Time-Course Transcriptome, Metabolome, and Weighted Gene Co-Expression Network Analysis Reveal the Roles of the OsBELH4A Gene in Regulating Leaf Senescence and Grain Yield of Rice
Abstract
1. Introduction
2. Results
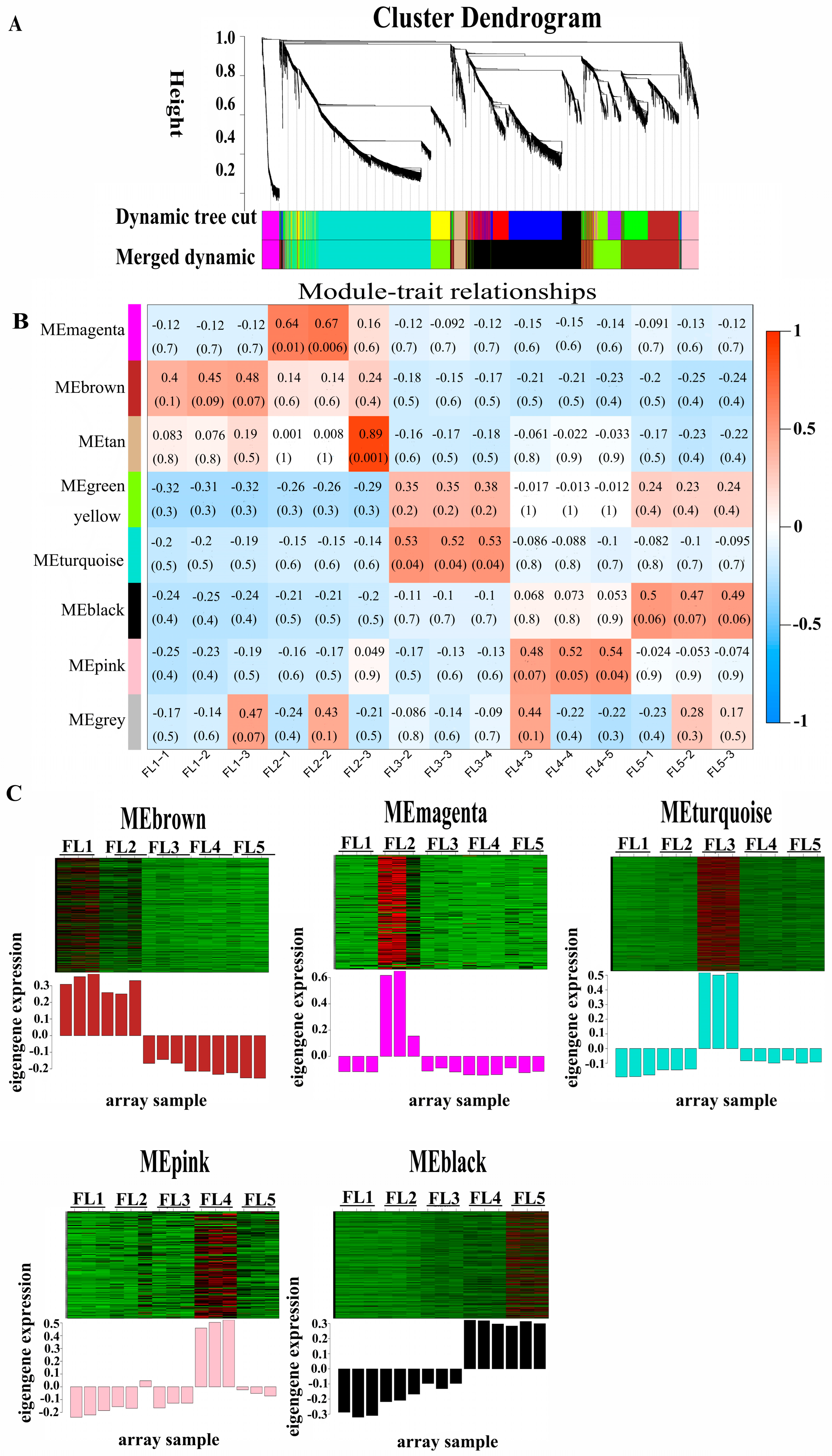
2.1. Senescence Feature, DEG Analysis and Weighted Gene Co-Expression Network Analysis
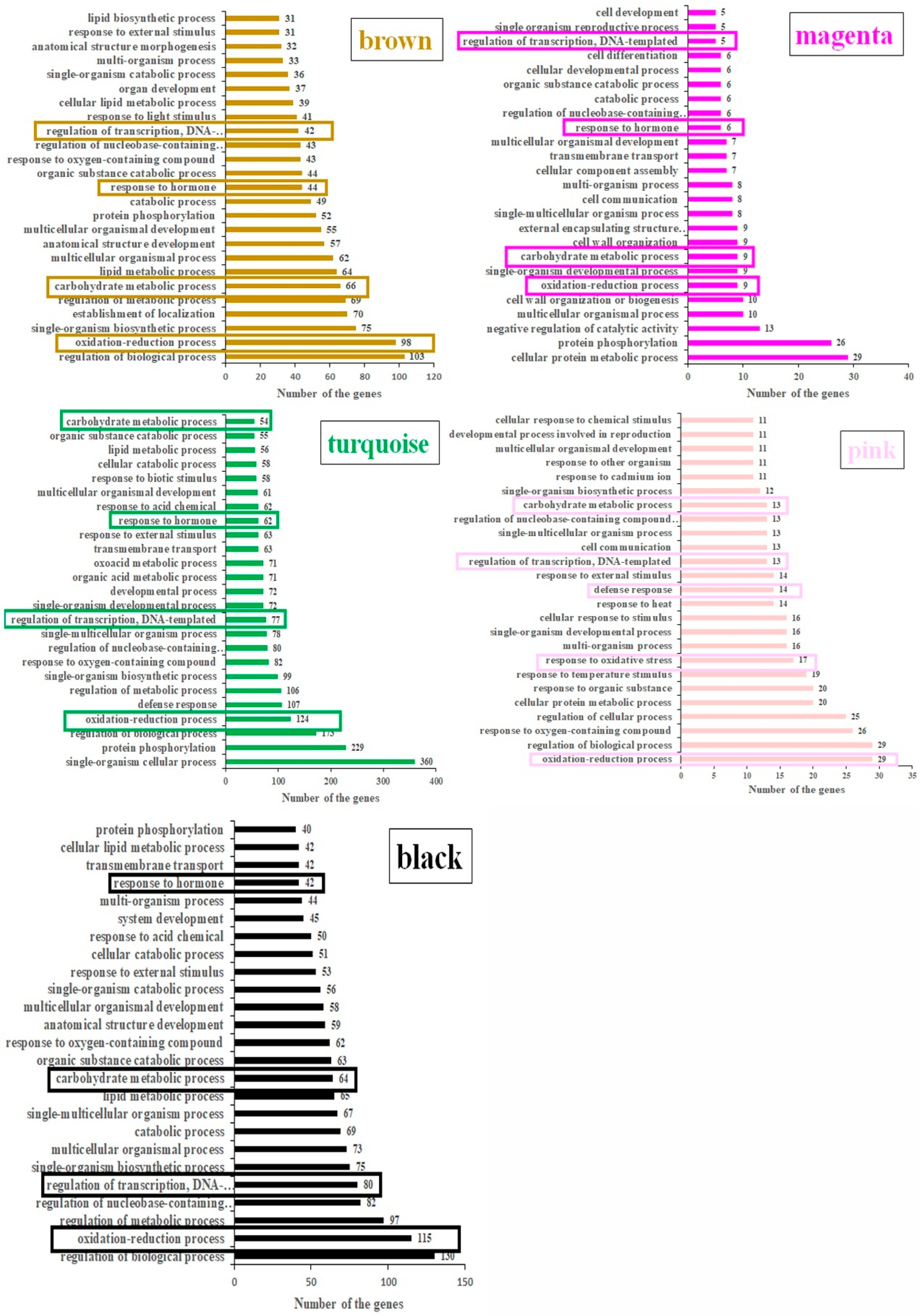
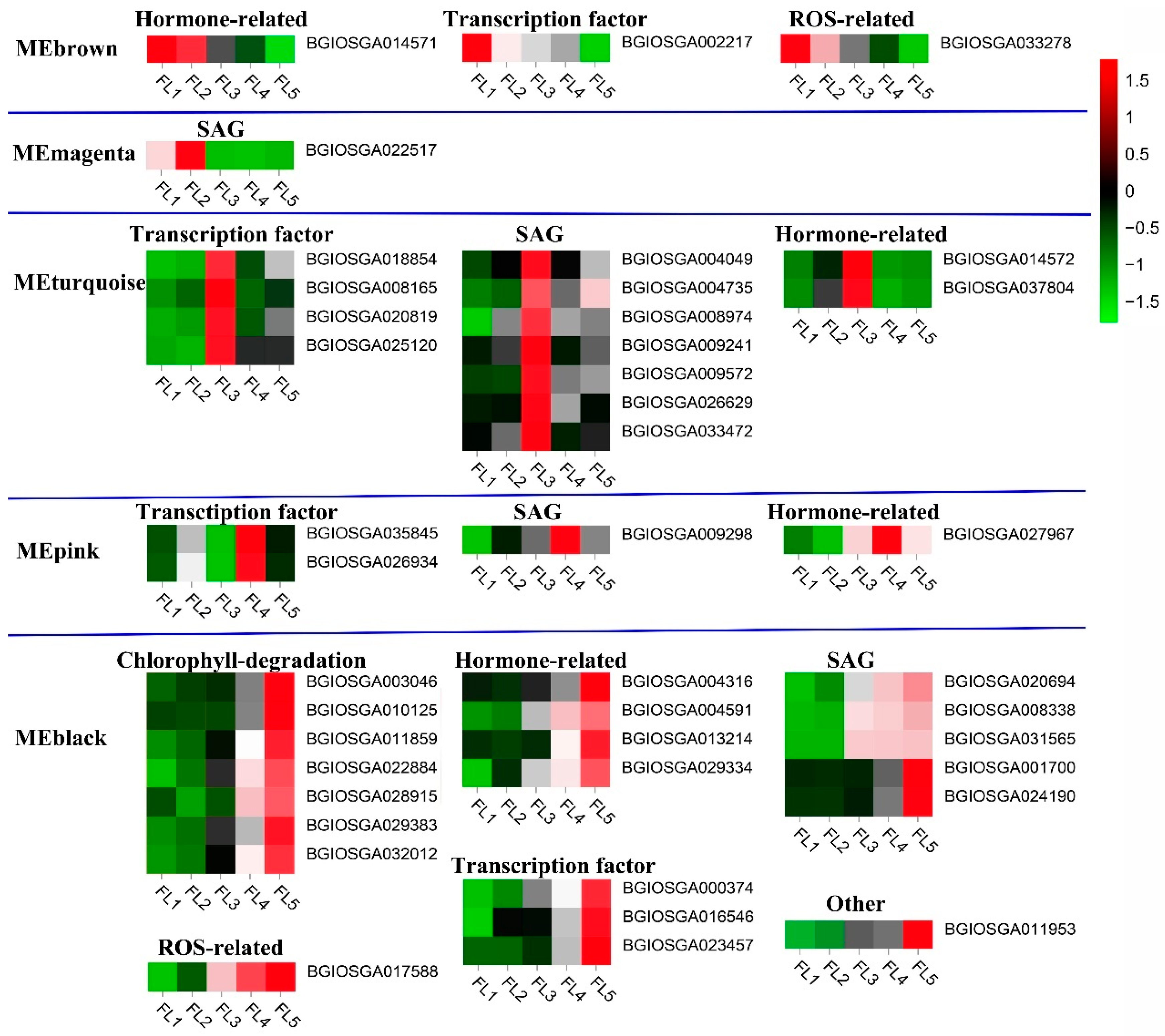
2.2. Identification of Transcription Factors in Five Stage-Specific Modules
2.3. Identification of Senescence-Related Genes in Five Stage-Specific Modules
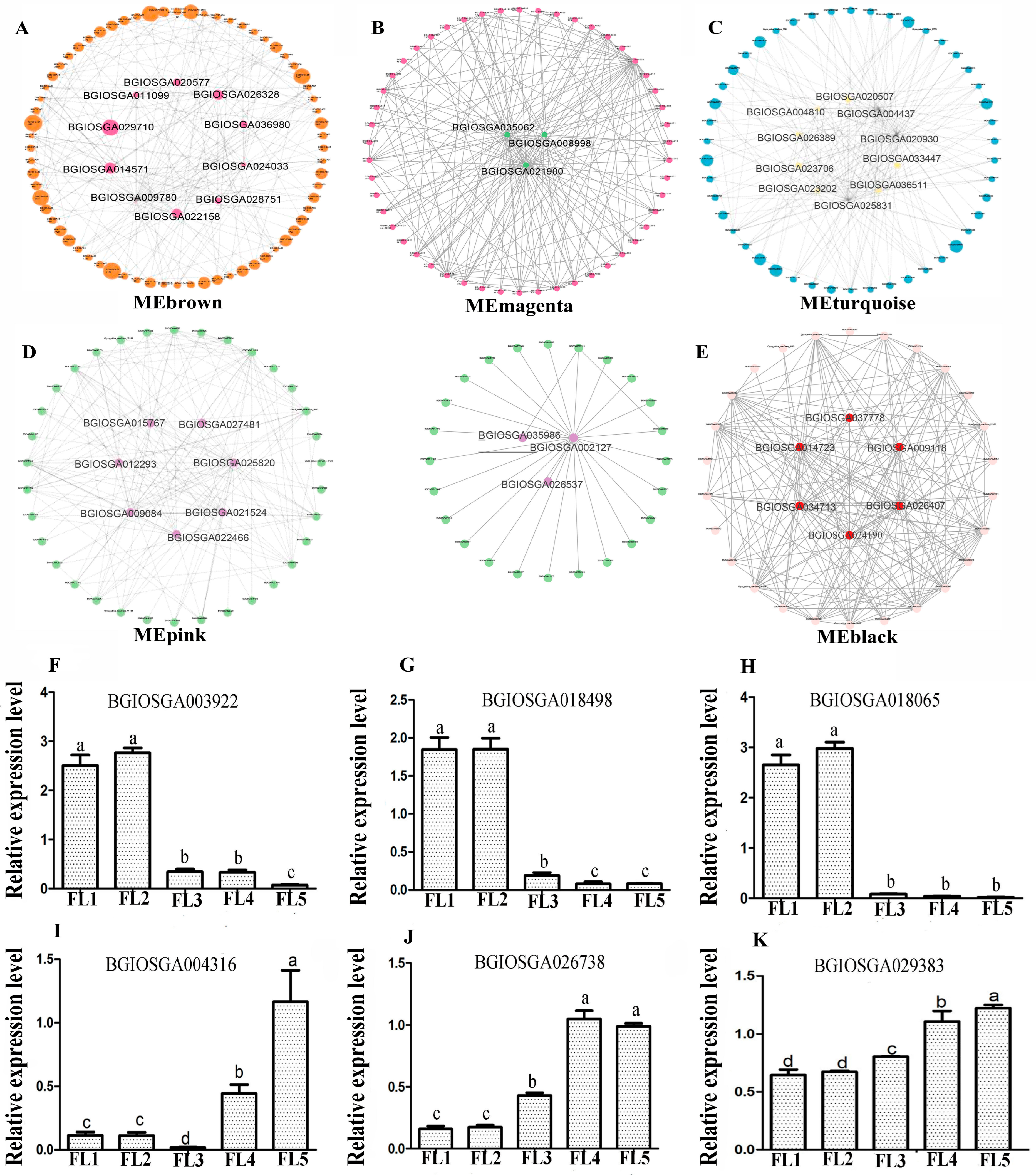
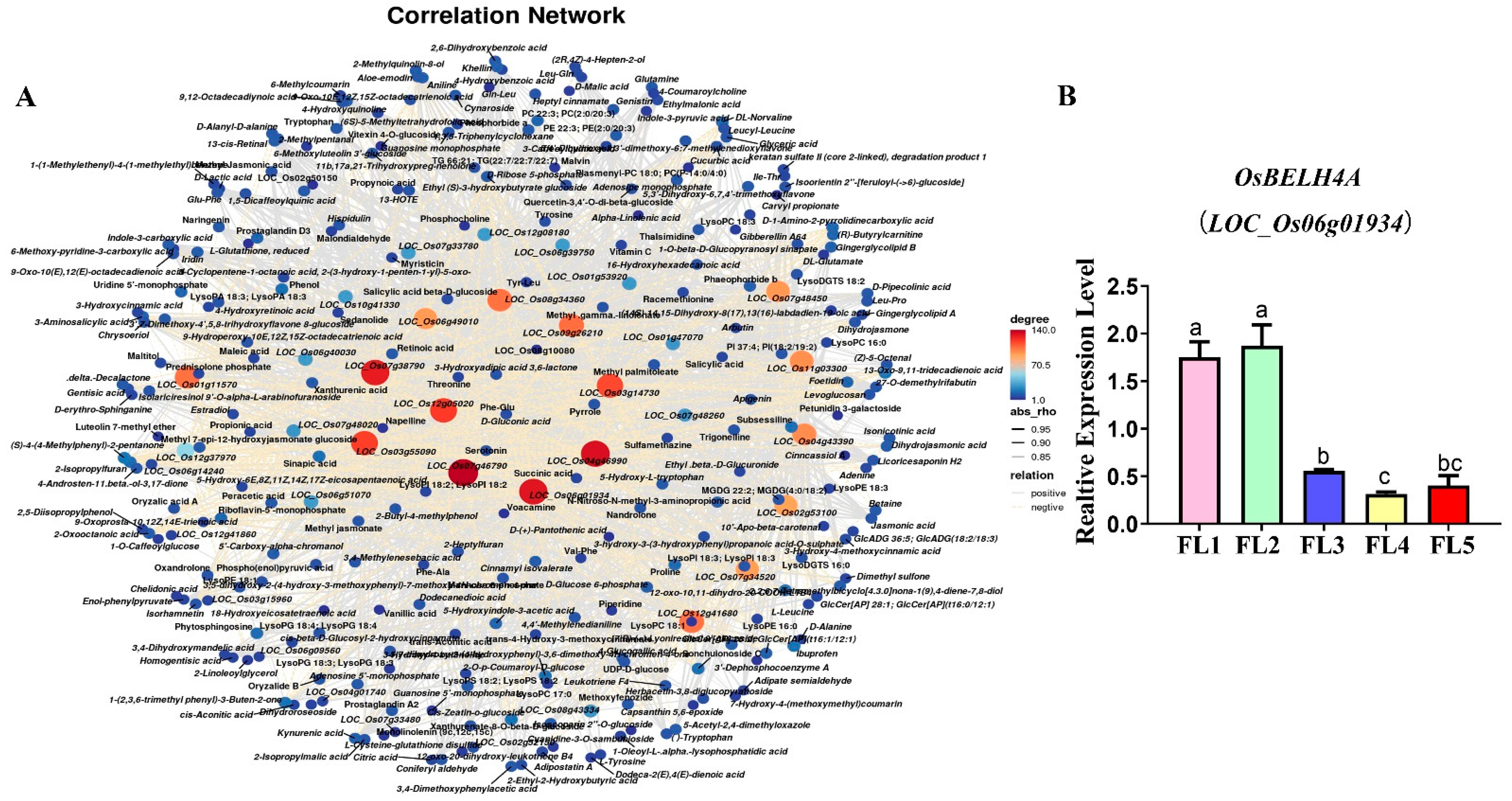
2.4. Identification and Visualization of Hub Genes
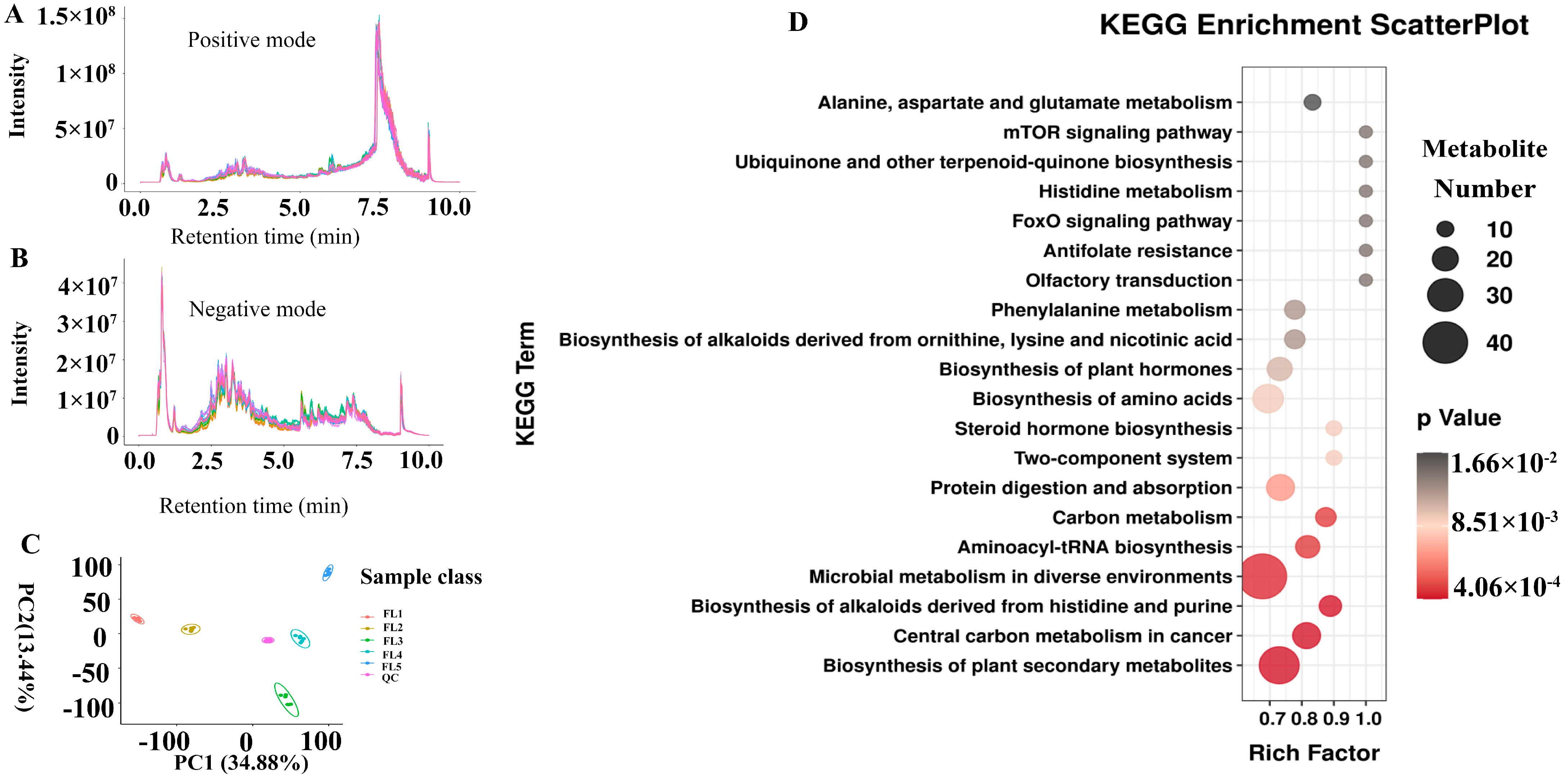
2.5. Integrative Transcriptome and Metabolome Analysis
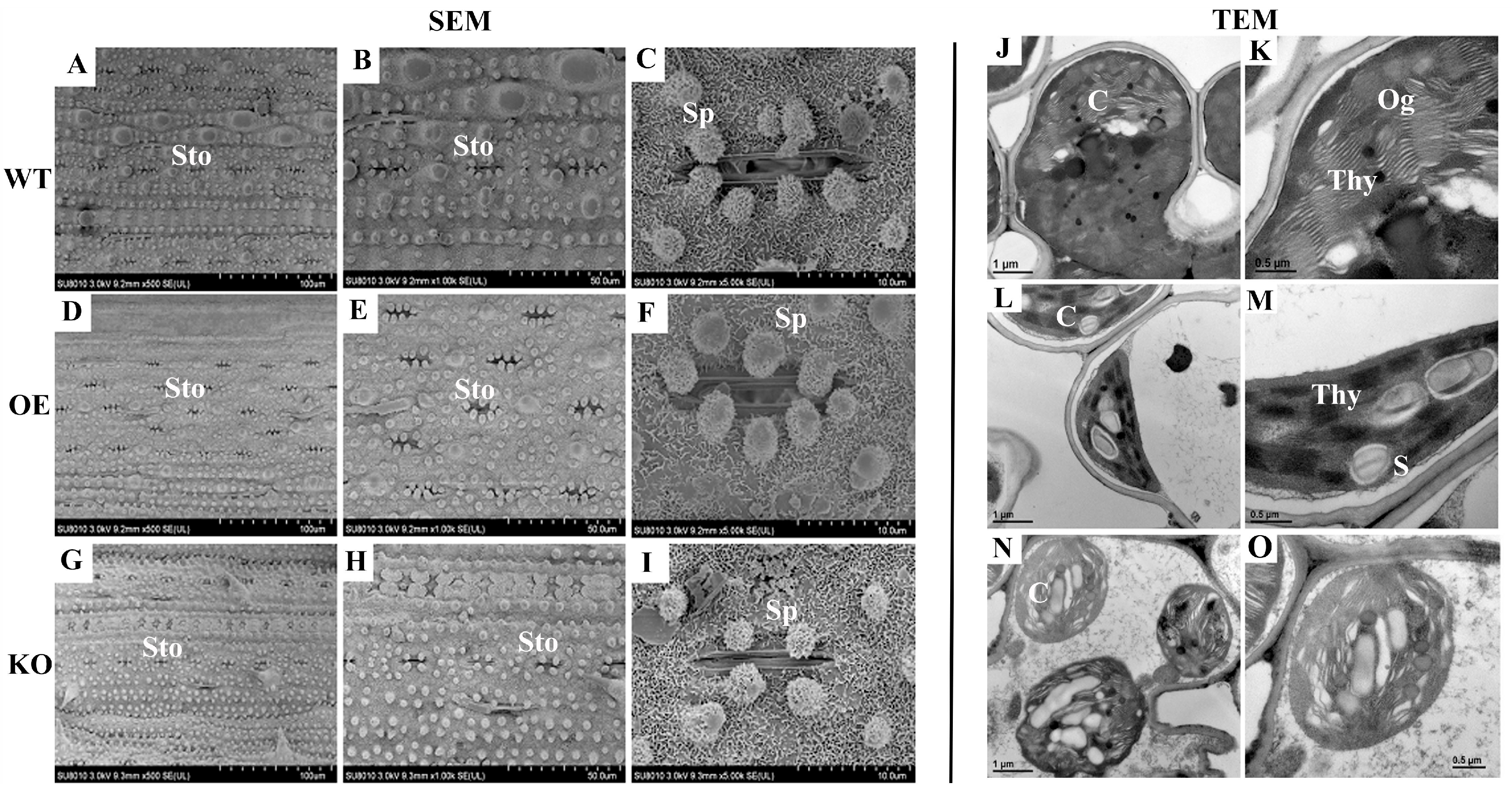
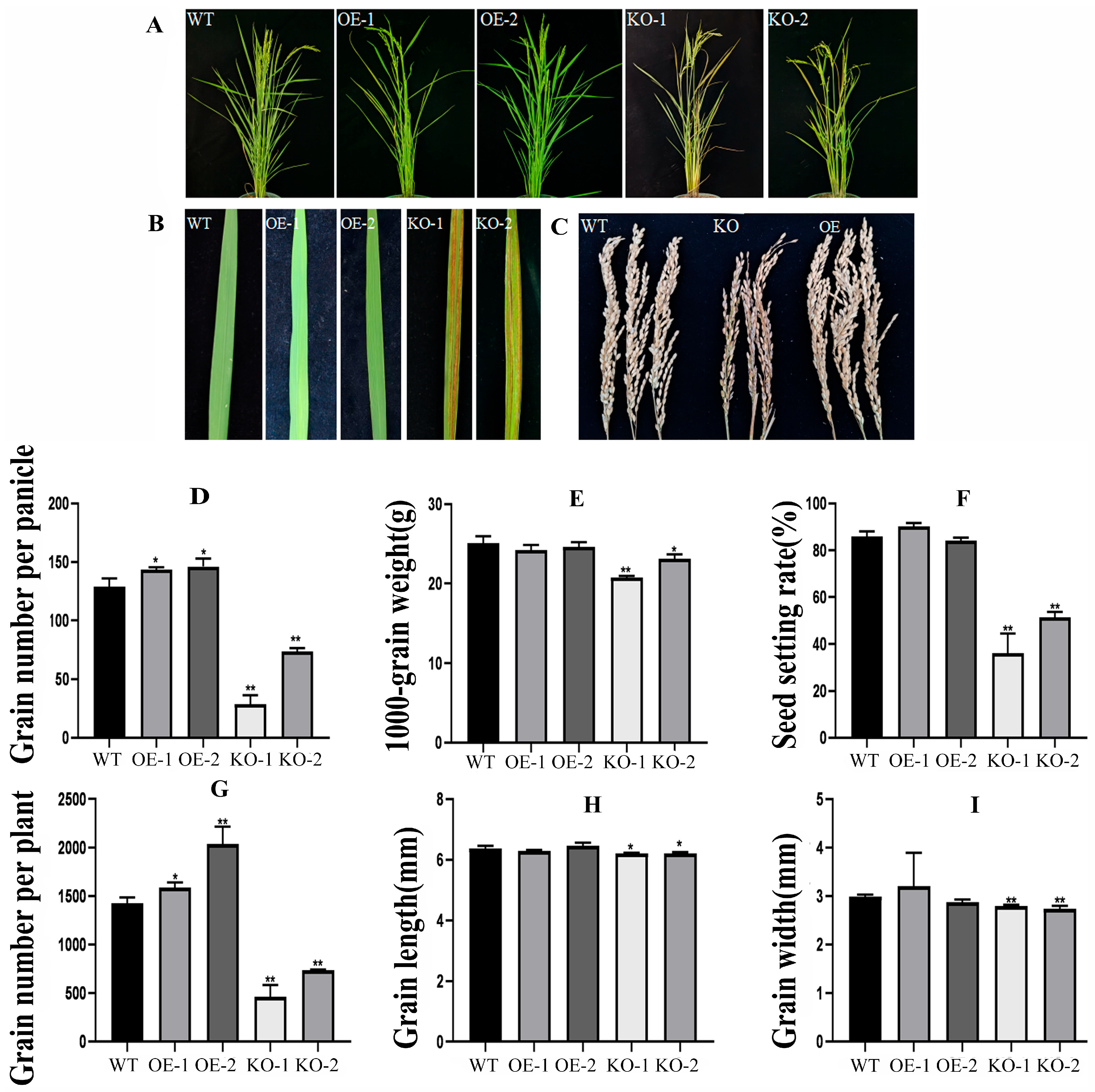
2.6. Overexpression of OsBELH4A Gene Delayed Leaf Senescence
2.7. OsBELH4A Gene Affected Rice Yield Traits in Varying Degrees
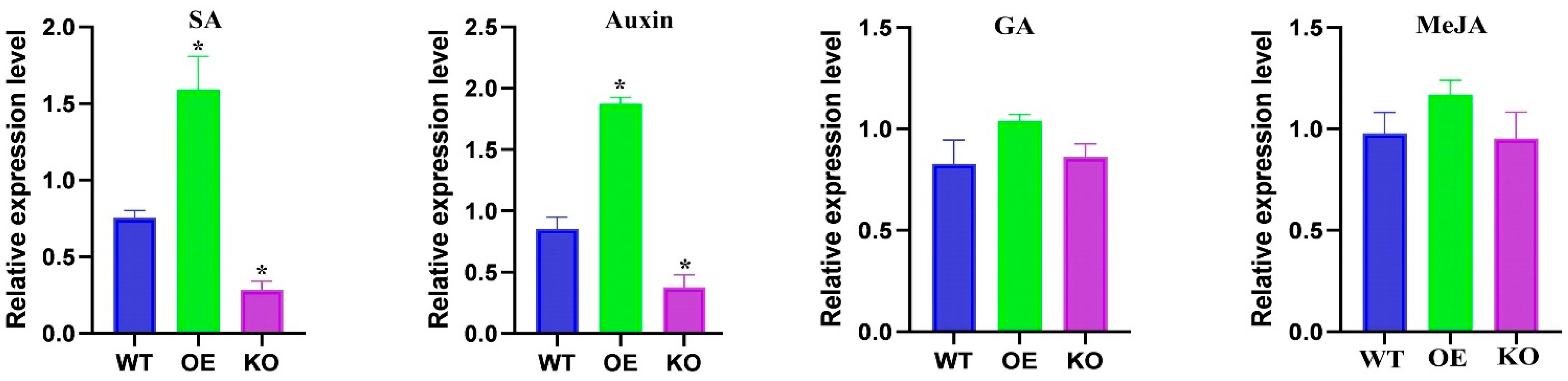
2.8. OsBELH4A Gene Expression Responded to Hormone SA and Auxin
3. Discussion
3.1. The Roles of Genes Related to Carbohydrate and Lipid Metabolism in the Leaf Senescence of Rice
3.2. The Roles of Transcription Factors in the Leaf Senescence of Rice
3.3. The Roles of Hormones in the Leaf Senescence of Rice
3.4. The Roles of Chlorophyll Degradation-Related Genes in the Leaf Senescence of Rice
3.5. The Roles of ROS-Related Genes in the Leaf Senescence of Rice
3.6. OsBELH4A Negatively Affected Leaf Senescence by Regulating Chlorophyll and ROS
3.7. OsBELH4A Negatively Affected Leaf Senescence by Regulating the Expression Levels of SAGs and Chlorophyll Degradation-Related and Photosynthetic System-Related Genes
4. Materials and Methods
4.1. Plant Material and Phenotypic Characterization of Senescence Stages
4.2. Transcriptome Data Collection, Differential Expression and Weighted Gene Co-Expression Network Analysis (WGCNA)
4.3. Quantitative Real-Time PCR Validation of Selected DEGs
4.4. Metabolome Analysis
4.5. Gene Cloning, Vector Construction and Rice Transformation
4.6. Detection of Physiological Indicators and Senescence Marker Genes in Transgenic Rice
4.7. Electron Microscopy Observation
4.8. Investigation of Leaf Phenotype and Agronomic Traits
4.9. Exogenous Phytohormone Treatment
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lim, P.O.; Kim, H.J.; Nam, H.G. Leaf senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef]
- Tan, S.Y.; Sha, Y.Q.; Sun, L.W.; Li, Z.H. Abiotic stress-induced leaf senescence: Regulatory mechanisms and application. Int. J. Mol. Sci. 2023, 24, 11996. [Google Scholar] [CrossRef]
- Lv, Z.; Zhao, W.Q.; Kong, S.X.; Li, L.; Lin, S.Y. Overview of molecular mechanisms of plant leaf development: A systematic review. Front. Plant Sci. 2023, 14, 1293424. [Google Scholar] [CrossRef] [PubMed]
- Gregersen, P.L.; Culetic, A.; Boschian, L.; Krupinska, K. Plant senescence and crop productivity. Plant Mol. Biol. 2013, 82, 603–622. [Google Scholar] [CrossRef]
- Li, Z.; Wang, F.; Lei, B.; Cao, Z.; Pan, G.; Cheng, F. Genotypic-dependent alteration in transcriptional expression of various CAT isoenzyme genes in esl mutant rice and its relation to H2O2-induced leaf senescence. Plant Growth Regul. 2014, 73, 237–248. [Google Scholar] [CrossRef]
- Han, R.C.; He, X.F.; Pan, X.H.; Shi, Q.H.; Wu, Z.M. Enhancing xanthine dehydrogenase activity is an effective way to delay leaf senescence and increase rice yield. Rice 2020, 13, 16. [Google Scholar] [CrossRef]
- Singh, S.; Singh, A.; Nandi, A.K. The rice OsSAG12-2 gene codes for a functional protease that negatively regulates stress-induced cell death. J. Biosci. 2016, 41, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Kamachi, K.; Yamaya, T.; Hayakawa, T.; Mae, T.; Ojima, K. Changes in cytosolic glutamine synthetase polypeptide and its mRNA in a leaf blade of rice plants during natural senescence. Plant Physiol. 1992, 98, 1323–1329. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Wang, Y.; Zhu, Y.; Tang, J.; Hu, B.; Liu, L.; Ou, S.; Wu, H.; Sun, X.; Chu, J.; et al. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 10013–10018. [Google Scholar] [CrossRef]
- Sun, L.N.; Xu, H.Q.; Song, J.; Yang, X.Y.; Wang, X.Y.; Liu, H.Y.; Pang, M.Z.; Hu, Y.C.; Yang, Q.; Ning, X.T.; et al. OsNAC103, a NAC transcription factor, positively regulates leaf senescence and plant architecture in rice. Rice 2024, 17, 15. [Google Scholar] [CrossRef]
- Xie, W.Y.; Li, X.R.; Wang, S.P.; Yuan, M. OsWRKY53 promotes abscisic acid accumulation to accelerate leaf senescence and inhibit seed germination by downregulating abscisic acid catabolic genes in rice. Front. Plant Sci. 2022, 12, 816156. [Google Scholar] [CrossRef]
- Li, Y.Y.; Liao, S.T.; Mei, P.Y.; Pan, Y.Y.; Zhang, Y.; Zheng, X.Z.; Xie, Y.K.; Miao, Y. OsWRKY93 dually functions between leaf senescence and in response to biotic stress in rice. Front. Plant Sci. 2021, 12, 643011. [Google Scholar] [CrossRef]
- Kusaba, M.; Ito, H.; Morita, R.; Iida, S.; Sato, Y.; Fujimoto, M.; Kawasaki, S.; Tanaka, R.; Hirochika, H.; Nishimura, M.; et al. Rice Non-yellow coloring1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 2007, 19, 1362–1375. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Morita, R.; Katsuma, S.; Nishimura, M.; Tanaka, A.; Kusaba, M. Two short-chain dehydrogenase/reductases, non-yellow coloring 1 and nyc1-like, are required for chlorophyll b and light-harvesting complexII degradation during senescence in rice. Plant J. 2009, 57, 120–131. [Google Scholar] [CrossRef]
- Yamatani, H.; Kohzuma, K.; Nakano, M.; Takami, T.; Kato, Y.; Hayashi, Y.; Monden, Y.; Okumoto, Y.; Abe, T.; Kumamaru, T.; et al. Impairment of Lhca4, a subunit of LHCI, causes high accumulation of chlorophyll and the stay-green phenotype in rice. J. Exp. Bot. 2018, 69, 1027–1035. [Google Scholar] [CrossRef]
- Shin, D.; Lee, S.; Kim, T.; Lee, J.; Park, J.; Lee, J.; Lee, J.Y.; Cho, L.; Choi, J.Y.; Lee, W.; et al. Natural variations at the stay-green gene promoter control lifespan and yield in rice cultivars. Nat. Commun. 2020, 11, 2819. [Google Scholar] [CrossRef]
- Hong, Y.; Zhang, Y.; Sinumporn, S.; Yu, N.; Zhan, X.; Shen, X.; Chen, D.; Yu, P.; Wu, W.; Liu, Q.; et al. Premature leaf senescence 3, encoding a methyltransferase, is required for melatonin biosynthesis in rice. Plant J. 2018, 95, 877–891. [Google Scholar] [CrossRef] [PubMed]
- Huangfu, L.; Chen, R.; Lu, Y.; Zhang, E.; Miao, J.; Zuo, Z.; Zhao, Y.; Zhu, M.; Zhang, Z.; Li, P.; et al. OsCOMT, encoding a caffeic acid O-methyltransferase in melatonin biosynthesis, increases rice grain yield through dual regulation of leaf senescence and vascular development. Plant Biotechnol. J. 2022, 20, 1122–1139. [Google Scholar] [CrossRef]
- Zhang, W.; Peng, K.; Cui, F.; Wang, D.; Zhao, J.; Zhang, Y.; Yu, N.; Wang, Y.; Zeng, D.; Wang, Y.; et al. Cytokinin oxidase/dehydrogenase OsCKX11 coordinates source and sink relationship in rice by simultaneous regulation of leaf senescence and grain number. Plant Biotechnol. J. 2021, 19, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Ji, Z.; Wang, C.; Xu, F.; Wang, F.; Zheng, Y.; Tang, Y.; Wei, Z.; Zhao, T.; Zhao, K. WATER-SOAKED SPOT1 controls chloroplast development and leaf senescence via regulating reactive oxygen species homeostasis in rice. Front. Plant Sci. 2022, 13, 918673. [Google Scholar] [CrossRef]
- Xiong, E.; Dong, G.; Chen, F.; Zhang, C.; Li, S.; Zhang, Y.; Shohag, J.I.; Yang, X.; Zhou, Y.; Qian, Q.; et al. Formyl tetrahydrofolate deformylase affects hydrogen peroxide accumulation and leaf senescence by regulating the folate status and redox homeostasis in rice. Sci. China Life Sci. 2021, 64, 720–738. [Google Scholar] [CrossRef]
- Xu, J.M.; Wang, C.L.; Wang, F.J.; Liu, Y.P.; Li, M.; Wang, H.J.; Zheng, Y.H.; Zhao, K.J.; Ji, Z.Y. PWL1, a G-type lectin receptor-like kinase, positively regulates leaf senescence and heat tolerance but negatively regulates resistance to Xanthomonas oryzae in rice. Plant Biotechnol. J. 2023, 21, 2525–2545. [Google Scholar] [CrossRef] [PubMed]
- Guo, N.; Qu, H.Y.; Zhi, Y.; Zhang, Y.Y.; Cheng, S.J.; Chu, J.F.; Zhang, Z.G.; Xu, G.H. Knockout of amino acid transporter gene OsLHT1 accelerates leaf senescence and enhances resistance to rice blast fungus. J. Exp. Bot. 2023, 74, 4143–4157. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, N.; Sun, X.W.; Tan, Y.N.; Sun, Z.Z.; Yu, D.; Liu, H.; Liu, C.T.; Liu, L.; Jin, L.; Zhao, B.R.; et al. Senescence-specific expression of RAmy1A accelerates non-structural carbohydrate remobilization and grain filling in rice (Oryza sativa L.). Front. Plant Sci. 2021, 12, 647574. [Google Scholar] [CrossRef]
- Chen, D.; Shi, Y.R.; Zhang, P.; Xie, W.Y.; Li, S.X.; Xiao, J.H.; Yuan, M. Deletion of the sugar importer gene OsSWEET1b accelerates sugar starvation-promoted leaf senescence in rice. Plant Physiol. 2024, 195, 2176–2194. [Google Scholar] [CrossRef]
- Xiao, P.; Qu, J.; Wang, Y.; Fang, T.; Xiao, W.; Wang, Y.; Zhang, Y.; Khan, M.; Chen, Q.; Xu, X.; et al. Transcriptome and metabolome atlas reveals contributions of sphingosine and chlorogenic acid to cold tolerance in Citrus. Plant Physiol. 2024, 196, 634–650. [Google Scholar] [CrossRef]
- Bao, S.; Li, J.; Wang, J.; Lan, T.; Wei, M.; Sun, X.; Fang, Y.; Ma, T. How does nature create the painting “gradient coloration of ‘Manicure Finger’ Grape”? Integrated omics unveil the pigments basis and metabolism networks of its formation. Food Front. 2024, 6, 921–939. [Google Scholar] [CrossRef]
- Liu, L.; Si, L.; Zhang, L.S.; Guo, R.; Wang, R.X.; Dong, H.M.; Guo, C.H. Metabolomics and transcriptomics analysis revealed the response mechanism of alfalfa to combined cold and saline-alkali stress. Plant J. 2024, 119, 1. [Google Scholar] [CrossRef]
- Nawade, B.; Shim, S.H.; Chu, S.H.; Zhao, W.G.; Lee, S.K.; Somsri, A.; Maung, T.Z.; Kang, K.K.; Kim, J.Y.; Lee, C.Y.; et al. Integrative transcriptogenomic analyses reveal a regulatory network of rice eating and cooking quality and unveil a role of alpha-globulin in modulating starch and sucrose metabolism. Plant Commun. 2025, 6, 101287. [Google Scholar] [CrossRef]
- Chen, H.; Rosin, F.M.; Prat, S.; Hannapel, D.J. Interacting transcription factors from the three-amino acid loop extension superclass regulate tuber formation. Plant Physiol. 2003, 132, 1391–1404. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.L.; Fu, D.Q. The Roles of BLH Transcription factors in plant development and environmental response. Int. J. Mol. Sci. 2022, 23, 3731. [Google Scholar] [CrossRef]
- Jeon, H.W.; Byrne, M.E. SAW homeodomain transcription factors regulate initiation of leaf margin serrations. J. Exp. Bot. 2021, 72, 1738–1747. [Google Scholar] [CrossRef]
- Niu, X.L.; Li, H.L.; Li, R.; Liu, G.S.; Peng, Z.Z.; Jia, W.; Ji, X.; Zhu, H.L.; Zhu, B.Z.; Grierson, D.; et al. Transcription factor SlBEL2 interferes with GOLDEN2-LIKE and influences green shoulder formation in tomato fruits. Plant J. 2022, 112, 982–997. [Google Scholar] [CrossRef]
- Jia, T.T.; Wang, H.Q.; Cui, S.Y.; Li, Z.H.; Shen, Y.C.; Li, H.B.; Xiao, G.H. Cotton BLH1 and KNOX6 antagonistically modulate fiber elongation via the regulation of linolenic acid biosynthesis. Plant Commun. 2024, 5, 100887. [Google Scholar] [CrossRef]
- An, G.; Yu, C.C.; Yan, C.H.; Wang, M.L.; Zhang, W.Y.; Jia, Y.; Shi, C.M.; Larkin, R.M.; Chen, J.J.; Lavelle, D.; et al. Loss-of-function of SAWTOOTH1 affects leaf dorsiventrality genes to promote leafy heads in lettuce. Plant Cell 2022, 34, 4329–4347. [Google Scholar] [CrossRef]
- Kong, Z.; Li, M.; Yang, W.; Xu, W.; Xue, Y. A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol. 2006, 141, 1376–1388. [Google Scholar] [CrossRef]
- Han, M.; Kim, C.; Lee, J.; Lee, S.K.; Jeon, J.S. OsWRKY42 represses OsMT1d and induces reactive oxygen species and leaf senescence in rice. Mol. Cells 2014, 37, 532–539. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Lu, S.; Lv, B.; Zhang, B.; Shen, J.; He, J.; Luo, L.; Xi, D.; Chen, X.; Ming, F. A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol. 2017, 174, 1747–1763. [Google Scholar] [CrossRef]
- El Mannai, Y.; Akabane, K.; Hiratsu, K.; Satoh-Nagasawa, N.; Wabiko, H. The NAC transcription factor gene OsY37 (ONAC011) promotes leaf senescence and accelerates heading time in rice. Int. J. Mol. Sci. 2017, 18, 2165. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.; Shim, Y.; Gi, E.; An, G.; Paek, N.C. Mutation of ONAC096 enhances grain yield by increasing panicle number and delaying leaf senescence during grain filling in rice. Int. J. Mol. Sci. 2019, 20, 5241. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Kang, K.; Kim, S.; An, G.; Paek, N.C. OsWRKY5 promotes rice leaf senescence via senescence-associated NAC and abscisic acid biosynthesis pathway. Int. J. Mol. Sci. 2019, 20, 4437. [Google Scholar] [CrossRef] [PubMed]
- Piao, W.; Kim, S.; Lee, B.; An, G.; Sakuraba, Y.; Paek, N.C. Rice transcription factor OsMYB102 delays leaf senescence by down-regulating abscisic acid accumulation and signaling. J. Exp. Bot. 2019, 70, 2699–2715. [Google Scholar] [CrossRef] [PubMed]
- Luan, W.; Shen, A.; Jin, Z.; Song, S.; Li, Z.; Sha, A. Knockdown of OsHox33, a member of the class III homeodomain-leucine zipper gene family, accelerates leaf senescence in rice. Sci. China Life Sci. 2013, 56, 1113–1123. [Google Scholar] [CrossRef]
- Nakashima, K.; Tran, L.P.; Van Nguyen, D.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007, 51, 617–630. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Piao, W.; Lim, J.; Han, S.H.; Kim, Y.S.; An, G.; Paek, N.C. Rice ONAC106 inhibits leaf senescence and increases salt tolerance and tiller angle. Plant Cell Physiol. 2015, 56, 2325–2339. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Guo, Y.; Liu, Y.; Zhang, F.; Wang, Z.; Wang, H.; Wang, F.; Li, D.; Mao, D.; Luan, S.; et al. 9-cis-epoxycarotenoid dioxygenase 3 regulates plant growth and enhances multi-abiotic stress tolerance in rice. Front. Plant Sci. 2018, 9, 162. [Google Scholar] [CrossRef]
- Huang, Y.; Jiao, Y.; Xie, N.; Guo, Y.; Zhang, F.; Xiang, Z.; Wang, R.; Wang, F.; Gao, Q.; Tian, L.; et al. OsNCED5, a 9-cis-epoxycarotenoid dioxygenase gene, regulates salt and water stress tolerance and leaf senescence in rice. Plant Sci. 2019, 287, 110188. [Google Scholar] [CrossRef]
- Fang, C.; Zhang, H.; Wan, J.; Wu, Y.; Li, K.; Jin, C.; Chen, W.; Wang, S.; Wang, W.; Zhang, H.; et al. Control of leaf senescence by an MeOH-jasmonates cascade that is epigenetically regulated by OsSRT1 in rice. Mol. Plant 2016, 9, 1366–1378. [Google Scholar] [CrossRef]
- Kant, S.; Bi, Y.; Zhu, T.; Rothstein, S.J. SAUR39, a small auxin-up RNA gene, acts as a negative regulator of auxin synthesis and transport in rice. Plant Physiol. 2009, 151, 691–701. [Google Scholar] [CrossRef]
- Zhang, W.; Zhou, X.; Wen, C. Modulation of ethylene responses by OsRTH1 overexpression reveals the biological significance of ethylene in rice seedling growth and development. J. Exp. Bot. 2012, 63, 4151–4164. [Google Scholar] [CrossRef]
- Kudo, T.; Makita, N.; Kojima, M.; Tokunaga, H.; Sakakibara, H. Cytokinin activity of cis-Zeatin and phenotypic alterations induced by overexpression of putative cis-Zeatin-O-glucosyltransferase in rice. Plant Physiol. 2012, 160, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.; Kim, Y.; Park, S.; Back, K. Senescence-induced serotonin biosynthesis and its role in delaying senescence in rice leaves. Plant Physiol. 2009, 150, 1380–1393. [Google Scholar] [CrossRef]
- Morita, R.; Sato, Y.; Masuda, Y.; Nishimura, M.; Kusaba, M. Defect in non-yellow coloring 3, an α/β hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J. 2009, 59, 940–952. [Google Scholar] [CrossRef]
- Tang, Y.Y.; Li, M.R.; Chen, Y.P.; Wu, P.Z.; Wu, G.J.; Jiang, H.W. Knockdown of OsPAO and OsRCCR1 cause different plant death phenotypes in rice. J. Plant Physiol. 2011, 168, 1952–1959. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.H.; Hsu, J.H.; Huang, H.J.; Lo, S.F.; Grace Chen, S.C. Alkaline alpha-galactosidase degrades thylakoid membranes in the chloroplast during leaf senescence in rice. New Phytol. 2009, 184, 596–606. [Google Scholar] [CrossRef]
- Ouyang, S.; Zhu, W.; Hamilton, J.; Lin, H.; Campbell, M.; Childs, K.; Thibaud-Nissen, F.; Malek, R.L.; Lee, Y.; Zheng, L.; et al. The TIGR rice genome annotation resource: Improvements and new features. Nucleic Acids Res. 2007, 35, D883–D887. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhou, Y.; Zhou, G.; Ye, R.; Zhao, L.; Li, X.; Lin, Y. Identification of early senescence-associated genes in rice flag leaves. Plant Mol. Biol. 2008, 67, 37–55. [Google Scholar] [CrossRef]
- Lee, R.H.; Wang, C.H.; Huang, L.T.; Chen, L.C. Leaf senescence in rice plants: Cloning and characterization of senescence up-regulated genes. J. Exp. Bot. 2001, 52, 1117–1121. [Google Scholar] [CrossRef]
- Chen, G.; Wu, C.; He, L.; Qiu, Z.; Zhang, S.; Zhang, Y.; Guo, L.; Zeng, D.; Hu, J.; Ren, D.; et al. Knocking out the gene RLS1 induces hypersensitivity to oxidative stress and premature leaf senescence in rice. Int. J. Mol. Sci. 2018, 19, 2853. [Google Scholar] [CrossRef]
- Zhou, Q.; Yu, Q.; Wang, Z.; Pan, Y.; Lv, W.; Zhu, L.; Chen, R.; He, G. Knockdown of GDCH gene reveals reactive oxygen species-induced leaf senescence in rice. Plant Cell Environ. 2013, 36, 1476–1489. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, Y.; Luo, W.; Li, W.; Chen, N.; Zhang, D.; Chong, K. The F-Box protein OsFBK12 targets OsSAMS1 for degradation and affects pleiotropic phenotypes, including leaf senescence, in rice. Plant Physiol. 2013, 163, 1673–1685. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhang, H.; Liao, J.; Wei, L.; Guo, R.; Xiao, W.; Kuang, W.; Huang, Y.; Wang, Z. Qualitative analysis of N-linked glycoproteome in senescent flag leaf of rice. Plant Growth Regul. 2019, 88, 309–326. [Google Scholar] [CrossRef]
- Veyres, N.; Danon, A.; Aono, M.; Galliot, S.; Karibasappa, Y.B.; Diet, A.; Grandmottet, F.; Tamaoki, M.; Lesur, D.; Pilard, S.; et al. The Arabidopsis sweetie mutant is affected in carbohydrate metabolism and defective in the control of growth, development and senescence. Plant J. 2008, 55, 665–686. [Google Scholar] [CrossRef] [PubMed]
- Izumi, M.; Ishida, H. The changes of leaf carbohydrate contents as a regulator of autophagic degradation of chloroplasts via Rubisco-containing bodies during leaf senescence. Plant Signal Behav. 2011, 6, 685–687. [Google Scholar]
- Troncoso-Ponce, M.A.; Cao, X.; Yang, Z.; Ohlrogge, J.B. Lipid turnover during senescence. Plant Sci 2013, 205–206, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Balazadeh, S.; Tohge, T.; Erban, A.; Giavalisco, P.; Kopka, J.; Mueller-Roeber, B.; Fernie, A.R.; Hoefgen, R. Comprehensive dissection of spatiotemporal metabolic shifts in primary, secondary, and lipid metabolism during developmental senescence in Arabidopsis. Plant Physiol. 2013, 162, 1290–1310. [Google Scholar] [CrossRef]
- Bengoa, L.S.; Astigueta, F.H.; Nicosia, S.; Sebastian, M.; Paula, F.; Ruth, H. Transcription factors associated with leaf senescence in crops. Plants 2019, 8, 411. [Google Scholar] [CrossRef]
- Woo, H.R.; Kim, H.J.; Lim, P.O.; Nam, H.G. Leaf senescence: Systems and dynamics aspects. Annu. Rev. Plant Biol. 2019, 70, 347–376. [Google Scholar] [CrossRef]
- Mueller-Roeber, B.; Balazadeh, S. Auxin and its role in plant senescence. J. Plant Growth Regul. 2014, 33, 21–33. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Sheng, Y.; Wang, Y.; Meijer, H.J.G.; Yang, X.Y.; Hua, C.L.; Ye, W.W.; Tao, K.; Liu, X.Y.; Govers, F.; Wang, Y.C. The heat shock transcription factor PsHSF1 of Phytophthora sojaeis required for oxidative stress tolerance and detoxifying the plant oxidative burst. Environ. Microbiol. 2015, 17, 1351–1364. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell 2005, 17, 1866–1875. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Wang, S.G.; Sun, D.Z. The research progress in the function and regulation of STAYGREEN in crops. J. Biol. 2019, 36, 83–87. (In Chinese) [Google Scholar]
- Lin, T.Z.; Sun, L.T.; Gong, H.B.; Liu, L.L.; Zhao, Z.G.; Dong, H.; Wang, Y.H.; Jiang, L.; Wan, J.M. Identification and gene mapping of a premature leaf senescence 5 mutant with starch accumulation in rice leaves. J. Nanjing Agric. Univ. 2020, 43, 414–422. (In Chinese) [Google Scholar]
- Clouse, S.D. Molecular genetic studies confirm the role of brassinosteroids in plant growth and development. Plant J. 1996, 10, 1–8. [Google Scholar] [CrossRef]
- Suzuki, Y.; Makino, A. Translational downregulation of RBCL is operative in the coordinated expression of rubisco genes in senescent leaves in rice. J. Exp. Bot. 2013, 64, 1145–1152. [Google Scholar] [CrossRef]
- Suzuki, Y.; Makino, A.; Mae, T. Changes in the turnover of Rubisco and levels of mRNAs of rbcL and rbcS in rice leaves from emergence to senescence. Plant Cell Environ. 2001, 24, 1353–1360. [Google Scholar] [CrossRef]
- Huang, X.P.; Zhang, H.Y.; Guo, R.; Wang, Q.; Liu, X.Z.; Kuang, W.G.; Song, H.Y.; Liao, J.L.; Huang, Y.J.; Wang, Z.H. Systematic identification and characterization of circular RNAs involved in flag leaf senescence of rice. Planta 2021, 253, 26. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Ren, G.; Fan, X.; Liu, T.; Wang, S.; Zhao, G. Genome-wide analysis of differentially expressed profiles of mRNAs, lncRNAs and circRNAs during Cryptosporidium baileyi infection. BMC Genom. 2018, 19, 356. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Jiang, C.; Guo, Y.; Li, C.; Zhao, W.; Nie, F.; Liu, Q. Suppression of OsSAUR2 gene expression immobilizes soil arsenic bioavailability by modulating root exudation and rhizosphere microbial assembly in rice. J. Hazard. Mater. 2024, 473, 134587. [Google Scholar] [CrossRef] [PubMed]

| Category | Gene ID | MSU-ID | Gene Name | Reference | Module |
|---|---|---|---|---|---|
| Transcription factors | BGIOSGA002217 | LOC_Os01g09620 | OsDOS | [36] | brown |
| BGIOSGA008165 | LOC_Os02g26430 | OsWRKY42 | [37] | turquoise | |
| BGIOSGA016546 | LOC_Os04g38720 | OsNAC2 | [38] | black | |
| BGIOSGA023457 | LOC_Os06g46270 | ONAC011 | [39] | black | |
| BGIOSGA025120 | LOC_Os07g04560 | ONAC096 | [40] | turquoise | |
| BGIOSGA018854 | LOC_Os05g04640 | OsWRKY5 | [41] | pink | |
| BGIOSGA020819 | LOC_Os06g43090 | OsMYB102 | [42] | turquoise | |
| BGIOSGA035845 | LOC_Os12g41860 | OsHox33 | [43] | turquoise | |
| BGIOSGA000374 | LOC_Os01g66120 | OsNAC6 | [44] | pink | |
| BGIOSGA026934 | LOC_Os08g33670 | ONAC106 | [45] | black | |
| Hormone-related genes | BGIOSGA013214 | LOC_Os03g44380 | OsNCED3 | [46] | black |
| BGIOSGA037804 | LOC_Os12g42280 | OsNCED5 | [47] | turquoise | |
| BGIOSGA004591 | LOC_Os01g57854 | OsPME1 | [48] | black | |
| BGIOSGA029334 | LOC_Os09g37330 | SAUR39 | [49] | black | |
| BGIOSGA004316 | LOC_Os01g51430 | OsRTH1 | [50] | black | |
| BGIOSGA014572 | LOC_Os04g46980 | cZOGT1 | [51] | turquoise | |
| BGIOSGA014571 | LOC_Os04g46990 | cZOGT2 | [51] | brown | |
| BGIOSGA027967 | LOC_Os08g04540 | TDC1 | [52] | pink | |
| Chlorophyll degradation-related genes | BGIOSGA003046 | LOC_Os01g12710 | NYC1 | [13] | black |
| BGIOSGA010125 | LOC_Os03g45194 | NOL | [14] | black | |
| BGIOSGA022884 | LOC_Os06g24730 | NYC3 | [53] | black | |
| BGIOSGA029383 | LOC_Os09g36200 | SGR | [16] | black | |
| BGIOSGA011859 | LOC_Os03g05310 | PAO | [54] | black | |
| BGIOSGA032012 | LOC_Os10g25030 | RCCR1 | [54] | black | |
| BGIOSGA028915 | LOC_Os08g38710 | OsAkaGal | [55] | black | |
| SAGs | BGIOSGA022517 | LOC_Os06g10560 | [56] | magenta | |
| BGIOSGA020694 | LOC_Os06g46160 | [57] | black | ||
| BGIOSGA008338 | LOC_Os02g32520 | [57] | black | ||
| BGIOSGA031565 | LOC_Os10g36848 | [57] | black | ||
| BGIOSGA009298 | LOC_Os02g57280 | [57] | pink | ||
| BGIOSGA004049 | LOC_Os01g44120 | [57] | turquoise | ||
| BGIOSGA004735 | LOC_Os01g61460 | [56] | turquoise | ||
| BGIOSGA009241 | LOC_Os02g56250 | [57] | turquoise | ||
| BGIOSGA009572 | LOC_Os03g60090 | [57] | turquoise | ||
| BGIOSGA026629 | LOC_Os08g41280 | [57] | turquoise | ||
| BGIOSGA008974 | LOC_Os02g49650 | [56] | turquoise | ||
| BGIOSGA033472 | LOC_Os10g41930 | [57] | turquoise | ||
| BGIOSGA001700 | LOC_Os01g24710 | [57] | black | ||
| BGIOSGA024190 | LOC_Os07g34520 | [58] | black | ||
| ROS | BGIOSGA017588 | LOC_Os05g48390 | RLS1 | [59] | black |
| BGIOSGA033278 | LOC_Os10g37180 | OsGDCH | [60] | brown | |
| Other | BGIOSGA011953 | LOC_Os03g07530 | OsFBK12 | [61] | black |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, R.; Chen, T.; Li, J.; Hu, C.; Yu, Z.; Zeng, Z.; Chen, Z.; Wang, L.; Xiang, T.; Huang, X. Time-Course Transcriptome, Metabolome, and Weighted Gene Co-Expression Network Analysis Reveal the Roles of the OsBELH4A Gene in Regulating Leaf Senescence and Grain Yield of Rice. Plants 2025, 14, 2973. https://doi.org/10.3390/plants14192973
Zheng R, Chen T, Li J, Hu C, Yu Z, Zeng Z, Chen Z, Wang L, Xiang T, Huang X. Time-Course Transcriptome, Metabolome, and Weighted Gene Co-Expression Network Analysis Reveal the Roles of the OsBELH4A Gene in Regulating Leaf Senescence and Grain Yield of Rice. Plants. 2025; 14(19):2973. https://doi.org/10.3390/plants14192973
Chicago/Turabian StyleZheng, Ruyi, Tianyu Chen, Jianjian Li, Chengcheng Hu, Zhiming Yu, Zhanghui Zeng, Zhehao Chen, Lilin Wang, Taihe Xiang, and Xiaoping Huang. 2025. "Time-Course Transcriptome, Metabolome, and Weighted Gene Co-Expression Network Analysis Reveal the Roles of the OsBELH4A Gene in Regulating Leaf Senescence and Grain Yield of Rice" Plants 14, no. 19: 2973. https://doi.org/10.3390/plants14192973
APA StyleZheng, R., Chen, T., Li, J., Hu, C., Yu, Z., Zeng, Z., Chen, Z., Wang, L., Xiang, T., & Huang, X. (2025). Time-Course Transcriptome, Metabolome, and Weighted Gene Co-Expression Network Analysis Reveal the Roles of the OsBELH4A Gene in Regulating Leaf Senescence and Grain Yield of Rice. Plants, 14(19), 2973. https://doi.org/10.3390/plants14192973







