Harnessing Genomics of Diaporthe amygdali for Improved Control of Peach Twig Canker and Shoot Blight (TCSB)
Abstract
1. Introduction
2. Results
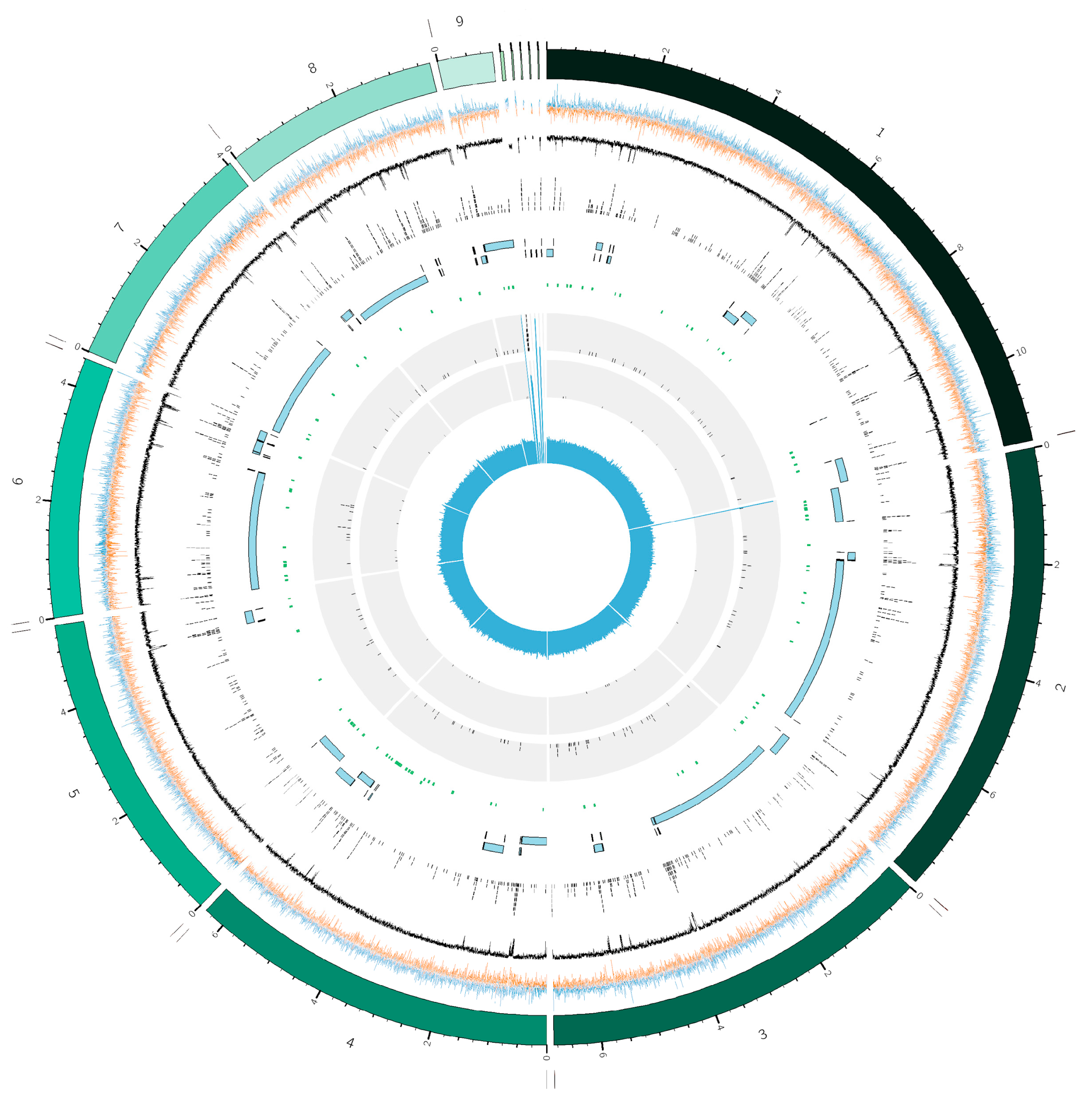
2.1. Assembly Metrics and Genomic Overview
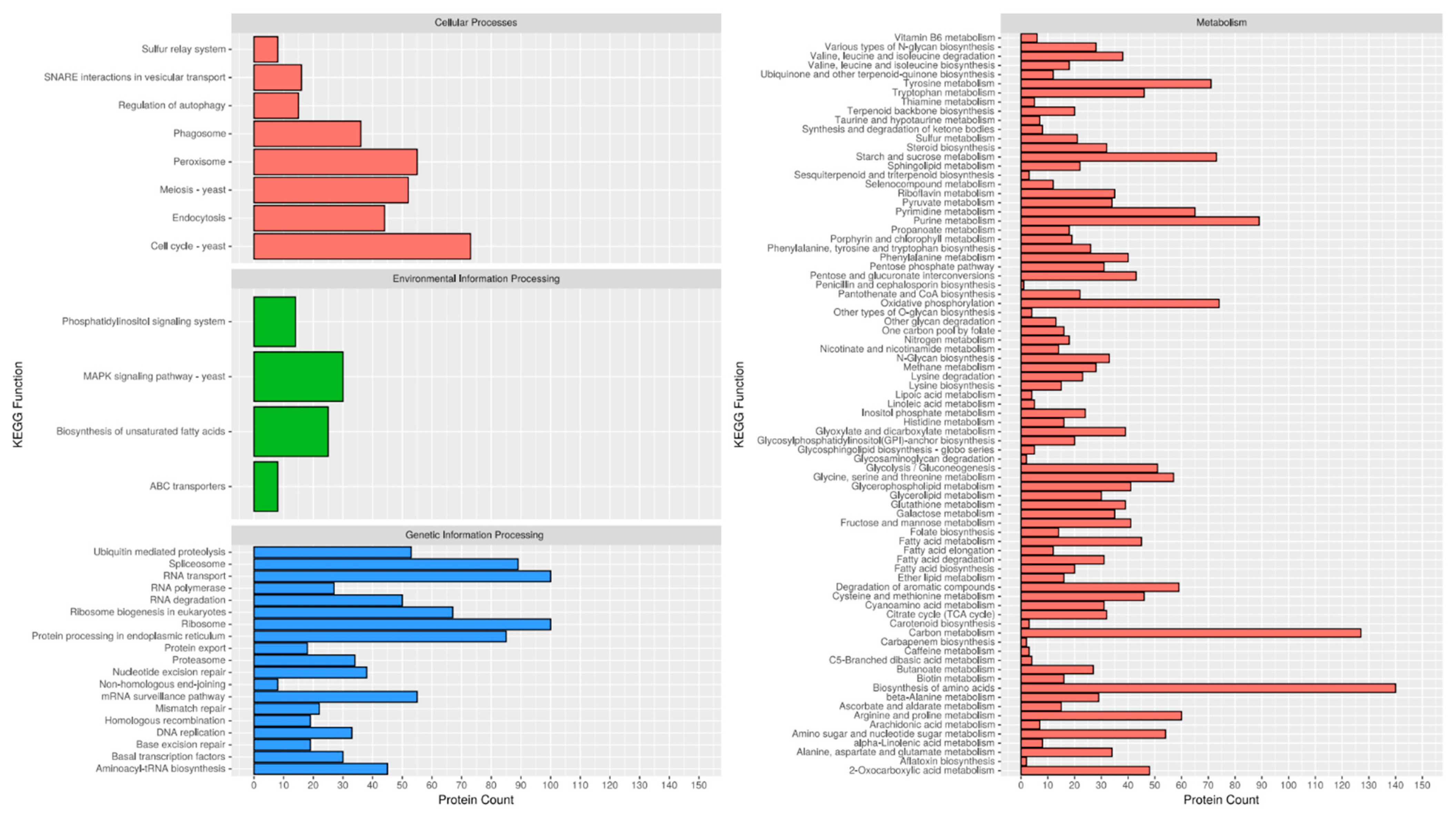
2.2. Functional Annotation
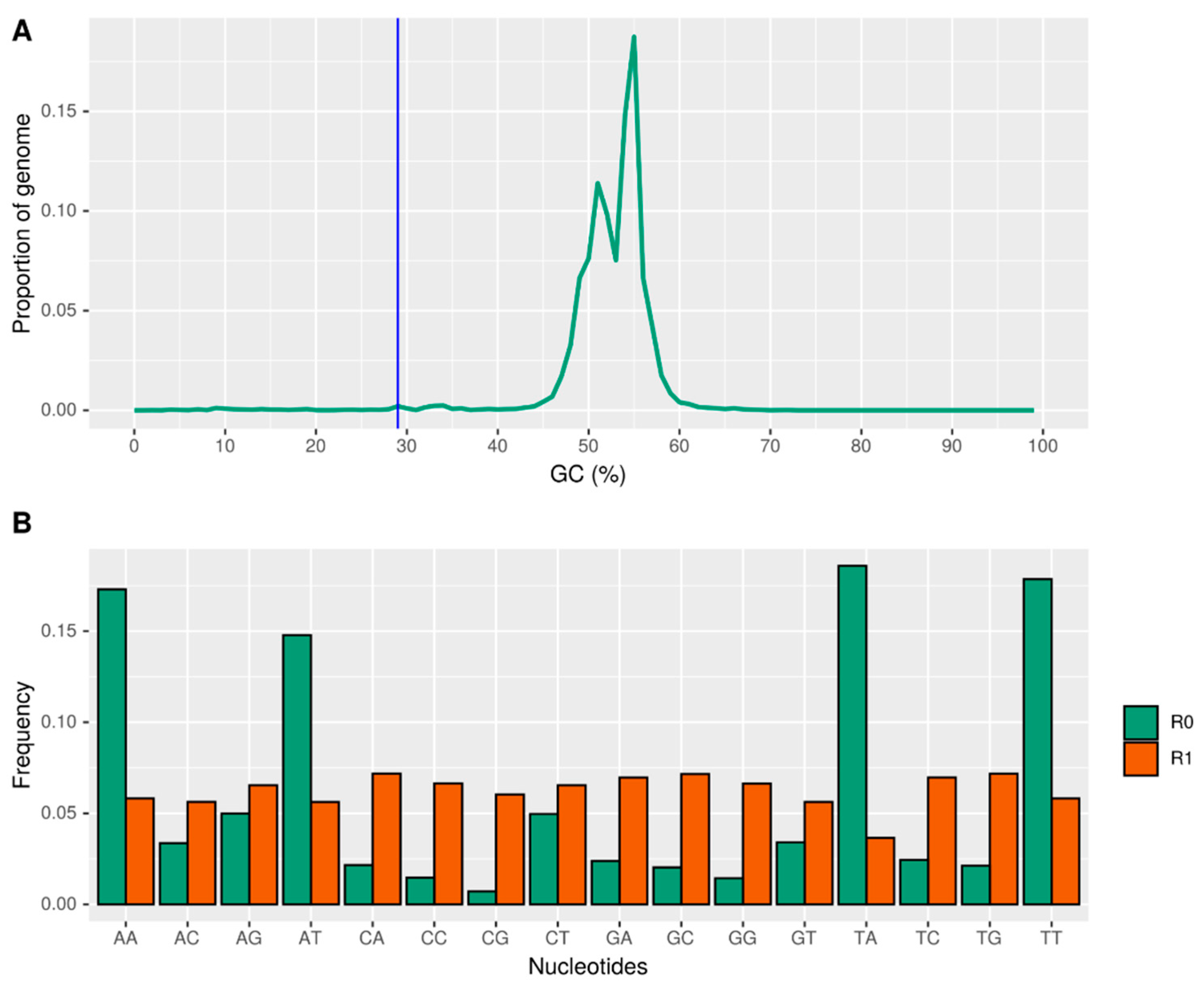
2.3. AT-Rich Regions
2.4. Secondary Metabolite Biosynthetic Gene Clusters
2.5. Identification of Features Related to Host Invasion and Colonization
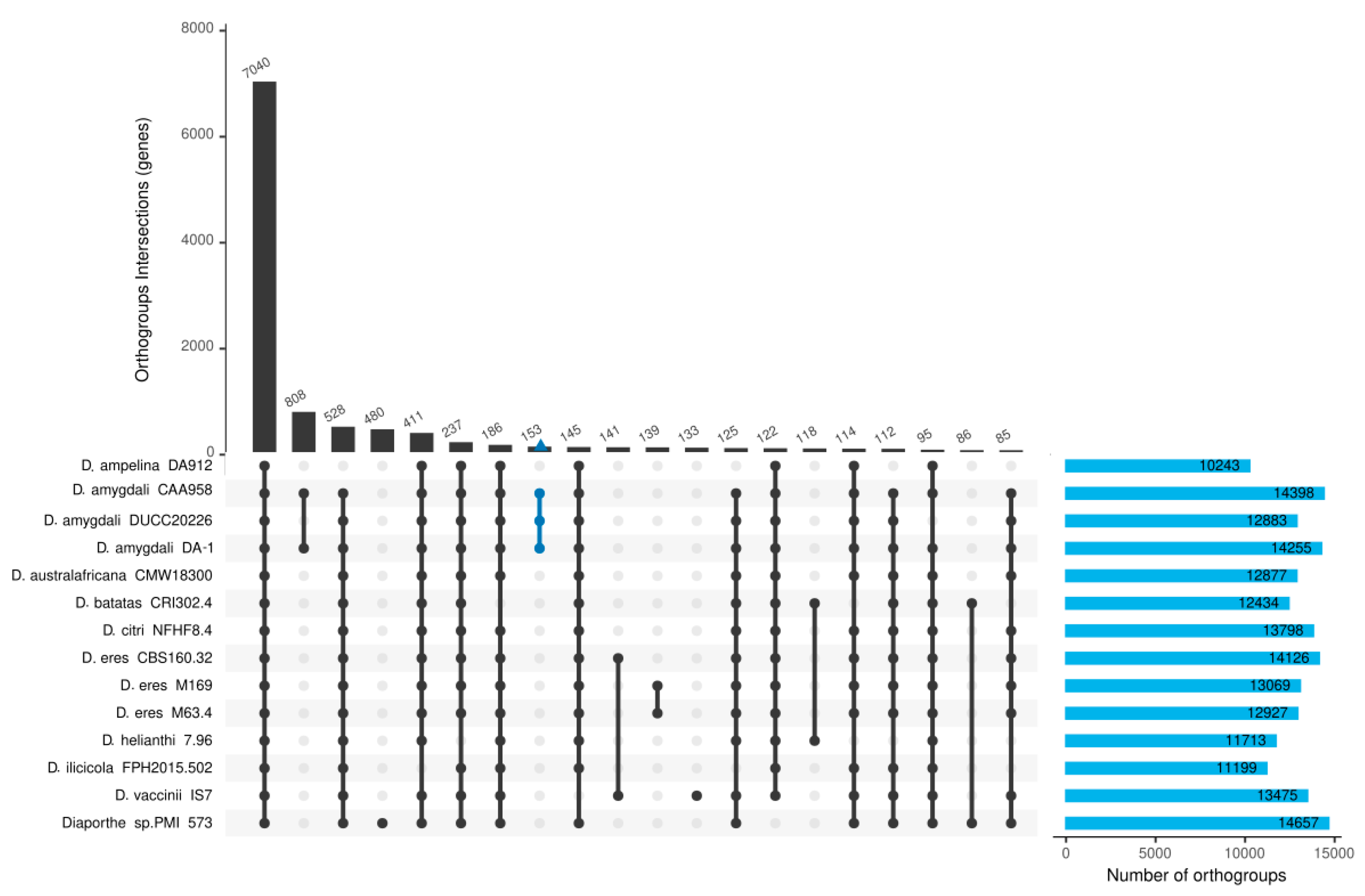
2.6. Comparative Genomics Analysis Among Diaporthe Isolates
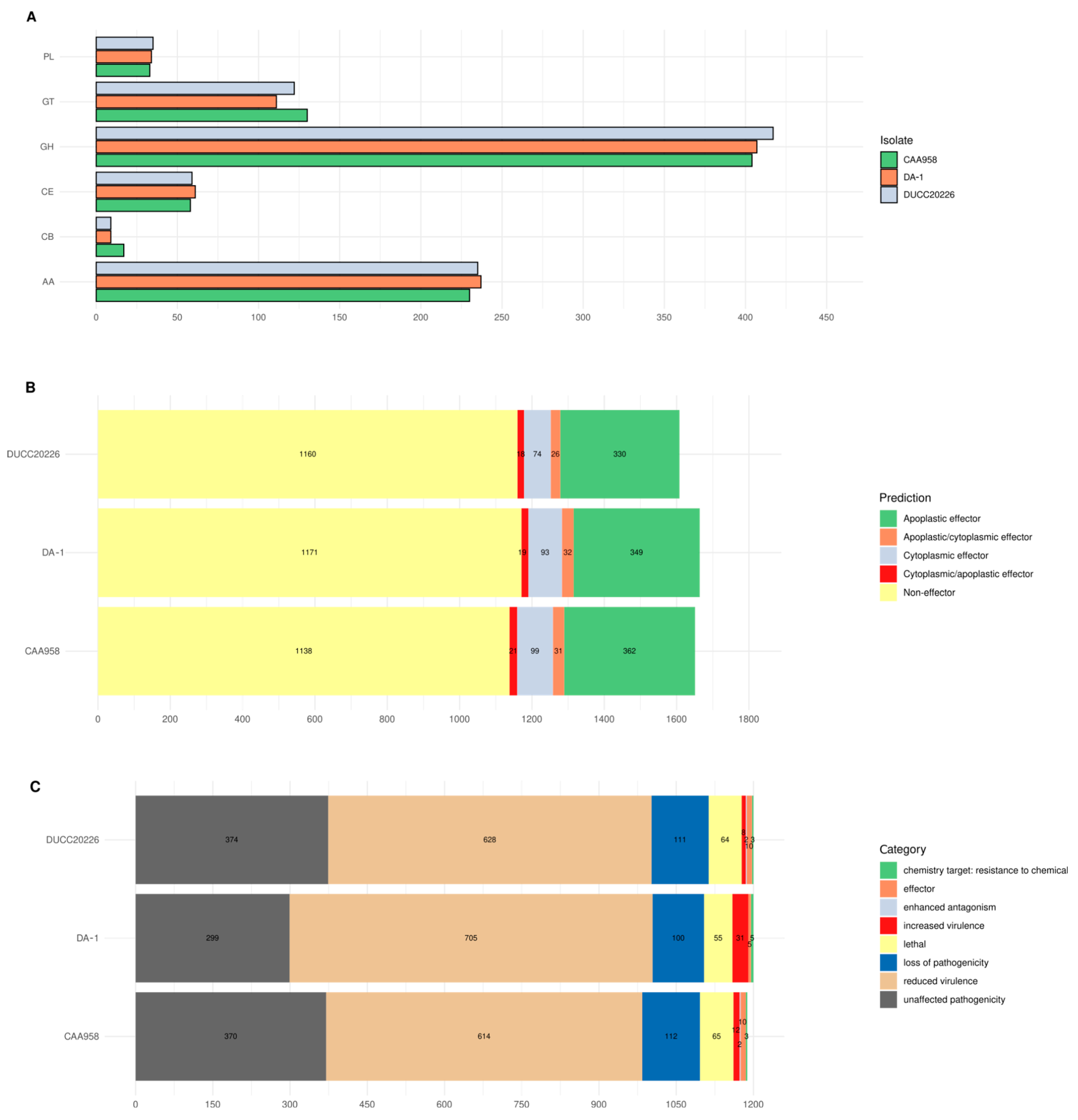
2.7. Effectors and Virulence-Related Features Among D. amygdali Isolates
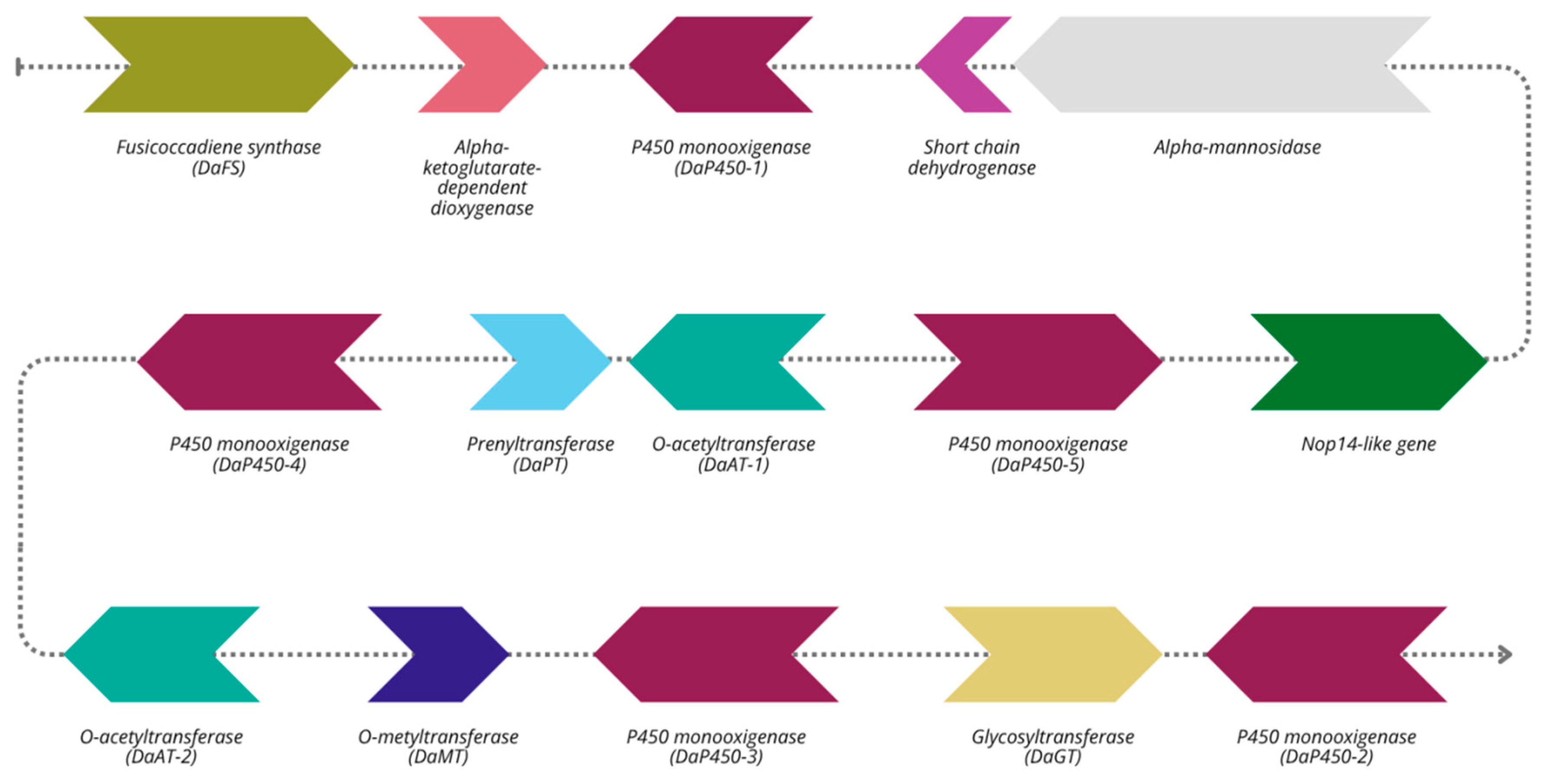
2.8. Fusicoccin Biosynthetic Gene Cluster
3. Discussion
4. Materials and Methods
4.1. Isolate Selection and HMW DNA Extraction
4.2. Whole-Genome Sequencing and Assembly
4.3. Structural and Functional Annotation
4.4. Pathogenicity-Related Features Among D. amygdali Isolates
4.5. Ortholog Identification and Phylogenetic Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gomes, R.R.; Glienke, C.; Videira, S.I.R.; Lombard, L.; Groenewald, J.Z.; Crous, P.W. Diaporthe: A Genus of Endophytic, Saprobic and Plant Pathogenic Fungi. Persoonia Mol. Phylogeny Evol. Fungi 2013, 31, 1–41. [Google Scholar] [CrossRef]
- Guarnaccia, V.; Crous, P.W. Emerging Citrus Diseases in Europe Caused by Species of Diaporthe. IMA Fungus 2017, 8, 317–334. [Google Scholar] [CrossRef]
- Gusella, G.; La Quatra, G.; Agustí-Brisach, C.; Trapero, A.; Polizzi, G. Elucidating the Almond Constriction Canker Caused by Diaporthe Amygdali in Sicily (South Italy). J. Plant Pathol. 2023, 105, 987–1000. [Google Scholar] [CrossRef]
- Brugneti, F.; Rossini, L.; Cirilli, M.; Nardecchia, A.; Onofri, M.; Mazzaglia, A.; Turco, S. Elucidation of ‘Twig Canker and Shoot Blight’ (TCSB) in Peach Caused by Diaporthe Amygdali in the North of Italy in Emilia-Romagna. Physiol. Plant. 2025. [CrossRef]
- Delacroix, G. Sur une maladie des Amendiers en Provence. Bull. De La Société Mycol. De Fr. 1905, 21, 180–185. [Google Scholar]
- Canonaco, A. 1936: Bollettino di Studi ed Informazioni, Reale Giardino Coloniale 14: - 20 -. Biota of NZ. Available online: https://biotanz.landcareresearch.co.nz/references/1cb0e6a4-36b9-11d5-9548-00d0592d548c (accessed on 22 July 2025).
- Diogo, E.L.F.; Santos, J.M.; Phillips, A.J.L. Phylogeny, Morphology and Pathogenicity of Diaporthe and Phomopsis Species on Almond in Portugal. Fungal Divers. 2010, 44, 107–115. [Google Scholar] [CrossRef]
- Zhao, Z.; Liu, H.; Wang, C.; Xu, J.-R. Comparative Analysis of Fungal Genomes Reveals Different Plant Cell Wall Degrading Capacity in Fungi. BMC Genom. 2013, 14, 274. [Google Scholar] [CrossRef] [PubMed]
- Udayanga, D.; Liu, X.; McKenzie, E.H.C.; Chukeatirote, E.; Bahkali, A.H.A.; Hyde, K.D. The Genus Phomopsis: Biology, Applications, Species Concepts and Names of Common Phytopathogens. Fungal Divers. 2011, 50, 189–225. [Google Scholar] [CrossRef]
- Hilário, S.; Pinto, G.; Monteiro, P.; Santos, L.; Alves, A. The Impact of Two Diaporthe Species on Vaccinium Corymbosum Physiological Performance under Different Water Availability Scenarios. Eur. J. Plant Pathol. 2023, 166, 161–177. [Google Scholar] [CrossRef]
- Beluzán, F.; Vicent, A.; Conesa, D.; Torrentí, F.; Olmo, D.; León, M.; Abad-Campos, P.; Armengol, J. Effect of Weather Variables on the Inoculum of Diaporthe Amygdali, the Causal Agent of Twig Canker and Shoot Blight in Almond Orchards. Phytopathology 2025, 115, 649–658. [Google Scholar] [CrossRef]
- León, M.; Berbegal, M.; Rodríguez-Reina, J.M.; Elena, G.; Abad-Campos, P.; Ramón-Albalat, A.; Olmo, D.; Vicent, A.; Luque, J.; Miarnau, X.; et al. Identification and Characterization of Diaporthe spp. Associated with Twig Cankers and Shoot Blight of Almonds in Spain. Agronomy 2020, 10, 1062. [Google Scholar] [CrossRef]
- Beluzán, F.; Miarnau, X.; Torguet, L.; Zazurca, L.; Abad-Campos, P.; Luque, J.; Armengol, J. Susceptibility of Almond (Prunus dulcis) Cultivars to Twig Canker and Shoot Blight Caused by Diaporthe Amygdali. Plant Dis. 2022, 106, 1890–1897. [Google Scholar] [CrossRef] [PubMed]
- Tuset, J.J.; Portilla, M.A.T. Taxonomic Status of Fusicoccum Amygdali and Phomopsis Amygdalina. Can. J. Bot. 1989, 67, 1275–1280. [Google Scholar] [CrossRef]
- Hilário, S.; Santos, L.; Alves, A. Diversity and Pathogenicity of Diaporthe Species Revealed from a Survey of Blueberry Orchards in Portugal. Agriculture 2021, 11, 1271. [Google Scholar] [CrossRef]
- Istituto Nazionale Di Statistica (ISTAT). Superficie e Produzione Delle Principali Coltivazioni Agricole. 2024. Available online: http://dati.istat.it/index.aspx?queryid=33706 (accessed on 5 July 2025).
- Franceschelli, F.; Scannavini, M.; Cavazza, F. Diffuso nella Pianura Romagnola e di Non Facile Contenimento, Questo Fungo può Compromettere la Produttività delle Piante e Portare alla loro Morte. Più Colpite le Percoche e Alcune Varietà di Pesche e Nettarine. Available online: https://agricoltura.regione.emilia-romagna.it/fitosanitario/avversita/schede/frutticole/pesco/2008-difficile-estirpare-il-fusicocco-del-pesco.pdf/@@download/file/2008%20Difficile%20estirpare%20il%20fusicocco%20del%20pesco%20.pdf (accessed on 5 July 2025).
- Fisher, M.C.; Henk, D.A.; Briggs, C.J.; Brownstein, J.S.; Madoff, L.C.; McCraw, S.L.; Gurr, S.J. Emerging Fungal Threats to Animal, Plant and Ecosystem Health. Nature 2012, 484, 186–194. [Google Scholar] [CrossRef]
- Studholme, D.J.; Glover, R.H.; Boonham, N. Application of High-Throughput DNA Sequencing in Phytopathology. Annu. Rev. Phytopathol. 2011, 49, 87–105. [Google Scholar] [CrossRef]
- Fagundes, W.C.; Huang, Y.-S.; Häußler, S.; Langner, T. From Lesions to Lessons: Two Decades of Filamentous Plant Pathogen Genomics. Mol. Plant Microbe Interact. 2025, 38, 187–205. [Google Scholar] [CrossRef]
- Klosterman, S.J.; Rollins, J.R.; Sudarshana, M.R.; Vinatzer, B.A. Disease management in the genomics era—Summaries of focus issue papers. Phytopathology 2016, 106, 1068–1070. [Google Scholar] [CrossRef][Green Version]
- Sarethy, I.P.; Saharan, A. Genomics, proteomics and transcriptomics in the biological control of plant pathogens: A review. Indian Phytopathol. 2021, 74, 3–12. [Google Scholar] [CrossRef]
- Capote, N.; Pastrana, A.M.; Aguado, A.; Sánchez-Torres, P. Molecular Tools for Detection of Plant Pathogenic Fungi and Fungicide Resistance. In Plant Pathology; Cumagun, C.J.R., Ed.; IntechOpen: Rijeka, Croatia, 2012. [Google Scholar] [CrossRef]
- Turco, S.; Zuppante, L.; Drais, M.I.; Mazzaglia, A. Dressing like a Pathogen: Comparative Analysis of Different Pseudomonas Genomospecies Wearing Different Features to Infect Corylus Avellana. J. Phytopathol. 2022, 170, 504–516. [Google Scholar] [CrossRef]
- Turco, S.; Brugneti, F.; Fiorenzani, C.; Baroncelli, R.; Mazzaglia, A. Hybrid de Novo Assembly of the Genome of Colletotrichum Acutatum Sensu Stricto Isolate COL14 from Olive Fruit in Central Italy. J. Plant Pathol. 2025, 107, 727–730. [Google Scholar] [CrossRef]
- Gonçalves, M.F.M. Plant Pathogenic Fungi Special Issue: Genetics and Genomics. Microorganisms 2025, 13, 925. [Google Scholar] [CrossRef] [PubMed]
- Wilson, H.; Bar, I.; Hobson, K.; Vaghefi, N.; Petronitis, T.; Ford, R. Integrated disease management, adaptation and genomics of fungal plant pathogens in cropping systems. Plant Pathol. 2025, 74, 1445–1469. [Google Scholar] [CrossRef]
- Gobeil-Richard, M.; Tremblay, D.-M.; Beaulieu, C.; Van der Heyden, H.; Carisse, O. A Pyrosequencing-Based Method to Quantify Genetic Substitutions Associated with Resistance to Succinate Dehydrogenase Inhibitor Fungicides in Botrytis spp. Populations. Pest Manag. Sci. 2016, 72, 566–573. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog Facilitate Examination of the Genomic Links among Antimicrobial Resistance, Stress Response, and Virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef]
- Samils, B.; Andersson, B.; Edin, E.; Elfstrand, M.; Rönneburg, T.; Bucur, D.; Hutton, F.; Heick, T.M.; Hellin, P.; Kildea, S. Development of a PacBio Long-Read Sequencing Assay for High Throughput Detection of Fungicide Resistance in Zymoseptoria Tritici. Front. Microbiol. 2021, 12, 692845. [Google Scholar] [CrossRef]
- Freitag, M.; Williams, R.L.; Kothe, G.O.; Selker, E.U. A Cytosine Methyltransferase Homologue Is Essential for Repeat-Induced Point Mutation in Neurospora crassa. Proc. Natl. Acad. Sci. USA 2002, 99, 8802–8807. [Google Scholar] [CrossRef]
- Rouxel, T.; Grandaubert, J.; James, K.H.; Claire, H.; Angela, P.V.D.W.; Arnaud, C.; Victoria, D.; Véronique, A.; Pascal, B.; Salim, B.; et al. Effector diversification within compartments of the Leptosphaeria maculans genome affected by Repeat-Induced Point mutations. Nat. Commun. 2011, 2, 202. [Google Scholar] [CrossRef]
- Baroncelli, R.; Amby, D.B.; Zapparata, A.; Sarrocco, S.; Vannacci, G.; Le Floch, G.; Harrison, R.J.; Holub, E.; Sukno, S.A.; Sreenivasaprasad, S.; et al. Gene Family Expansions and Contractions Are Associated with Host Range in Plant Pathogens of the Genus Colletotrichum. BMC Genom. 2016, 17, 555. [Google Scholar] [CrossRef]
- Ferrara, M.; Perrone, G.; Gallo, A. Recent Advances in Biosynthesis and Regulatory Mechanisms of Principal Mycotoxins. Curr. Opin. Food Sci. 2022, 48, 100923. [Google Scholar] [CrossRef]
- Toyomasu, T.; Tsukahara, M.; Kaneko, A.; Niida, R.; Mitsuhashi, W.; Dairi, T.; Kato, N.; Sassa, T. Fusicoccins are biosynthesized by an unusual chimera diterpene synthase in fungi. Proc. Natl. Acad. Sci. USA 2007, 104, 3084–3088. [Google Scholar] [CrossRef] [PubMed]
- Toyomasu, T. Recent Advances Regarding Diterpene Cyclase Genes in Higher Plants and Fungi. Biosci. Biotechnol. Biochem. 2008, 72, 1168–1175. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Chou, W.K.W.; Toyomasu, T.; Cane, D.E.; Christianson, D.W. Structure and Function of Fusicoccadiene Synthase, a Hexameric Bifunctional Diterpene Synthase. ACS Chem. Biol. 2016, 11, 889–899. [Google Scholar] [CrossRef]
- Hilário, S.; Gonçalves, M.F.M.; Fidalgo, C.; Tacão, M.; Alves, A. Genome Analyses of Two Blueberry Pathogens: Diaporthe amygdali CAA958 and Diaporthe Eres CBS 160.32. J. Fungi 2022, 8, 804. [Google Scholar] [CrossRef]
- de Boer, A.H. The Fusicoccin Story Revisited. J. Exp. Bot. 2024, 75, 5531–5546. [Google Scholar] [CrossRef]
- Du, J.; Li, Y.; Yin, Z.; Wang, H.; Zhang, X.; Ding, X. High-Throughput Customization of Plant Microbiomes for Sustainable Agriculture. Front. Plant Sci. 2020, 11, 569742. [Google Scholar] [CrossRef]
- Gardy, J.L.; Loman, N.J. Towards a Genomics-Informed, Real-Time, Global Pathogen Surveillance System. Nat. Rev. Genet. 2018, 19, 9–20. [Google Scholar] [CrossRef]
- Weisberg, A.J.; Grünwald, N.J.; Savory, E.A.; Putnam, M.L.; Chang, J.H. Genomic Approaches to Plant-Pathogen Epidemiology and Diagnostics. Annu. Rev. Phytopathol. 2021, 59, 311–332. [Google Scholar] [CrossRef]
- Cardacino, A.; Tastekin, T.; Brugneti, F.; Cirilli, M.; Mazzaglia, A.; Turco, S. Peach Buds’ Microbiome Profiling Reveals Cultivar-Specific Signatures Associated with TCSB Susceptibility. Stresses 2025, 5, 60. [Google Scholar] [CrossRef]
- He, L.; Zhou, Y.; Mo, Q.; Huang, Y.; Tang, X. Spray-induced gene silencing in phytopathogen: Mechanisms, applications, and progress. Adv. Agrochem 2024, 3, 289–297. [Google Scholar] [CrossRef]
- Babraham Bioinformatics—FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 23 July 2025).
- Cheng, H.; Concepcion, G.T.; Feng, X.; Zhang, H.; Li, H. Haplotype-Resolved de Novo Assembly Using Phased Assembly Graphs with Hifiasm. Nat. Methods 2021, 18, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows–Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Uliano-Silva, M.; Ferreira, J.G.R.N.; Krasheninnikova, K.; Blaxter, M.; Mieszkowska, N.; Hall, N.; Holland, P.; Durbin, R.; Richards, T.; Kersey, P.; et al. MitoHiFi: A Python Pipeline for Mitochondrial Genome Assembly from PacBio High Fidelity Reads. BMC Bioinform. 2023, 24, 288. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- García-Alcalde, F.; Okonechnikov, K.; Carbonell, J.; Cruz, L.M.; Götz, S.; Tarazona, S.; Dopazo, J.; Meyer, T.F.; Conesa, A. Qualimap: Evaluating next-Generation Sequencing Alignment Data. Bioinformatics 2012, 28, 2678–2679. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, İ.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Hu, K.; Ni, P.; Xu, M.; Zou, Y.; Chang, J.; Gao, X.; Li, Y.; Ruan, J.; Hu, B.; Wang, J. HiTE: A Fast and Accurate Dynamic Boundary Adjustment Approach for Full-Length Transposable Element Detection and Annotation. Nat. Commun. 2024, 15, 5573. [Google Scholar] [CrossRef]
- Burge, C.; Karlin, S. Prediction of Complete Gene Structures in Human Genomic DNA1. J. Mol. Biol. 1997, 268, 78–94. [Google Scholar] [CrossRef]
- Stanke, M.; Waack, S. Gene Prediction with a Hidden Markov Model and a New Intron Submodel. Bioinformatics 2003, 19 (Suppl. 2), ii215–ii225. [Google Scholar] [CrossRef] [PubMed]
- Majoros, W.H.; Pertea, M.; Salzberg, S.L. TigrScan and GlimmerHMM: Two Open Source Ab Initio Eukaryotic Gene-FInders. Bioinformatics 2004, 20, 2878–2879. [Google Scholar] [CrossRef] [PubMed]
- Blanco, E.; Parra, G.; Guigó, R. Using Geneid to Identify Genes. Curr. Protoc. Bioinform. 2003, 64, e56. [Google Scholar] [CrossRef]
- Korf, I. Gene Finding in Novel Genomes. BMC Bioinform. 2004, 5, 59. [Google Scholar] [CrossRef] [PubMed]
- Keilwagen, J.; Hartung, F.; Grau, J. GeMoMa: Homology-Based Gene Prediction Utilizing Intron Position Conservation and RNA-Seq Data. Gene Predict. Methods Protoc. 2019, 1962, 161–177. [Google Scholar] [CrossRef]
- Haas, B.J.; Salzberg, S.L.; Zhu, W.; Pertea, M.; Allen, J.E.; Orvis, J.; White, O.; Buell, C.R.; Wortman, J.R. Automated Eukaryotic Gene Structure Annotation Using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008, 9, R7. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A Program for Improved Detection of Transfer RNA Genes in Genomic Sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-Fold Faster RNA Homology Searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Burge, S.W.; Bateman, A.; Daub, J.; Eberhardt, R.Y.; Eddy, S.R.; Floden, E.W.; Gardner, P.P.; Jones, T.A.; Tate, J.; et al. Rfam 12.0: Updates to the RNA Families Database. Nucleic Acids Res. 2015, 43, D130–D137. [Google Scholar] [CrossRef]
- Hane, J.K.; Oliver, R.P. RIPCAL: A Tool for Alignment-Based Analysis of Repeat-Induced Point Mutations in Fungal Genomic Sequences. BMC Bioinform. 2008, 9, 478. [Google Scholar] [CrossRef]
- Testa, A.C.; Oliver, R.P.; Hane, J.K. OcculterCut: A Comprehensive Survey of AT-Rich Regions in FungalGenomes. Genome Biol. Evol. 2016, 8, 2044–2064. [Google Scholar] [CrossRef]
- Tatusov, R.L. The COG Database: A Tool for Genome-Scale Analysis of Protein Functions and Evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG Resource for Deciphering the Genome. Nucleic Acids Res. 2004, 32 (Suppl. 1), D277–D280. [Google Scholar] [CrossRef] [PubMed]
- Boeckmann, B. The SWISS-PROT Protein Knowledgebase and Its Supplement TrEMBL in 2003. Nucleic Acids Res. 2003, 31, 365–370. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam Protein Families Database: Towards a More Sustainable Future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A Universal Tool for Annotation, Visualization and Analysis in Functional Genomics Research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving Cluster Detection and Comparison Capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Saier, M.H. TCDB: The Transporter Classification Database for Membrane Transport Protein Analyses and Information. Nucleic Acids Res. 2006, 34, D181–D186. [Google Scholar] [CrossRef]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Fischer, M.; Knoll, M.; Sirim, D.; Wagner, F.; Funke, S.; Pleiss, J. The Cytochrome P450 Engineering Database: A navigation and prediction tool for the cytochrome P450 protein family. Bioinformatics 2007, 23, 2015–2017. [Google Scholar] [CrossRef]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and taxonomy in diagnostics for food security: Soft-rotting enterobacterial plant pathogens. Anal. Methods 2016, 8, 12–24. [Google Scholar] [CrossRef]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A Meta Server for Automated Carbohydrate-Active Enzyme Annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef]
- Sperschneider, J.; Dodds, P.N. EffectorP 3.0: Prediction of apoplastic and cytoplasmic effectors in fungi and oomycetes. Mol. Plant-Microbe Interact. 2022, 35, 146–156. [Google Scholar] [CrossRef]
- Urban, M.; Cuzick, A.; Seager, J.; Wood, V.; Rutherford, K.; Venkatesh, S.Y.; De Silva, N.; Martinez, M.C.; Pedro, H.; Yates, A.D.; et al. PHI-Base: The Pathogen–Host Interactions Database. Nucleic Acids Res. 2020, 48, D613–D620. [Google Scholar] [CrossRef] [PubMed]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic Orthology Inference for Comparative Genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R Package for the Visualization of Intersecting Sets and Their Properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef] [PubMed]
| Genomic Metrics | D. amygdali DA-1 |
|---|---|
| # Contigs | 14 |
| Genome size | 51,431,816 |
| Largest Contig | 11,470,755 |
| N50 | 6,815,508 |
| N90 | 4,044,788 |
| L50 | 3 |
| L90 | 7 |
| GC content % | 52.64 |
| # N’s per 100 kbp | 0 |
| # N’s | 0 |
| Genome coverage | 76.42x |
| BUSCO completeness | 99.2% |
| Complete | 253 |
| Single copy | 253 |
| Duplicated | 0 |
| Fragmented | 2 |
| Missing | 0 |
| Predicted genes | 15,206 |
| rRNAs | 51 |
| tRNAs | 160 |
| ncRNAs | 84 |
| Signal peptide | 1799 |
| Retroelements | 663 |
| DNA transposons | 37 |
| Simple repeats | 11,811 |
| Accession Number | Organism Name | Strain | Host |
|---|---|---|---|
| GCA_026229845.1 | Diaporthe amygdali | CAA958 | Vaccinium corymbosum |
| GCA_019321695.1 | Diaporthe batatas | CRI 302-4 | Ipomoea batatas |
| GCA_014595645.1 | Diaporthe citri | NFHF-8-4 | Citrus reticulata cv. ‘Nanfengmiju’ |
| GCA_023242295.1 | Diaporthe ilicicola | FPH2015-502 | Ilex verticillata |
| GCA_001006365.1 | Diaporthe ampelina | DA912 | Vitis vinifera |
| GCA_032433605.1 | Diaporthe amygdali | DUCC20226 | Apple tree |
| GCA_022570775.2 | Diaporthe eres | M63-4 | Malus sp. |
| GCA_022570805.2 | Diaporthe eres | M169 | Prunus armeniaca |
| GCA_024867555.1 | Diaporthe eres | CBS 160.32 | Vaccinium corymbosum |
| GCA_021399435.1 | Diaporthe sp. | PMI_573 | not applicable |
| GCA_042826995.1 | Diaporthe vaccinii | IS7 | Vaccinium macrocarpon cv. ‘Stevens’ |
| GCA_001702395.2 | Diaporthe helianthi | Jul-96 | Helianthus sp. |
| GCA_042257625.1 | Diaporthe australafricana | CMW 18300 | Elegia capensis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turco, S.; Brugneti, F.; Cardacino, A.; Mazzaglia, A. Harnessing Genomics of Diaporthe amygdali for Improved Control of Peach Twig Canker and Shoot Blight (TCSB). Plants 2025, 14, 2960. https://doi.org/10.3390/plants14192960
Turco S, Brugneti F, Cardacino A, Mazzaglia A. Harnessing Genomics of Diaporthe amygdali for Improved Control of Peach Twig Canker and Shoot Blight (TCSB). Plants. 2025; 14(19):2960. https://doi.org/10.3390/plants14192960
Chicago/Turabian StyleTurco, Silvia, Federico Brugneti, Antonella Cardacino, and Angelo Mazzaglia. 2025. "Harnessing Genomics of Diaporthe amygdali for Improved Control of Peach Twig Canker and Shoot Blight (TCSB)" Plants 14, no. 19: 2960. https://doi.org/10.3390/plants14192960
APA StyleTurco, S., Brugneti, F., Cardacino, A., & Mazzaglia, A. (2025). Harnessing Genomics of Diaporthe amygdali for Improved Control of Peach Twig Canker and Shoot Blight (TCSB). Plants, 14(19), 2960. https://doi.org/10.3390/plants14192960






