mb), QDDRW.daas-1BL (586.3–609.0 Mb), and QDDRW.daas-5D (378.9–393.5 Mb) are different from the reported plant height and vernalization genes.
Notably, compared with previous results and meta-analyses, QDDRW.daas-1BL, QDNRT.daas-4DS, and QDDRW.daas-5D were novel. We have presented the linkage mapping results for agronomic traits in the Zhoumai16/DK171 RIL population [15] and pinpointed several genomic regions linked to both drought root system architecture-related traits and agronomic traits. Specifically, QDDRW.daas-1BL (586.3–609.0 Mb) overlaps with a QTL cluster (556–654 Mb) influencing kernel number per spike (KNS), PH, and flag leaf width (FLW), and QDDRW.daas-5D (378.9–393.5 Mb) co-locates with a QTL cluster (277.0–491.2 Mb) related to KNS, FLW, TKW, and heading date [15]. These findings suggest that the loci associated with drought response and survival mechanisms may also serve as targets for enhancing yield potential and stability.
The limitation raised regarding the absence of control experiments in this study, which currently prevents definitive verification of whether the identified QTLs are specific to drought conditions. Although these QTLs demonstrated significant effects under drought stress, their potential functionality under non-stress conditions remains unexamined. Unfortunately, insufficient seed availability precluded the inclusion of control treatments in the present experiment. To address this gap, subsequent phenotyping of root-related traits will be conducted under optimal growing conditions. The acquired data will support two analytical approaches: direct QTL mapping under control conditions to compare with drought-induced QTLs, and mapping based on trait ratios between stress and control conditions. These analyses will help distinguish QTLs unique to drought response from those constitutive to plant growth—a distinction critical for breeding applications. While the current study provides directly relevant insights for drought tolerance breeding and genetic dissection under water-limited environments, further validation under controlled conditions will significantly enhance the biological interpretation and practical utility of these loci.
A total of eight candidate genes were identified, primarily implicated in the biological metabolism of plant hormones and calcium-dependent lipid-binding domain proteins. Among these, TraesCS4D01G001900 (QDNRT.daas-4DS) encodes a CDPK-related kinase, playing a crucial role in diverse signaling pathways for root growth and development [33,34], such as root hair growth and cell length [35]. Additionally, TraesCS1B01G373900, associated with QDDRW.daas-1BL, encodes an ABC transporter family protein essential for primary root growth and shoot development. Root development is governed by various plant hormones [36]. TraesCS4A01G326400 (QDTRS.daas-4AL) and TraesCS4D01G002400 (QDNRT.daas-4DS) encode ethylene-responsive transcription factors [37]. Ethylene plays diverse roles in growth, development, signal transduction, and cell differentiation, including root growth [8,9], and influences drought root system architecture-related traits like root hair and cluster root formation [25]. TraesCS1B01G356600 (QDDRW.daas-1BL), TraesCS4D01G002400 (QDNRT.daas-4DS), and TraesCS5D01G285900 (QDDRW.daas-5D) encode the auxin-responsive proteins, auxin transport protein, and auxin-induced protein 12 in root cultures. Auxin, a core regulator, integrates with other plant hormones to regulate root development. The biosynthesis, transport, and signaling pathways of auxin, particularly indole-3-acetic acid, are crucial for plant root development [20]. TraesCS3A01G406200 (QDTRL.daas-3AL) encodes the gibberellin 20 oxidase 2 (GA20ox), a key enzyme in gibberellin synthesis [36,37], which regulate various stages of plant growth and development, promoting seed germination, plant growth, flowering induction, and other biological functions.
In this study, the candidate genes were preliminarily screened through bioinformatic annotation and expression profiling analyses. These candidate genes currently serve only as reference targets as their biological functions remain to be experimentally validated. To systematically characterize these candidate genes, the following research pipeline were applied: (1) construction of a secondary mapping population coupled with KASP marker development for high-resolution genetic mapping; (2) comprehensive identification of target genes through integrated transcriptomic and genomic variation analyses; (3) functional validation employing both gene editing (e.g., CRISPR/Cas9) and transgenic complementation approaches. It should be emphasized that the KASP markers utilized in this study were specifically designed as genetic linkage markers rather than functional markers.
Traditional wheat breeding primarily focuses on yield and disease-related traits, with root system architecture closely linked to yield traits. Although traditional breeding has improved root system characteristics, the selection process remains lengthy and less efficient due to the challenges in field measurement of drought root system architecture-related traits [19]. Moreover, seedling root development is crucial for early wheat growth. KASP markers have been widely adopted for detecting genetic variations in wheat, enabling high-throughput genotyping. By utilizing genotype data from wheat SNP arrays for QTL mapping and genome-wide association studies, linked SNPs can be converted into KASP markers, which can then be directly applied in marker-assisted selection breeding programs. This approach facilitates the efficient identification and selection of desirable traits in wheat breeding efforts [19]. KASP markers are extensively applied in the improvement of yield, disease resistance, and quality traits in wheat. In this study, we successfully developed based on tightly linked SNP markers. The accuracy rates of Kasp_1BL_DTRS, Kasp_3AL_DTRS, Kasp_DTRS_4A, Kasp_DDRW_5D, and Kasp_DNRT_4D in detecting superior and non-superior phenotypes for DTRS, DDRW, and DNRT were 66.0% and 75.5%; 90.6% and 92.5%; 60.4% and 73.6%; 58.5% and 79.2%; 52.8% and 66.0%, respectively. Thus, these KASP markers could be used as valuable tools in MAS breeding programs. Additionally, accessions carrying more favorable alleles and exhibiting superior DRSA traits along with desirable agronomic characteristics, such as Jinmai 61, Liangxing 99, Yumai 35, Yumai 47, Liangxing 66, Bainong 64, Lumai 8, Yanzhan 4110, Zhengmai 366, Jimai 22, and Aikang 58, are recommended as parental lines for the improvement of drought root system architecture traits.
4. Materials and Methods
4.1. Plant Materials and Phenotypic Traits
Zhoumai16 is a high-yield winter wheat variety in the Huang-Huai wheat region. It is suitable for high-fertilizer and high-water cultivation and has moderate drought tolerance. DK171 is a newly developed high-yield and stress-tolerant winter wheat variety, with higher drought tolerance. This study utilized a Zhoumai16/DK171 F2:6~ RIL population to conduct hydroponic experiments under greenhouse drought stress conditions, measuring DRSA-related traits with three replicates.
The standard Hoagland nutrient solution includes: Macronutrients (mg/L): Calcium Nitrate (Ca(NO3)2·4H2O) at 945 mg/L, Potassium Nitrate (KNO3) at 607 mg/L, Ammonium Dihydrogen Phosphate (NH4H2PO4) at 115 mg/L, Magnesium Sulfate (MgSO4·7H2O) at 493 mg/L. Micronutrients (mg/L): EDTA-Iron (Fe-EDTA) at 20 mg/L, Boric Acid (H3BO3) at 2.86 mg/L (or Borax), Manganese Sulfate (MnSO4·H2O) at 2.13 mg/L, Zinc Sulfate (ZnSO4·7H2O) at 0.22 mg/L, Copper Sulfate (CuSO4·5H2O) at 0.08 mg/L, and Ammonium Molybdate ((NH4)6Mo7O24·4H2O) at 0.02 mg/L. The solution pH should be adjusted to 5.5–6.5 using acid/base before use. Prepare 1× full-strength Hoagland solution in advance and adjust its pH to 5.8–6.0. Weigh 5.0 g, 7.5 g, 10.0 g, 12.5 g, 15.0 g, 17.5 g, and 20.0 g of PEG 6000 into seven clean beakers. Add about 60 mL of the pre-warmed (50–60 °C) 1× Hoagland solution to each beaker and stir gently on a magnetic stirrer until the PEG 6000 is completely dissolved. Transfer each solution to its corresponding 100 mL volumetric flask, bring the volume to the mark with additional 1× Hoagland solution, and mix thoroughly by inverting the flasks.
The root dry weights of Zhoumai16 plants subjected to 5%, 7.5%, 10%, 12.5%, 15%, 17.5%, and 20% PEG6000 were 0.0128 g, 0.0122 g, 0.0098 g, 0.0090 g, 0.0065 g, 0.0060 g, 0.0052 g, and 0.0026 g, respectively. A 12.5% concentration of PEG 6000 was added to the culture medium to simulate drought conditions. The methodology is as follows: 20 wheat seeds from each line were randomly selected and surface-sterilized with 10% H2O2 for 20 min, then placed in Petri dishes containing moist filter paper. When the coleoptile length reached approximately 2 cm, the seedlings were transferred to plastic trays (53 × 27 cm) with Hoagland nutrient solution supplemented with 12.5% PEG 6000 to induce osmotic stress.
These trays were then placed in a constant temperature culture room maintained at 25 °C, with 16 h light and 8 h darkness. After three weeks of growth in the greenhouse, seedling DRSA-related traits, including DDRW, DTRL, DTRA, and DNRT, were measured using the WinRHIZO software V1.0 (https://www.quantitative-plant.org/software/winrhizo, accessed on 24 June 2025) (Regent, Vancouver, BC, Canada). The specific method was as follows: thoroughly washed wheat roots were neatly arranged in a scanning tray, and scanned images were obtained using an Expression 11000XL scanner (Seiko Epson Corporation, Nagano Prefecture, Japan). The scanned images were analyzed using WinRHIZO software. Five plants were measured for each line, with three biological replicates.
This study used 108 varieties to validate the effects of KASP markers. The phenotypic values of DDRW, DTRL, DTRA, and DNRT for this validation population were also uniformly identified using the aforementioned method.
4.2. Genome Wide Linkage Mapping
The Zhoumai16/DK171 RIL population was genotyped using the wheat 90K SNP chip (CapitalBio Corporation, Beijing, China). The quality control followed: SNPs were set at missing data exceeding 20% or minor allele frequency (MAF) below 0.5. Filtered SNPs were classified into bin markers by IciMapping v4.2 [17]. Subsequently, the regression mapping algorithm in JoinMap v4.0 was used to calculate linkage distances for the obtained BIN markers and construct a higher-density linkage map. The successfully constructed linkage map has been previously reported by Wen et al. [18] and Li et al. [15]. Based on the constructed high-density SNP genetic map and the obtained root DRSA-related traits under drought, genome-wide linkage mapping was conducted by the inclusive composite interval mapping (ICIM-add) using IciMapping v4.1 [17]. The logarithm of odds (LOD) threshold for significant QTLs was determined to be 2.60 based on 1000 permutations. The physical positions were determined by IWGSC v1.0.
4.3. Identification of Candidate Genes for Drought-Related Traits
To identify the candidate genes associated with drought-related trait QTLs detected in the Zhoumai16/DK171 RIL population, high-confidence annotation genes within the LD block region surrounding each QTL peak SNP were extracted from the wheat IWGSC v1.0 [21]. Combining annotation, high-confidence genes with relevant annotated functions and differences in the coding regions between the two parents were screened as candidate genes. To investigate the expression patterns and identify candidate genes with notable transcription levels in seedlings or root tissues, we utilized the publicly accessible Triticum aestivum gene expression database [12] (http://wheat-expression.com/, accessed on 24 June 2025). After phenotyping, roots were sampled, and RNA was extracted from the root samples using the TRIzol method. cDNA was synthesized using the HiScript II cDNA Synthesis Kit. Primers for qRT-PCR were designed with Primer Premier 5.0. The reaction mixture consisted of 20 µL, including 2 µL of cDNA, 10 µL of ChamQ Universal SYBR qPCR Master Mix, and 0.4 µL of each primer. TaActin1 was used as an internal control to normalize the expression levels of different samples. The gene expression levels were analyzed using the 2−ΔΔCT method. All assays were performed with two biological replicates and three technical replicates.
4.4. KASP Marker Development and Validation
For all loci, flanking SNPs were converted to KASP markers [19], designed using PolyMarker (http://www.polymarker.info/, accessed on 24 June 2025). The 384-well plates were analyzed on a PHERAstarplus SNP, and genotyping was conducted by KlusterCaller (LGC) (https://www.lgcstandards.com/, accessed on 24 June 2025) (London, UK). All developed KASP markers required genetic effect validation using 108 varieties mainly from the Yellow and Huai Wheat Region [20]. These 108 materials primarily include the main popularized varieties, key backbone parents, and representative lines from the Yellow and Huai River Valleys Wheat Zone.
5. Conclusions
In conclusion, this study highlights the critical role of drought-related root system architecture in enhancing wheat resilience to drought. By analyzing the Zhoumai16/DK171 RIL population, five QTLs associated with DRSA-related traits were identified, including novel loci QDDRW.daas-1BL, QDNRT.daas-4DS, and QDDRW.daas-5D. These QTLs explained 6.1% to 18.9% of the phenotypic variances, with favorable alleles contributed by both Zhoumai16 and DK171. Additionally, five KASP markers were developed and validated, providing valuable tools for MAS breeding. The identification of eight candidate genes further enriches the genetic resources for wheat drought tolerance. This research advances our understanding of the genetic basis of drought resistance and offers practical tools for breeding drought-tolerant wheat cultivars.
Author Contributions
Y.J. carried out the experimental and wrote the paper. G.C., X.Q., F.W., and H.J. participated in field trials. L.Z., C.L., J.L., and W.L. contributed to data analysis. P.L. designed the experiment and assisted in writing the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financially supported by the Shandong Province Wheat Industry Technology System (SDAIT-01-23); Shandong Agricultural Seeds Engineering Project (2023LZGC009, 2021LZGC013); Shandong Province Key R&D Plan (2022LZG002-4); Taishan Scholarship (tspd20221108); Jinan 20 New Universities Project (202228067).
Institutional Review Board Statement
We declare that these experiments complied with the ethical standards in China.
Data Availability Statement
All datasets generated for this study are included in the article; further inquiries can be directed to the first author.
Conflicts of Interest
The authors declare that they have no competing interests.
Appendix A

Table A1.
The drought root system architecture-related traits in Zhoumai16/DK171 RIL population.
Table A1.
The drought root system architecture-related traits in Zhoumai16/DK171 RIL population.
| Accessions | Total Root Length-Drought (mm) | Total Root Surface-Drought (cm2) | Total Root Volume-Drought (mm3) | Dry Root Weight-Drought (g) |
|---|---|---|---|---|
| LINE1 | 26.5 | 5.8 | 201.0 | 0.0203 |
| LINE2 | 25.8 | 6.0 | 24.7 | 0.0169 |
| LINE3 | 24.1 | 5.4 | 35.7 | 0.0181 |
| LINE4 | 27.6 | 6.5 | 48.3 | 0.0277 |
| LINE5 | 35.0 | 7.6 | 45.3 | 0.0276 |
| LINE6 | 42.5 | 8.6 | 73.3 | 0.0254 |
| LINE7 | 35.5 | 8.7 | 94.3 | 0.0295 |
| LINE8 | 38.1 | 6.1 | 81.0 | 0.0271 |
| LINE9 | 35.5 | 6.9 | 105.7 | 0.0213 |
| LINE10 | 50.0 | 10.3 | 79.3 | 0.0268 |
| LINE11 | 26.8 | 5.7 | 108.0 | 0.0235 |
| LINE12 | 24.6 | 6.5 | 57.3 | 0.0176 |
| LINE13 | 35.9 | 8.1 | 48.7 | 0.0267 |
| LINE14 | 37.9 | 6.4 | 63.0 | 0.0209 |
| LINE15 | 60.6 | 8.5 | 70.7 | 0.0244 |
| LINE16 | 40.5 | 9.2 | 136.7 | 0.0237 |
| LINE17 | 42.5 | 6.7 | 99.0 | 0.0279 |
| LINE18 | 36.2 | 7.2 | 137.3 | 0.0248 |
| LINE19 | 31.9 | 6.2 | 86.0 | 0.0246 |
| LINE20 | NN | NN | NN | NN |
| LINE21 | 49.1 | 8.1 | 79.3 | 0.0232 |
| LINE22 | 42.8 | 7.9 | 119.7 | 0.0284 |
| LINE23 | 41.8 | 7.4 | 104.7 | 0.0305 |
| LINE24 | 40.8 | 6.1 | 147.0 | 0.0233 |
| LINE25 | 44.1 | 7.8 | 99.7 | 0.0286 |
| LINE26 | 40.6 | 8.1 | 84.7 | 0.0237 |
| LINE27 | 45.3 | 8.3 | 90.0 | 0.0226 |
| LINE28 | 43.6 | 10.7 | 85.7 | 0.0271 |
| LINE29 | 39.4 | 8.4 | 103.3 | 0.0278 |
| LINE30 | 46.7 | 8.9 | 140.0 | 0.0255 |
| LINE31 | NN | NN | NN | NN |
| LINE32 | 41.9 | 8.3 | 128.3 | 0.0273 |
| LINE33 | 35.2 | 7.6 | 198.7 | 0.0310 |
| LINE34 | 34.5 | 7.0 | 96.7 | 0.0315 |
| LINE35 | 41.6 | 8.2 | 116.7 | 0.0231 |
| LINE36 | 38.0 | 7.2 | 156.7 | 0.0206 |
| LINE37 | 37.4 | 6.9 | 84.0 | 0.0342 |
| LINE38 | 40.5 | 10.4 | 99.0 | 0.0235 |
| LINE39 | 35.9 | 6.0 | 129.3 | 0.0298 |
| LINE40 | 39.3 | 8.2 | 82.3 | 0.0346 |
| LINE41 | 35.0 | 8.6 | 87.0 | 0.0272 |
| LINE42 | 40.7 | 8.3 | 91.0 | 0.0259 |
| LINE43 | 37.0 | 7.4 | 163.0 | 0.0246 |
| LINE44 | 47.5 | 9.0 | 105.0 | 0.0285 |
| LINE45 | 6.8 | 103.7 | 0.0297 | |
| LINE46 | 40.4 | 7.7 | 124.7 | 0.0292 |
| LINE47 | 47.3 | 7.9 | 101.7 | 0.0313 |
| LINE48 | 35.9 | 9.9 | 134.3 | 0.0331 |
| LINE49 | 35.0 | 6.0 | 88.0 | 0.0238 |
| LINE50 | 48.5 | 9.8 | 114.0 | 0.0284 |
| LINE51 | 36.1 | 8.4 | 117.0 | 0.0273 |
| LINE52 | 52.8 | 7.6 | 79.0 | 0.0296 |
| LINE53 | 36.2 | 7.8 | 114.0 | 0.0304 |
| LINE54 | 56.6 | 10.3 | 72.0 | 0.0298 |
| LINE55 | 42.5 | 8.1 | 176.3 | 0.0285 |
| LINE56 | 41.9 | 8.2 | 128.0 | 0.0261 |
| LINE57 | 35.7 | 7.1 | 113.3 | 0.0275 |
| LINE58 | 36.9 | 6.9 | 96.0 | 0.0303 |
| LINE59 | 48.0 | 7.5 | 126.0 | 0.0332 |
| LINE60 | 32.2 | 7.8 | 116.3 | 0.0278 |
| LINE61 | 47.5 | 8.6 | 77.3 | 0.0266 |
| LINE62 | 40.3 | 8.1 | 129.0 | 0.0245 |
| LINE63 | 37.1 | 7.1 | 99.0 | 0.0267 |
| LINE64 | 44.5 | 7.5 | 95.7 | 0.0299 |
| LINE65 | 47.8 | 6.9 | 139.0 | 0.0310 |
| LINE66 | 40.1 | 7.0 | 117.7 | 0.0364 |
| LINE67 | 41.2 | 9.2 | 122.3 | 0.0295 |
| LINE68 | 42.8 | 7.7 | 141.0 | 0.0288 |
| LINE69 | 39.7 | 6.2 | 151.7 | 0.0304 |
| LINE70 | NN | NN | NN | NN |
| LINE71 | 37.1 | 9.2 | 114.7 | 0.0278 |
| LINE72 | 41.9 | 7.8 | 109.0 | 0.0315 |
| LINE73 | 38.6 | 7.3 | 112.7 | 0.0289 |
| LINE74 | 47.3 | 9.0 | 95.0 | 0.0370 |
| LINE75 | 42.1 | 10.6 | 122.0 | 0.0221 |
| LINE76 | 37.1 | 6.8 | 144.3 | 0.0374 |
| LINE77 | NN | NN | NN | NN |
| LINE78 | 39.0 | 7.8 | 89.7 | 0.0274 |
| LINE79 | 39.6 | 6.9 | 109.7 | 0.0301 |
| LINE80 | 43.8 | 8.5 | 135.0 | 0.0255 |
| LINE81 | 43.1 | 8.4 | 102.7 | 0.0276 |
| LINE82 | 45.0 | 7.1 | 100.0 | 0.0283 |
| LINE83 | 40.3 | 7.8 | 123.3 | 0.0256 |
| LINE84 | 30.8 | 6.8 | 82.0 | 0.0317 |
| LINE85 | 34.2 | 7.8 | 62.3 | 0.0278 |
| LINE86 | NN | NN | NN | NN |
| LINE87 | 41.1 | 6.2 | 93.0 | 0.0282 |
| LINE88 | 37.3 | 9.5 | 136.3 | 0.0236 |
| LINE89 | 50.6 | 9.1 | 143.3 | 0.0286 |
| LINE90 | 29.8 | 6.4 | 98.3 | 0.0271 |
| LINE91 | 30.4 | 8.7 | 96.7 | 0.0274 |
| LINE92 | 45.5 | 9.3 | 100.7 | 0.0308 |
| LINE93 | 38.1 | 7.0 | 114.0 | 0.0342 |
| LINE94 | 44.4 | 7.9 | 141.7 | 0.0282 |
| LINE95 | 46.9 | 8.7 | 139.0 | 0.0363 |
| LINE96 | 38.1 | 7.4 | 153.3 | 0.0301 |
| LINE97 | 27.7 | 6.8 | 99.7 | 0.0247 |
| LINE98 | 32.8 | 6.4 | 53.3 | 0.0245 |
| LINE99 | 34.9 | 8.2 | 156.3 | 0.0309 |
| LINE100 | 46.1 | 8.0 | 99.7 | 0.0298 |
| LINE101 | 42.4 | 7.4 | 117.3 | 0.0323 |
| LINE102 | 48.4 | 8.1 | 68.3 | 0.0345 |
| LINE103 | 50.4 | 9.2 | 117.0 | 0.0272 |
| LINE104 | 47.3 | 8.6 | 121.3 | 0.0431 |
| LINE105 | 40.3 | 7.7 | 164.7 | 0.0297 |
| LINE106 | 45.8 | 6.0 | 80.0 | 0.0305 |
| LINE107 | 35.7 | 7.0 | 145.0 | 0.0329 |
| LINE108 | 50.4 | 8.1 | 87.7 | 0.0296 |
| LINE109 | 43.2 | 8.2 | 136.3 | 0.0287 |
| LINE110 | NN | NN | NN | 0.0286 |
| LINE111 | 41.0 | 6.4 | 71.7 | 0.0307 |
| LINE112 | NN | NN | NN | 0.0274 |
| LINE113 | NN | NN | NN | 0.0284 |
| LINE114 | NN | NN | NN | 0.0413 |
| LINE115 | NN | NN | NN | 0.0302 |
| LINE116 | 54.5 | 10.3 | 97.0 | 0.0320 |
| LINE117 | 44.6 | 7.7 | 109.7 | 0.0328 |
| LINE118 | NN | NN | NN | 0.0256 |
| LINE119 | NN | NN | NN | 0.0312 |
| LINE120 | 41.3 | 7.0 | 104.3 | 0.0272 |
| LINE121 | 38.3 | 6.0 | 91.0 | 0.0278 |
| LINE122 | 50.3 | 7.6 | 77.7 | 0.0325 |
| LINE123 | 45.1 | 8.3 | 165.3 | 0.0317 |
| LINE124 | 44.7 | 8.2 | 77.0 | 0.0306 |
| LINE125 | 37.8 | 7.2 | 101.7 | 0.0328 |
| LINE126 | 38.2 | 6.2 | 117.0 | 0.0227 |
| LINE127 | 30.7 | 5.7 | 164.7 | 0.0206 |
| LINE128 | 43.8 | 7.9 | 55.0 | 0.0288 |
| LINE129 | 57.4 | 7.7 | 102.7 | 0.0327 |
| LINE130 | 49.5 | 7.8 | 147.7 | 0.0345 |
| LINE131 | 44.4 | 7.6 | 125.3 | 0.0354 |
| LINE132 | 38.6 | 7.3 | 145.3 | 0.0292 |
| LINE133 | 45.8 | 8.6 | 97.7 | 0.0278 |
| LINE134 | 44.3 | 9.0 | 67.3 | 0.0286 |
| LINE135 | 36.1 | 6.7 | 117.0 | 0.0296 |
| LINE136 | 34.8 | 7.3 | 126.3 | 0.0322 |
| LINE137 | 42.3 | 8.2 | 143.3 | 0.0294 |
| LINE138 | 27.3 | 6.3 | 151.3 | 0.0264 |
| LINE139 | 36.2 | 7.5 | 68.0 | 0.0308 |
| LINE140 | 42.1 | 7.9 | 137.0 | 0.0299 |
| LINE141 | NN | NN | NN | NN |
| LINE142 | 50.2 | 8.3 | 204.0 | 0.0304 |
| LINE143 | 28.0 | 6.7 | 98.7 | 0.0162 |
| LINE144 | 36.1 | 7.1 | 66.3 | 0.0284 |
| LINE145 | 32.9 | 6.4 | 70.0 | 0.0258 |
| LINE146 | 38.6 | 9.3 | 83.3 | 0.0221 |
| LINE147 | 45.2 | 8.6 | 88.7 | 0.0280 |
| LINE148 | 44.6 | 8.9 | 196.7 | 0.0278 |
| LINE149 | 40.5 | 8.2 | 187.3 | 0.0326 |
| LINE150 | 46.6 | 9.4 | 126.7 | 0.0308 |
| LINE151 | 45.4 | 7.7 | 150.7 | 0.0287 |
| LINE152 | 41.3 | 8.8 | 115.3 | 0.0255 |
| LINE153 | 37.4 | 6.8 | 113.3 | 0.0251 |
| LINE154 | 41.0 | 7.4 | 89.3 | 0.0330 |
| LINE155 | 39.5 | 6.2 | 108.0 | 0.0243 |
| LINE156 | 56.6 | 7.9 | 127.3 | 0.0207 |
| LINE157 | 30.6 | 7.0 | 142.0 | 0.0236 |
| LINE158 | 41.0 | 9.0 | 48.0 | 0.0318 |
| LINE159 | 49.9 | 9.9 | 165.3 | 0.0323 |
| LINE160 | 46.7 | 7.4 | 213.7 | 0.0372 |
| LINE161 | 46.6 | 8.7 | 87.7 | 0.0268 |
| LINE162 | 33.0 | 7.0 | 115.0 | 0.0293 |
| LINE163 | 39.9 | 8.5 | 86.0 | 0.0242 |
| LINE164 | 37.1 | 7.1 | 67.7 | 0.0267 |
| LINE165 | 43.7 | 8.6 | 60.3 | 0.0350 |
| LINE166 | 44.4 | 8.6 | 119.3 | 0.0293 |
| LINE167 | 63.8 | 9.2 | 145.0 | 0.0306 |
| LINE168 | 41.0 | 7.6 | 136.0 | 0.0279 |
| LINE169 | 39.0 | 9.0 | 122.7 | 0.0287 |
| LINE170 | 35.7 | 7.7 | 180.0 | 0.0241 |
| LINE171 | 52.4 | 9.0 | 59.7 | 0.0230 |
| LINE172 | 49.4 | 10.1 | 172.7 | 0.0343 |
| LINE173 | 41.9 | 7.2 | 142.3 | 0.0279 |
| LINE174 | 44.7 | 9.2 | 101.7 | 0.0271 |
| LINE175 | 36.8 | 8.6 | 121.0 | 0.0268 |
| LINE176 | 49.0 | 8.6 | 81.0 | 0.0346 |
| LINE177 | 48.7 | 8.8 | 131.3 | 0.0304 |
| LINE178 | 45.5 | 9.1 | 202.0 | 0.0301 |
| LINE179 | 44.1 | 8.2 | 85.7 | 0.0301 |
| LINE180 | 32.6 | 6.7 | 86.7 | 0.0253 |
| LINE181 | 33.7 | 7.8 | 53.0 | 0.0259 |
| LINE182 | 33.8 | 6.4 | 72.0 | 0.0230 |
| LINE183 | 50.8 | 9.7 | 94.0 | 0.0212 |
| LINE184 | 40.6 | 7.6 | 71.3 | 0.0324 |
| LINE185 | 40.5 | 8.9 | 107.0 | 0.0257 |
| LINE186 | 41.9 | 8.2 | 109.3 | 0.0277 |
| LINE187 | 45.8 | 9.3 | 96.3 | 0.0314 |
| LINE188 | 34.6 | 7.0 | 138.0 | 0.0316 |
| LINE189 | 35.0 | 8.0 | 114.0 | 0.0317 |
| LINE190 | 32.0 | 7.3 | 120.0 | 0.0203 |
| LINE191 | 35.3 | 7.1 | 51.3 | 0.0314 |
| LINE192 | 41.6 | 7.2 | 116.7 | 0.0311 |
| LINE193 | 43.9 | 7.7 | 58.0 | 0.0369 |
| LINE194 | 44.7 | 8.5 | 124.0 | 0.0280 |
| LINE195 | 42.2 | 9.3 | 117.3 | 0.0323 |
| LINE196 | 39.1 | 7.3 | 189.3 | 0.0292 |
| LINE197 | NN | NN | NN | NN |
| LINE198 | 43.5 | 7.9 | 85.7 | 0.0321 |
| LINE199 | 29.3 | 7.1 | 130.0 | 0.0195 |
| LINE200 | 51.9 | 8.3 | 77.3 | 0.0233 |
| LINE201 | 39.5 | 8.5 | 65.7 | 0.0246 |
| LINE202 | 40.1 | 7.1 | 44.7 | 0.0212 |
| LINE203 | 40.3 | 8.2 | 87.7 | 0.0284 |
| LINE204 | 38.6 | 8.3 | 96.7 | 0.0217 |
| LINE205 | 39.2 | 8.5 | 65.0 | 0.0263 |
| LINE206 | 39.2 | 8.2 | 82.7 | 0.0229 |
| LINE207 | 49.0 | 7.9 | 72.7 | 0.0232 |
| LINE208 | 35.8 | 7.4 | 102.0 | 0.0221 |
| LINE209 | NN | NN | NN | NN |
| LINE210 | 39.1 | 7.3 | 56.7 | 0.0234 |
| LINE211 | 44.0 | 7.9 | 68.3 | 0.0306 |
| LINE212 | NN | NN | NN | NN |
| LINE213 | 34.0 | 7.8 | 133.7 | 0.0309 |
| LINE214 | 51.4 | 7.2 | 87.0 | 0.0331 |
| LINE215 | 46.3 | 9.4 | 97.7 | 0.0306 |
| LINE216 | 46.8 | 7.8 | 61.7 | 0.0338 |
| LINE217 | 40.2 | 9.2 | 107.0 | 0.0297 |
| LINE218 | 40.5 | 8.2 | 76.3 | 0.0265 |
| LINE219 | 36.5 | 9.1 | 73.7 | 0.0258 |
| LINE220 | 41.4 | 7.4 | 67.0 | 0.0327 |
| LINE221 | 47.1 | 9.4 | 86.0 | 0.0304 |
| LINE222 | 38.9 | 9.6 | 126.0 | 0.0276 |
| LINE223 | 38.5 | 6.7 | 103.0 | 0.0285 |
| LINE224 | 39.2 | 7.3 | 67.3 | 0.0306 |
| LINE225 | 35.0 | 7.1 | 80.3 | 0.0281 |
| LINE226 | 37.4 | 7.8 | 82.7 | 0.0276 |
| LINE227 | 30.5 | 7.0 | 84.7 | 0.0182 |
| LINE228 | 33.5 | 7.2 | 27.7 | 0.0280 |
| LINE229 | 43.4 | 7.1 | 81.7 | 0.0302 |
| LINE230 | 35.5 | 7.7 | 74.7 | 0.0294 |
| LINE231 | NN | 0.0 | NN | NN |
| LINE232 | 42.3 | 9.1 | 114.7 | 0.0268 |
| LINE233 | 39.7 | 9.2 | 102.0 | 0.0287 |
| LINE234 | NN | NN | NN | NN |
| LINE235 | 33.2 | 7.8 | 95.7 | 0.0335 |
| LINE236 | 33.6 | 7.1 | 79.3 | 0.0251 |
| LINE237 | 41.8 | 7.3 | 68.3 | 0.0303 |
| LINE238 | 42.4 | 8.1 | 95.0 | 0.0284 |
| LINE239 | 47.0 | 8.5 | 173.7 | 0.0278 |
| LINE240 | 38.1 | 8.1 | 79.3 | 0.0322 |
| LINE241 | 33.2 | 6.0 | 80.0 | 0.0304 |
| LINE242 | 27.6 | 6.0 | 113.3 | 0.0344 |
| LINE243 | 33.0 | 6.1 | 105.3 | 0.0262 |
| LINE244 | 36.8 | 8.3 | 157.0 | 0.0285 |
| LINE245 | 40.2 | 7.3 | 102.0 | 0.0418 |
| LINE246 | 32.2 | 7.4 | 213.7 | 0.0227 |
| LINE247 | 33.1 | 6.1 | 112.7 | 0.0311 |
| LINE248 | 29.7 | 8.2 | 85.3 | 0.0264 |
| LINE249 | 26.2 | 5.6 | 85.7 | 0.0235 |
| LINE250 | 37.7 | 7.8 | 57.0 | 0.0247 |
| LINE251 | 41.7 | 6.8 | 77.0 | 0.0312 |
| LINE252 | 28.7 | 6.9 | 159.0 | 0.0230 |
| LINE253 | 35.1 | 7.5 | 79.7 | 0.0315 |
| LINE254 | 38.5 | 7.1 | 150.0 | 0.0302 |
| LINE255 | 28.6 | 7.2 | 126.7 | 0.0294 |
| LINE256 | 38.9 | 7.5 | 108.7 | 0.0273 |
| LINE257 | 37.0 | 7.0 | 155.0 | 0.0250 |
| LINE258 | 41.8 | 9.0 | 88.3 | 0.0348 |
| LINE259 | 44.1 | 8.0 | 118.7 | 0.0262 |
| LINE260 | 44.1 | 9.5 | 87.3 | 0.0281 |
| LINE261 | 34.6 | 7.0 | 165.7 | 0.0234 |
| LINE262 | 39.7 | 6.4 | 78.7 | 0.0380 |

Table A2.
The qRT-PCR primers for the candidate genes.
Table A2.
The qRT-PCR primers for the candidate genes.
| Candidate Gene (IWGSC1.0) | Candidate Gene (IWGSC2.1) | Forward Primers (5′-3′) | Reverse Primers (5′-3′) |
|---|---|---|---|
| TraesCS1B01G356600 | TraesCS1B03G0973300 | AGGCTTTCTTGAAGCCCAGA | GACCAGCATCCTGTCTCCTT |
| TraesCS1B01G373900 | TraesCS1B03G1018500 | GCCCTCAAGGTGAAAGGTCT | CTTCCAGAGCTGCAAGTCG |
| TraesCS3A01G406200 | TraesCS3A03G0950800 | CATGCGGTGCAACTACTACC | GTGTCGCCGATGTTGATGAC |
| TraesCS3A01G416300 | TraesCS3A03G0969200 | CATGAGCCTCAACGAGAAGC | CTCTCATGCTGGTACGACGA |
| TraesCS4A01G326400 | TraesCS4A03G0812600 | AGCAGAACCACCACCTACC | CTGGTGTCGAACGGAAGAAC |
| TraesCS4D01G001900 | TraesCS4D03G0002800 | AGGAATTTGAGGCTGAAGCG | GGGACGTTGGAGTCGTAGTA |
| TraesCS4D01G002400 | TraesCS4D03G0003700 | GACACTGGAGTGGAACTGGA | CTAACAGCGGTCATCAAGGC |
| TraesCS5D01G285900 | TraesCS5D03G0654200 | CTGAGTCTGGTAGCGTGGAG | CCGTCGTTGCCCACGCCGTC |

Table A3.
The DRSA-related traits in the 108 wheat varieties and the genotype data for the developed KASP markers.
Table A3.
The DRSA-related traits in the 108 wheat varieties and the genotype data for the developed KASP markers.
| Name | Kasp_1BL_DDRW | DDRW (g) | Kasp_5D_DDRW | Kasp_3AL_DTRL | DDRT (mm) | Kasp_4DS_DNRT | DNRT | Kasp_4AL_DTRS | DTRS (cm2) |
|---|---|---|---|---|---|---|---|---|---|
| Aikang 58 | GG | 0.0304 | CC | AA | 74.0 | CC | 100.2 | GG | 7.7 |
| Bainong 3217 | AA | 0.0316 | TT | AA | 54.1 | CC | 18.6 | AA | 3.1 |
| Bainong 64 | AA | 0.0358 | TT | AA | 88.3 | TT | 115.2 | GG | 9.2 |
| Bima 4 | GG | 0.0198 | TT | GG | 48.9 | TT | 11.7 | GG | 2.4 |
| Gaoyou 503 | AA | 0.0291 | TT | AA | 58.6 | CC | 57.6 | GG | 5.2 |
| Gaocheng 8901 | AA | 0.0316 | TT | AA | 90.5 | CC | 123.5 | GG | 10.4 |
| Huapei 5 | GG | 0.0235 | TT | AA | 61.5 | CC | 22.7 | GG | 3.6 |
| Huaimai 18 | AA | 0.0251 | TT | GG | 64.5 | TC | 69.5 | GG | 5.9 |
| Huaimai 20 | GG | 0.0287 | TT | AA | 80.2 | CC | 48.8 | AA | 6.1 |
| Huaimai 21 | GG | 0.0345 | CC | GG | 64.0 | TT | 16.7 | AA | 3.2 |
| Jimai 19 | GG | 0.0313 | CC | GG | 86.1 | CC | 170.8 | AA | 11.4 |
| Jimai 20 | AA | 0.0324 | TT | GG | 62.9 | CC | 69.8 | GG | 6 |
| Jimai 21 | GG | 0.0222 | TT | AA | 46.8 | CC | 10.8 | AA | 2.2 |
| Jimai 22 | GG | 0.0307 | CC | GG | 59.6 | CC | 103.9 | AA | 6.3 |
| Jinan 13 | AA | 0.0299 | TT | AA | 77.9 | TT | 23 | AA | 4.1 |
| Jinan 17 | GG | 0.0233 | TT | AA | 36.8 | CC | 14.4 | AA | 2.1 |
| Jining 16 | AA | 0.0196 | TT | AA | 44.8 | CC | 10.3 | AA | 2.1 |
| Jishi 02-1 | AA | 0.0259 | TT | GG | 36.0 | CC | 8 | GG | 1.7 |
| Jinmai 61 | AA | 0.0384 | TT | AA | 66.5 | CC | 44.9 | GG | 4.8 |
| Lankao 24 | GG | 0.0279 | TT | GG | 61.6 | TT | 30.9 | AA | 4.2 |
| Lankao 2 | GG | 0.0252 | TT | AA | 69.5 | TT | 162.5 | AA | 9.4 |
| Lankao 906 | AA | 0.0326 | TT | AA | 54.9 | TT | 58.3 | AA | 5.3 |
| Liangxing 66 | AA | 0.0361 | CC | AA | 92.7 | CC | 234 | AA | 12.8 |
| Liangxing 99 | GG | 0.0364 | CC | GG | 61.1 | CC | 18.6 | GG | 3.3 |
| Linhan 2 | AA | 0.0247 | TT | AA | 79.5 | TT | 49.6 | GG | 5.8 |
| Linkang 12 | AA | 0.0265 | TT | AA | 63.9 | TT | 62 | AA | 5.6 |
| Linmai 2 | GG | 0.0184 | TT | GG | 58.6 | CC | 55.8 | GG | 5 |
| Linmai 4 | AA | 0.0245 | TT | AA | 45.1 | CC | 47.8 | AA | 4 |
| Lumai 15 | AA | 0.0271 | TT | GG | 57.9 | CC | 23.7 | GG | 3.6 |
| Lumai 21 | GG | 0.0203 | TT | AA | 54.3 | CC | 79.4 | GG | 5.8 |
| Lumai 23 | AA | 0.0283 | TT | AA | 76.1 | CC | 70.1 | AA | 6.8 |
| Lumai 5 | GG | 0.0118 | CC | GG | 60.7 | TT | 23.9 | AG | 3.6 |
| Lumai 6 | AA | 0.024 | TT | AA | 63.1 | CC | 61.7 | AA | 5.6 |
| Lumai 7 | AA | 0.0297 | TT | AA | 70.3 | CC | 68 | GG | 6.6 |
| Lumai 8 | AA | 0.0357 | TT | GG | 62.0 | CC | 24.8 | GG | 3.9 |
| Lumai 9 | GG | 0.0184 | TT | AA | 81.6 | CC | 150.8 | GG | 10.9 |
| Luyuan 502 | AA | 0.0243 | TT | AA | 51.8 | CC | 129.4 | GG | 7.1 |
| Luohan 2 | AA | 0.023 | TT | GG | 61.9 | TT | 79 | AA | 6.8 |
| Luomai 21 | GG | 0.0271 | TT | AA | 66.7 | CC | 122.5 | GG | 8.9 |
| Neixiang 188 | AA | 0.028 | TT | GG | 58.1 | TT | 85 | GG | 5.9 |
| Neixiang 5 | AA | 0.019 | TT | AA | 63.2 | CC | 77.8 | AA | 6.8 |
| Shannong 20 | AA | 0.0232 | TT | GG | 63.7 | TT | 166.8 | GG | 9.5 |
| Shan 150 | AA | 0.028 | TT | AA | 67.1 | TT | 72.2 | GG | 6.8 |
| Shan 229 | AA | 0.0294 | TT | AA | 89.5 | CC | 140 | GG | 10.5 |
| Shan 253 | AA | 0.0202 | TT | AA | 69.5 | CC | 295.4 | GG | 12.2 |
| Shan 354 | AA | 0.022 | TT | GG | 55.0 | CC | 66.9 | AA | 5.9 |
| Shan 512 | AA | 0.0226 | TT | AA | 47.3 | CC | 71 | AA | 5 |
| Shan 715 | GG | 0.0316 | CC | GG | 72.0 | CC | 80 | AA | 7 |
| Shanmai 94 | AA | 0.0158 | TT | AA | 59.7 | TT | 224.4 | AA | 11.3 |
| Shannong 78-59 | AA | 0.0279 | TT | GG | 45.2 | TT | 64.4 | AA | 5 |
| Shanyou 225 | AA | 0.0219 | TT | GG | 68.0 | TT | 90.7 | AA | 7.1 |
| DK171 | GG | 0.0214 | TT | AA | 62.3 | CC | 54.5 | GG | 5.4 |
| Shijiazhuang 15 | AA | 0.0252 | TT | AA | 65.4 | CC | 31.4 | AA | 4.3 |
| Shixin 733 | GG | 0.0199 | TT | GG | 63.0 | TT | 60.5 | AA | 5.9 |
| Shiyou 17 | AA | 0.0246 | TT | AA | 68.2 | CC | 83.2 | GG | 7.1 |
| Sunong 6 | AA | 0.0237 | TT | AA | 69.8 | CC | 199.5 | GG | 11.6 |
| Taishan 5 | GG | 0.0288 | TT | AA | 64.5 | TT | 177.3 | GG | 8.8 |
| Wanmai 19 | AA | 0.0183 | TT | AA | 62.5 | CC | 228.1 | GG | 11.3 |
| Wanmai 29 | AA | 0.026 | TT | AA | 47.6 | CC | 235.6 | GG | 8.4 |
| Wanmai 38 | AA | 0.0208 | TT | AA | 77.1 | CC | 190.9 | GG | 11.8 |
| Wanmai 50 | AA | 0.017 | TT | AA | 70.0 | CC | 293.3 | GG | 13.8 |
| Wanmai 52 | GG | 0.0212 | TT | AA | 58.6 | CC | 99 | GG | 7.1 |
| Wanmai 53 | AA | 0.0213 | TT | AA | 71.6 | CC | 140.1 | GG | 9.5 |
| Wennong 14 | AA | 0.0211 | TT | AA | 47.8 | CC | 156.2 | AA | 7.7 |
| Wennong 5 | AA | 0.025 | TT | AA | 71.3 | CC | 159.4 | GG | 10.2 |
| Wunong 148 | AA | 0.023 | TT | AA | 50.1 | TT | 23.7 | AA | 3.2 |
| Xinnong 291 | AA | 0.0372 | TT | AA | 71.3 | TT | 39 | GG | 5 |
| Xiaoyan 22 | AA | 0.0224 | TT | GG | 63.6 | CC | 128.8 | AA | 8.1 |
| Xiaoyan 54 | GG | 0.023 | TT | AA | 60.2 | CC | 102.1 | AA | 5.6 |
| Xiaoyan 6 | AA | 0.0287 | TT | AA | 46.3 | CC | 119.9 | AA | 8.6 |
| Xiaoyan 81 | AA | 0.026 | TT | AA | 75.6 | CC | 91.2 | AA | 6.9 |
| Xinmai 19 | AA | 0.0231 | TT | AA | 68.7 | CC | 108 | GG | 9 |
| Xinmai 9 | GG | 0.0268 | CC | GG | 81.0 | TT | 12.7 | AG | 2.8 |
| Yannong 18 | AA | 0.0223 | TT | AA | 61.3 | TT | 68.1 | GG | 6 |
| Yannong 19 | GG | 0.0244 | TT | AA | 64.9 | CC | 99.2 | GG | 6 |
| Yanzhan 4110 | GG | 0.0356 | CC | GG | 52.6 | TC | 54.6 | GG | 5.8 |
| Yumai 18 | AA | 0.0268 | TT | AA | 67.6 | CC | 155.8 | AA | 8.6 |
| Yumai 21 | AA | 0.0293 | TT | AA | 58.9 | CC | 42.7 | AA | 5.3 |
| Yumai 2 | AA | 0.0224 | TT | AA | 75.3 | CC | 62.4 | AA | 5.5 |
| Yumai 34 | AA | 0.0254 | TT | AA | 60.6 | TT | 168.9 | GG | 9.2 |
| Yumai 35 | AA | 0.0363 | TT | AA | 61.2 | TT | 44.9 | GG | 5.4 |
| Yumai 47 | GG | 0.0363 | CC | GG | 68.8 | CC | 76.1 | AG | 5.6 |
| Yumai 49 | AA | 0.0194 | TT | AA | 48.2 | TT | 169.7 | GG | 7.1 |
| Yumai 50 | AA | 0.0151 | TT | AA | 49.3 | CC | 9.7 | AA | 1.9 |
| Yumai 63 | AA | 0.02 | TT | GG | 37.9 | CC | 97.3 | AA | 7.2 |
| Yumai 7 | AA | 0.0218 | TT | AA | 59.1 | CC | 80.5 | GG | 6.2 |
| Zheng 9023 | AA | 0.0227 | TT | AA | 53.5 | CC | 58.9 | AA | 4.5 |
| Zhengmai 366 | GG | 0.0318 | TT | GG | 41.8 | CC | 74.1 | GG | 5.6 |
| Zhengyin 1 | AA | 0.0237 | TT | AA | 56.8 | TT | 181.6 | GG | 8.6 |
| Zhengzhou 3 | AA | 0.0302 | TT | AA | 63.4 | TC | 140.2 | AA | 9.4 |
| Zhongmai 871 | AA | 0.0235 | TT | GG | 77.7 | CC | 306.2 | GG | 10.8 |
| Zhongmai 875 | AA | 0.0266 | TT | AA | 52.3 | CC | 93.6 | GG | 7 |
| Zhongmai 895 | GG | 0.026 | TT | GG | 61.3 | CC | 148.6 | GG | 9.2 |
| Zhongyu 5 | AA | 0.0259 | TT | AA | 65.3 | TT | 94.4 | GG | 7.1 |
| Zhou 8425B | AA | 0.0262 | TT | AA | 58.1 | TT | 68.7 | GG | 6 |
| Zhoumai16 | GG | 0.0182 | TT | AA | 57.9 | CC | 58 | GG | 5.5 |
| Zhoumai 18 | AA | 0.0236 | TT | AA | 59.1 | CC | 13.3 | GG | 2.6 |
| Zhoumai 19 | AA | 0.0252 | TT | GG | 52.4 | CC | 40.5 | GG | 3.9 |
| Zhoumai 22 | AA | 0.0197 | TT | GG | 47.5 | TT | 60.9 | GG | 4.7 |
| Zhoumai 23 | GG | 0.0181 | TT | GG | 46.7 | CC | 88.5 | GG | 5.3 |
| Zhoumai 25 | AA | 0.0155 | TT | GG | 47.8 | TT | 74.1 | GG | 4.5 |
| Zhoumai 26 | GG | 0.0166 | CC | GG | 40.6 | CC | 133.9 | GG | 8.4 |
| Zhoumai 28 | AA | 0.0195 | TT | AA | 56.7 | CC | 94.6 | GG | 5.6 |
| Zhoumai 30 | GG | 0.015 | TT | GG | 46.2 | TT | 107.8 | AA | 5.5 |
| Zhoumai 31 | AA | 0.0162 | TT | GG | 44.4 | CC | 36.8 | GG | 3.8 |
| Zhoumai 32 | GG | 0.0178 | TT | AA | 48.1 | CC | 127 | AA | 9 |
| Zimai 12 | AA | 0.0263 | TT | AA | 73.7 | TT | 50 | AA | 5.6 |
| Zixuan 2 | AA | 0.0224 | TT | AA | 67.8 | CC | 141.5 | GG | 7.9 |
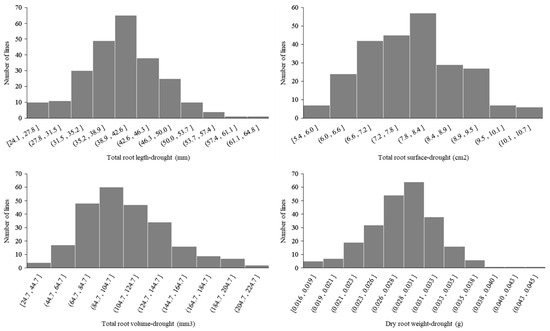
Figure A1.
Frequency distribution of the DRSA-related traits in the Zhoumai16/DK171 RIL population.
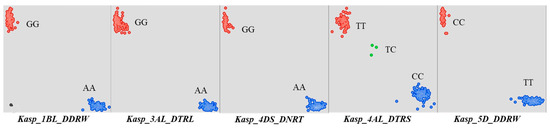
Figure A2.
The genotype of the KASP markers.
References
- Kadam, N.N.; Tamilselvan, A.; Lawas, L.M.; Quinones, C.; Bahuguna, R.N.; Thomson, M.J.; Dingkuhn, M.; Muthurajan, R.; Struik, P.C.; Yin, X.; et al. Genetic control of plasticity in root morphology and anatomy of rice in response to water deficit. Plant Physiol. 2017, 174, 2302–2315. [Google Scholar] [CrossRef]
- Ayalew, H.; Liu, H.; Börner, A.; Kobiljski, B.; Liu, C.J.; Yan, G.J. Genome-wide association mapping of major root length QTLs under PEG-induced water stress in wheat. Front. Plant Sci. 2018, 9, 1759. [Google Scholar] [CrossRef]
- Mohammadi, R. Breeding for increased drought tolerance in wheat: A review. Crop Pasture Sci. 2018, 69, 223–241. [Google Scholar] [CrossRef]
- Bapela, T.; Shimelis, H.; Tsilo, T.J.; Mathew, I. Genetic improvement of wheat for drought tolerance: Progress, challenges and opportunities. Plants 2022, 11, 1331. [Google Scholar] [CrossRef] [PubMed]
- Beyer, S.; Daba, S.; Tyagi, P.; Bockelman, H.; Brown-Guedira, G.; IWGSC; Mohammadi, M. Loci and candidate genes controlling root traits in wheat seedlings—A wheat root GWAS. Funct. Integr. Genom. 2019, 19, 91–107. [Google Scholar] [CrossRef]
- Mathew, I.; Shimelis, H.; Shayanowako, A.I.T.; Laing, M.; Chaplot, V. Genome-wide association study of drought tolerance and biomass allocation in wheat. PLoS ONE 2019, 14, e0225383. [Google Scholar] [CrossRef]
- Sabar, M.; Shabir, G.; Shah, S.M.; Aslam, K.; Naveed, S.A.; Arif, M. Identification and mapping of QTLs associated with drought tolerance traits in rice by a cross between Super Basmati and IR55419-04. Breed. Sci. 2019, 69, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.X.; Wei, J.T.; Tian, R.; Zeng, Z.Y.; Tang, H.P.; Liu, Y.L.; Xu, Q.; Deng, M.; Jiang, Q.T.; Chen, G.Y.; et al. A major quantitative trait locus for wheat total root length associated with precipitation distribution. Front. Plant Sci. 2022, 13, 995183. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.J.; Wang, C.R.; Hassan, M.A.; Wu, Y.Y.; Xia, X.C.; Shi, S.B.; Xiao, Y.G.; He, Z.H. QTL mapping of seedling biomass and root traits under different nitrogen conditions in bread wheat (Triticum aestivum L.). J. Integr. Agric. 2021, 20, 1180–1192. [Google Scholar] [CrossRef]
- Wang, X.; Wang, H.; Liu, S.; Zhang, B.; Zhang, J.; Li, Z.; Wang, Y.; Xie, C.; Huang, L.; Cao, T.; et al. Genetic mapping of quantitative trait loci for yield and yield components in an advanced backcross population derived from a cultivated × wild soybean cross. Theor. Appl. Genet. 2014, 127, 581–589. [Google Scholar]
- Soriano, J.M.; Alvaro, F. Discovering consensus genomic regions in wheat for root-related traits by QTL meta-analysis. Sci. Rep. 2019, 9, 10537. [Google Scholar] [CrossRef]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
- Wen, W.; He, Z.H.; Gao, F.M.; Liu, J.D.; Jin, H.; Zhai, S.N.; Qu, Y.Y.; Xia, X.C. A high-density consensus map of common wheat integrating four mapping populations scanned by the 90K SNP array. Front. Plant Sci. 2017, 8, 1389. [Google Scholar] [CrossRef]
- Rasheed, A.; Hao, Y.; Xia, X.; Khan, A.; Xu, Y.; Varshney, R.K.; He, Z. Crop breeding chips and genotyping platforms: Progress, challenges, and perspectives. Mol. Plant 2017, 10, 1047–1064. [Google Scholar] [CrossRef]
- Liu, R.X.; Wu, F.K.; Xin, Y.; Yu, L.; Wang, Z.Q.; Liu, S.H.; Deng, M.; Ma, J.; Wei, Y.M.; Zhang, Y.L.; et al. Quantitative trait loci analysis for root traits in synthetic hexaploid wheat under drought stress conditions. J. Integr. Agric. 2020, 19, 1947–1960. [Google Scholar] [CrossRef]
- International Wheat Genome Sequencing Consortium (IWGSC). The Wheat Genome Reference Sequence. Available online: https://www.wheatgenome.org/projects/reference-genome-project/refseq-v1.0/ (accessed on 1 January 2023).
- Wheat Expression Database. Available online: www.wheat-expression.com (accessed on 1 January 2023).
- Collard, B.C.Y.; Jahufer, M.Z.Z.; Brouwer, J.B.; Pang, E.C.K. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica 2005, 142, 169–196. [Google Scholar] [CrossRef]
- Nehe, A.S.; Foulkes, M.J.; Ozturk, I.; Rasheed, A.; York, L.; Kefauver, S.C.; Ozdemir, F.; Morgounov, A. Root and canopy traits and adaptability genes explain drought tolerance responses in winter wheat. PLoS ONE 2021, 16, e0242472. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.R.; Wang, Y.M.; Liu, J.D.; Wang, F.Y.; Qiu, X.D.; Liu, P. Genome-wide linkage mapping of root system architecture-related traits in common wheat (Triticum aestivum L.). Front. Plant Sci. 2023, 14, 1274392. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.X.; Wang, W.Q.; Wang, W.L.; Zhang, G.Q.; Liu, Y.; Wang, Y.; Wang, W. Wheat F-box protein gene TaFBA1 is involved in plant tolerance to heat stress. Front. Plant Sci. 2018, 9, 521. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Roorkiwal, M.; Valliyodan, B.; Zhou, L.J.; Chen, P.Y.; Varshney, R.K.; Nguyen, H.T. Genetic diversity of root system architecture in response to drought stress in grain legumes. J. Exp. Bot. 2018, 69, 3267–3277. [Google Scholar] [CrossRef]
- Ibrahim, S.E.; Schubert, A.; Pillen, K.; Léon, J. QTL analysis of drought tolerance for seedling root morphological traits in an advanced backcross population of spring wheat. Theor. Appl. Genet. 2012, 124, 533–545. [Google Scholar]
- Liu, W.X.; Wang, J.R.; Wang, C.Y.; Ma, G.; Wei, Q.R.; Lu, H.F.; Xie, Y.X.; Ma, D.Y.; Kang, G.Z. Root growth, water and nitrogen use efficiencies in winter wheat under different irrigation and nitrogen regimes in North China Plain. Front. Plant Sci. 2018, 9, 1798. [Google Scholar] [CrossRef] [PubMed]
- Li, C.N.; Li, L.; Reynolds, M.P.; Wang, J.Y.; Chang, X.P.; Mao, X.G.; Jing, R.L. Recognizing the hidden half in wheat: Root system attributes associated with drought tolerance. J. Exp. Bot. 2021, 72, 5117–5133. [Google Scholar] [CrossRef]
- Alahmad, S.; El Hassouni, K.; Bassi, F.M.; Dinglasan, E.; Youssef, C.; Quarry, G.; Aksoy, A.; Mazzucotelli, E.; Juhász, A.; Able, J.A.; et al. A major root architecture QTL responding to water limitation in durum wheat. Front. Plant Sci. 2019, 10, 436. [Google Scholar] [CrossRef]
- Rufo, R.; Salvi, S.; Royo, C.; Soriano, J.M. Exploring the genetic architecture of root-related traits in Mediterranean bread wheat landraces by genome-wide association analysis. Agronomy 2020, 10, 613. [Google Scholar] [CrossRef]
- Xu, P.; Zhao, P.X.; Cai, X.T.; Mao, J.L.; Miao, Z.Q.; Xiang, C.B. Integration of jasmonic acid and ethylene into auxin signaling in root development. Front. Plant Sci. 2020, 11, 271. [Google Scholar] [CrossRef]
- Tian, X.L.; Xia, X.C.; Xu, D.G.; Liu, Y.Q.; Xie, L.; Hassan, M.A.; Song, J.; Li, F.J.; Wang, D.S.; Zhang, Y.; et al. Rht24b, an ancient variation of TaGA2ox-A9, reduces plant height without yield penalty in wheat. New Phytol. 2022, 233, 738–750. [Google Scholar] [CrossRef]
- Li, A.L.; Zhu, Y.F.; Tan, X.M.; Wang, X.; Wei, B.; Guo, H.Z.; Zhang, Z.L.; Chen, X.B.; Zhao, G.Y.; Kong, X.Y.; et al. Evolutionary and functional study of the CDPK gene family in wheat (Triticum aestivum L.). Plant Mol. Biol. 2008, 66, 429–443. [Google Scholar] [CrossRef]
- Sun, T.; Wang, Y.; Wang, M.; Li, T.T.; Zhou, Y.; Wang, X.T.; Wei, S.Y.; He, G.Y.; Yang, G.X. Identification and comprehensive analyses of the CBL and CIPK gene families in wheat (Triticum aestivum L.). BMC Plant Biol. 2015, 15, 269. [Google Scholar] [CrossRef] [PubMed]
- Ivashuta, S.I.; Petrick, J.S.; Heisel, S.E.; Zhang, Y.J.; Guo, L.; Reynolds, T.L.; Rice, J.F.; Allen, E.; Roberts, J.K. Endogenous small RNAs in grain: Semi-quantification and sequence homology to human and animal genes. Food Chem. Toxicol. 2009, 47, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Petricka, J.J.; Winter, C.M.; Benfey, P.N. Control of Arabidopsis root development. Annu. Rev. Plant Biol. 2012, 63, 563–590. [Google Scholar] [CrossRef] [PubMed]
- Miao, Z.Q.; Zhao, P.X.; Mao, J.L.; Yu, L.H.; Yuan, Y.; Tang, H.; Liu, Z.B.; Xiang, C.B. HOMEOBOX PROTEIN52 mediates the crosstalk between ethylene and auxin signaling during primary root elongation by modulating auxin transport-related gene expression. Plant Cell 2018, 30, 2761–2778. [Google Scholar] [CrossRef] [PubMed]
- Divte, P.; Yadav, P.; Jain, P.K.; Paul, S.; Singh, B. Ethylene regulation of root growth and phytosiderophore biosynthesis determines iron deficiency tolerance in wheat (Triticum spp.). Environ. Exp. Bot. 2019, 162, 1–13. [Google Scholar] [CrossRef]
- Hu, Z.R.; Wang, R.; Zheng, M.; Liu, X.B.; Meng, F.; Wu, H.L.; Yao, Y.Y.; Xin, M.M.; Peng, H.R.; Ni, Z.F.; et al. TaWRKY51 promotes lateral root formation through negative regulation of ethylene biosynthesis in wheat (Triticum aestivum L.). Plant J. 2018, 96, 372–388. [Google Scholar] [CrossRef]
- Zhao, Y.D. Essential roles of local auxin biosynthesis in plant development and in adaptation to environmental changes. Annu. Rev. Plant Biol. 2018, 69, 417–435. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).