The Association Between DNA Methylation and Three-Dimensional Genome During Whole Genome Doubling in Arabidopsis thaliana
Abstract
1. Introduction
2. Results
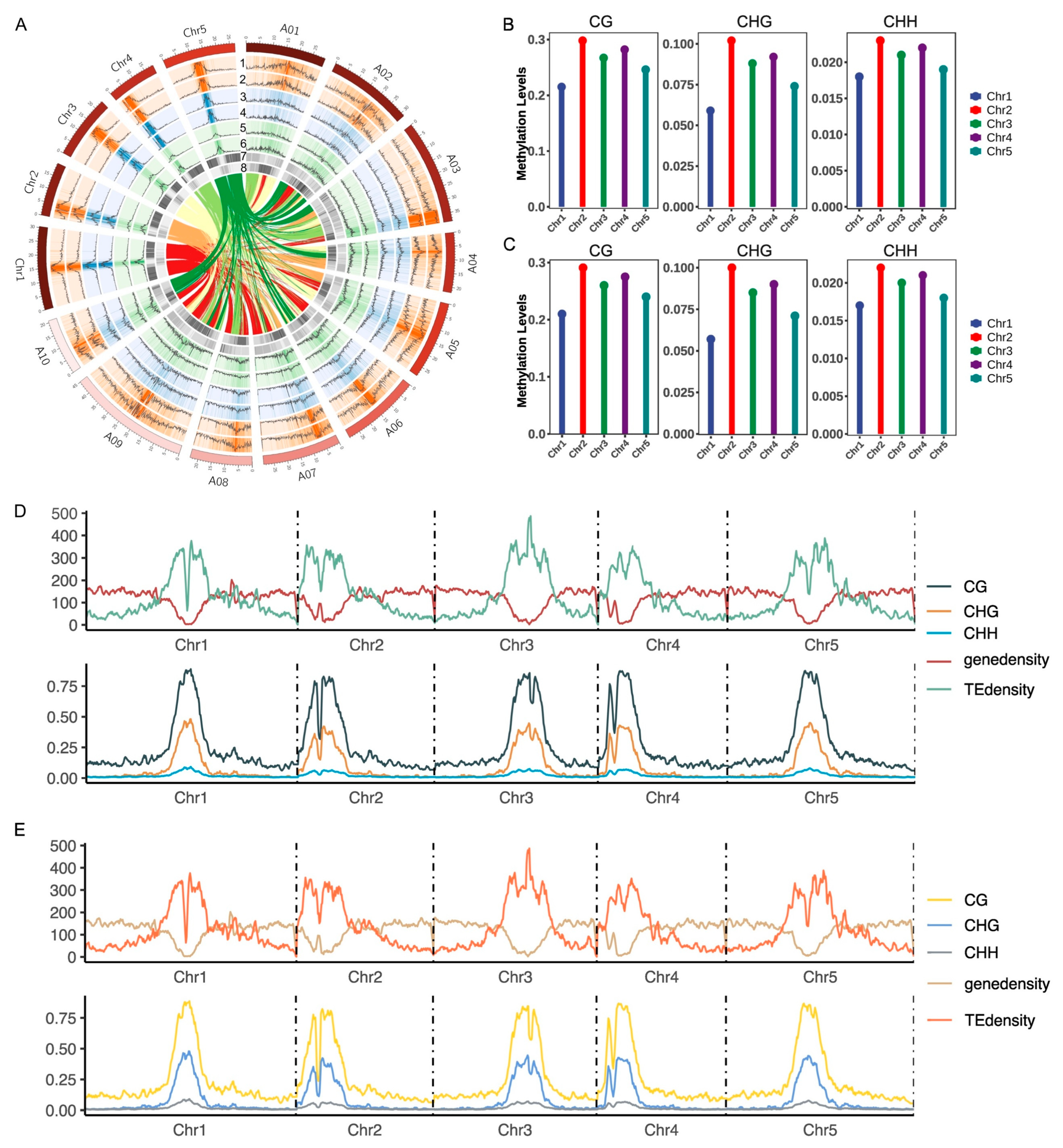
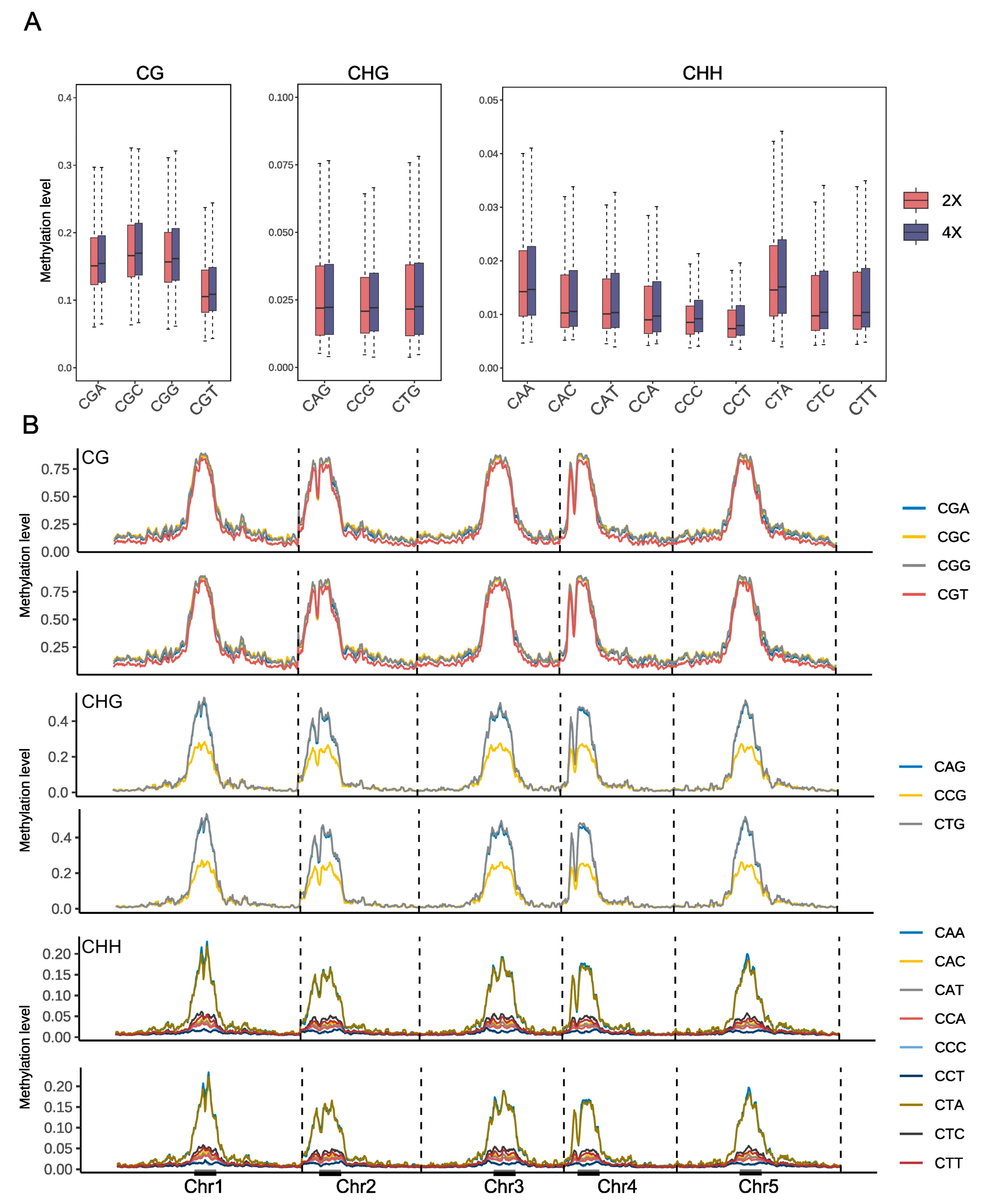
2.1. Genome-Wide DNA Methylation Landscapes in Arabidopsis thaliana After Whole Genome Doubling
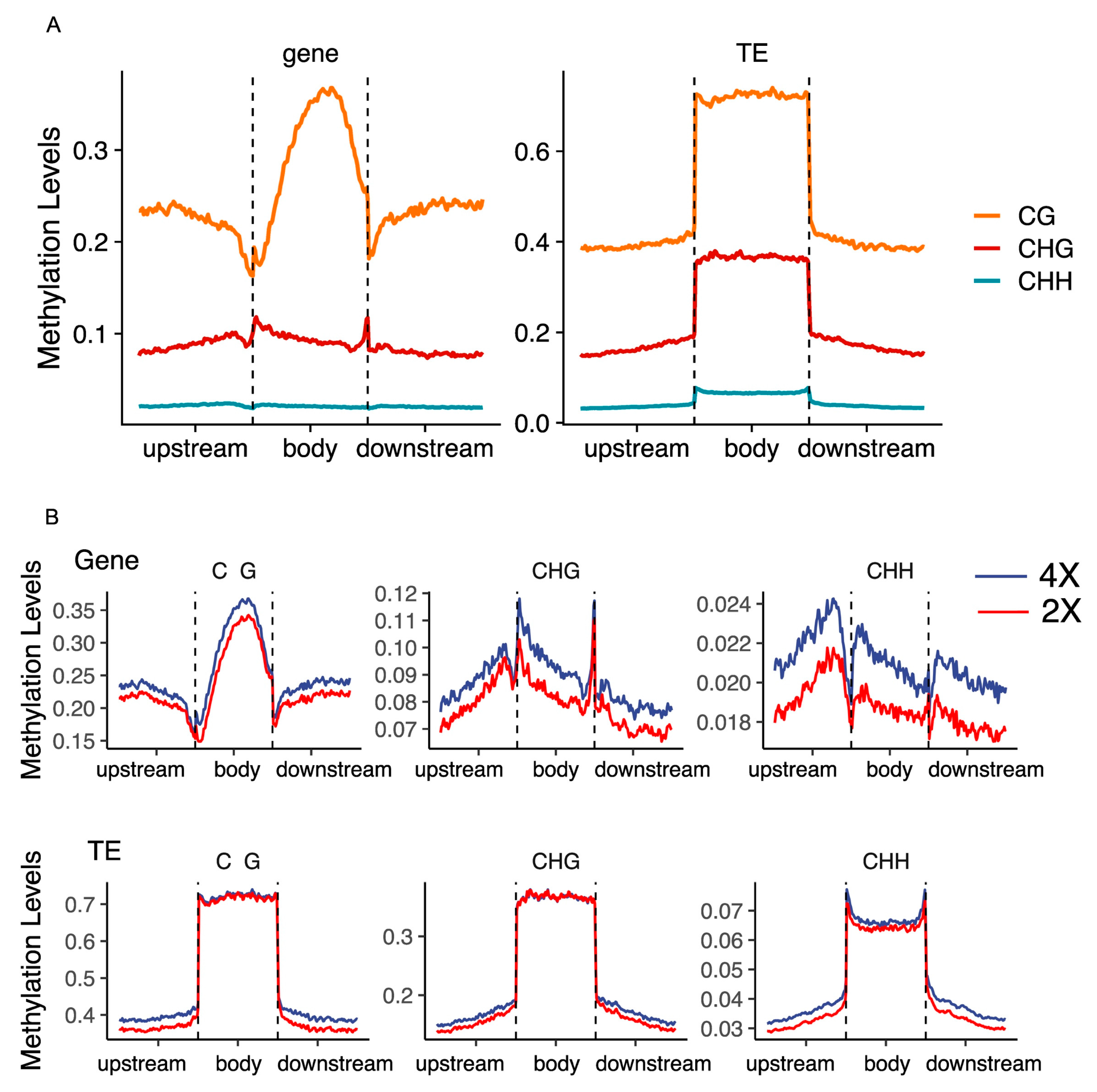
2.2. DNA Methylation Changes upon Genome Duplication Around Gene and TE
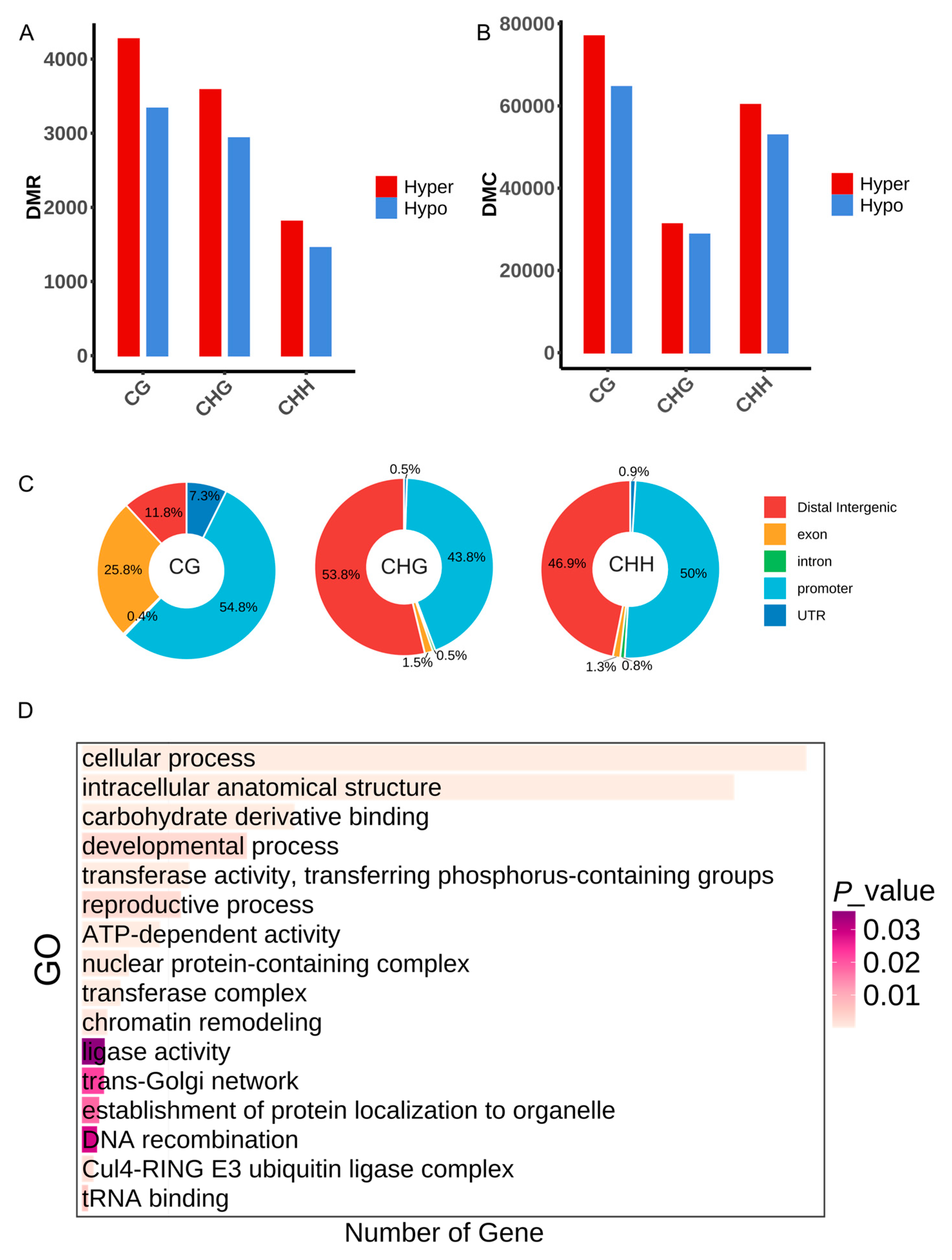
2.3. Characterization of Differential Methylation Sites (DMCs) and Differential Methylation Regions (DMRs) at Diploid and Autotetraploid Arabidopsis
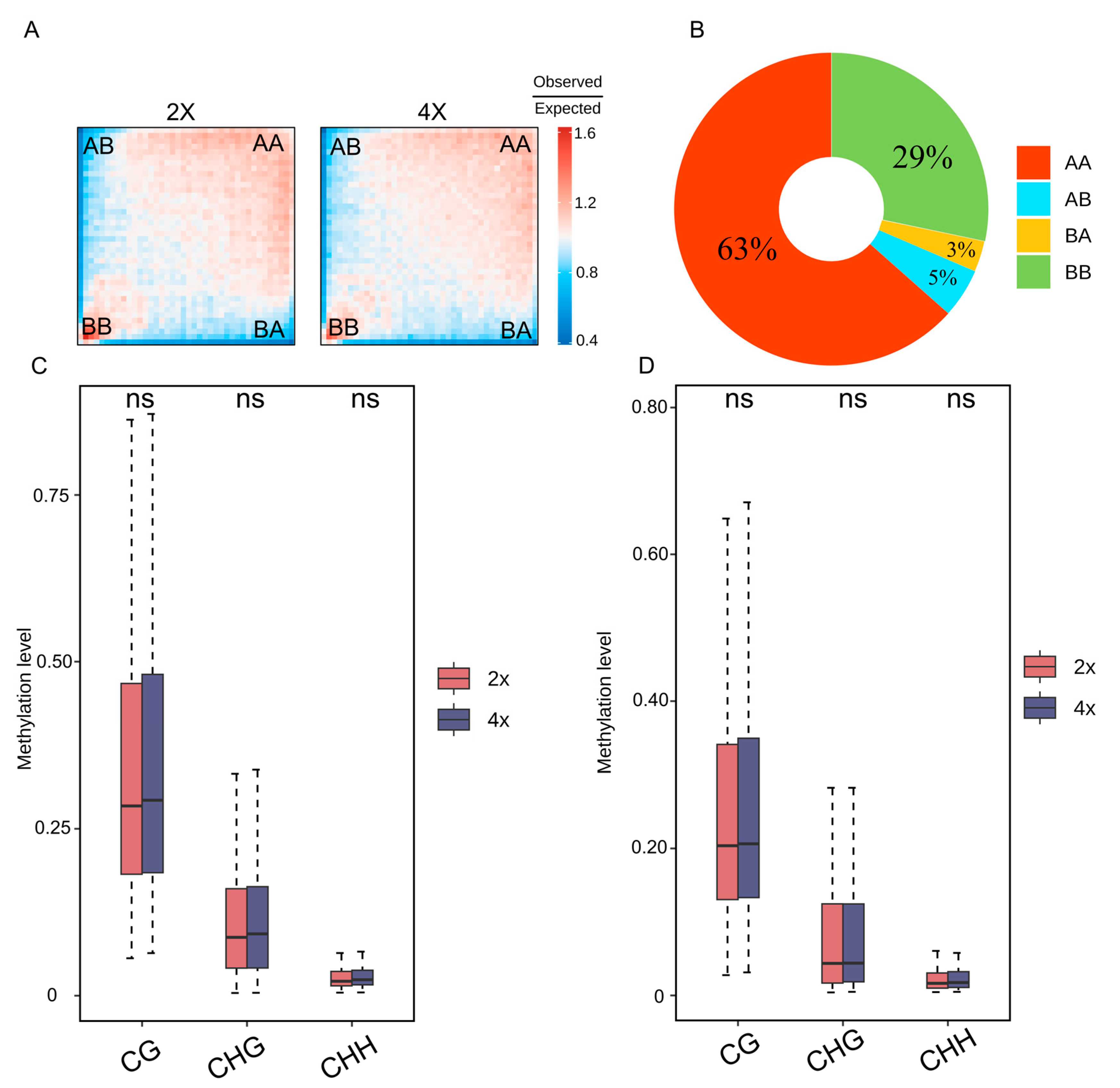
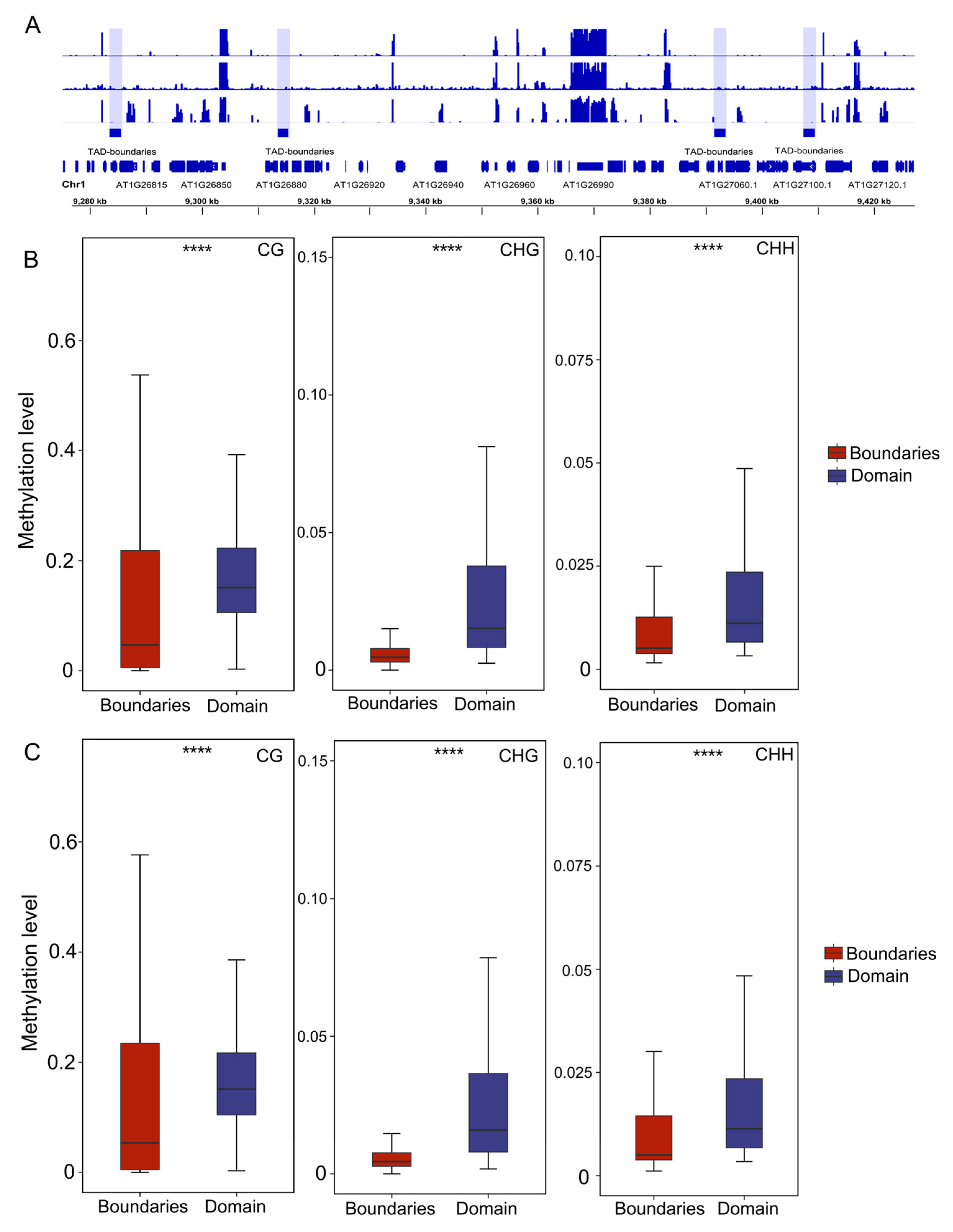
2.4. Methylation Levels Remain Stable Across Varying Compartments and TADs
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. RNA-Seq Analysis
4.3. WGBS and Analysis
4.4. Hi-C Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Hi-C | High-resolution chromosome conformation capture |
| Ara | Arabidopsis thaliana |
| Bra | Brassica rapa |
| 2x | diploid |
| 4x | autotetraploid |
| TAD | topologically associating domain |
| DMGs | Different methylation genes |
References
- Doyle, J.J.; Coate, J.E. Polyploidy, the Nucleotype, and Novelty: The Impact of Genome Doubling on the Biology of the Cell. Int. J. Plant Sci. 2019, 180, 1–52. [Google Scholar] [CrossRef]
- Jiao, Y.N.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.Y.; Soltis, P.S.; et al. Ancestral polyploidy in seed plants and angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Barker, M.S.; Arrigo, N.; Baniaga, A.E.; Li, Z.; Levin, D.A. On the relative abundance of autopolyploids and allopolyploids. New Phytol. 2016, 210, 391–398. [Google Scholar] [CrossRef]
- Corneillie, S.; De Storme, N.; Van Acker, R.; Fangel, J.U.; De Bruyne, M.; De Rycke, R.; Geelen, D.; Willats, W.G.T.; Vanholme, B.; Boerjan, W. Polyploidy Affects Plant Growth and Alters Cell Wall Composition. Plant Physiol. 2019, 179, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zuo, L.; Wei, T.; Zhang, Y.; Zhang, Y.; Ming, R.; Bachar, D.; Xiao, W.; Madiha, K.; Chen, C.; et al. CHH methylation of genes associated with fatty acid and jasmonate biosynthesis contributes to cold tolerance in autotetraploids of Poncirus trifoliata. J. Integr. Plant Biol. 2022, 64, 2327–2343. [Google Scholar] [CrossRef]
- Sun, Z.F.; Wang, Y.L.; Song, Z.J.; Zhang, H.; Wang, Y.D.; Liu, K.P.; Ma, M.; Wang, P.; Fang, Y.P.; Cai, D.T.; et al. DNA methylation in transposable elements buffers the connection between three-dimensional chromatin organization and gene transcription upon rice genome duplication. J. Adv. Res. 2022, 42, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Zhang, M.; Wang, T.T.; Duan, Y.Y.; Ren, J.; Gao, H.; Fan, Y.J.; Xia, Q.M.; Cao, H.X.; Xie, K.D.; et al. Polyploidization leads to salt stress resilience via ethylene signaling in citrus plants. New Phytol. 2025, 246, 176–191. [Google Scholar] [CrossRef]
- Song, X.M.; Wang, J.P.; Sun, P.C.; Ma, X.; Yang, Q.H.; Hu, J.J.; Sun, S.R.; Li, Y.X.; Yu, J.G.; Feng, S.Y.; et al. Preferential gene retention increases the robustness of cold regulation in Brassicaceae and other plants after polyploidization. Hortic. Res. 2020, 7, 20. [Google Scholar] [CrossRef]
- Zhang, H.; Zheng, R.; Wang, Y.; Zhang, Y.; Hong, P.; Fang, Y.; Li, G.; Fang, Y. The effects of Arabidopsis genome duplication on the chromatin organization and transcriptional regulation. Nucleic Acids Res. 2019, 47, 7857–7869. [Google Scholar] [CrossRef]
- Stroud, H.; Do, T.; Du, J.M.; Zhong, X.H.; Feng, S.H.; Johnson, L.; Patel, D.J.; Jacobsen, S.E. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 2014, 21, 64–72. [Google Scholar] [CrossRef]
- Liu, P.P.; Liu, R.E.; Xu, Y.P.; Zhang, C.X.; Niu, Q.F.; Lang, Z.B. DNA cytosine methylation dynamics and functional roles in horticultural crops. Hortic. Res. 2023, 10, uhad170. [Google Scholar] [CrossRef]
- Ma, L.; Xing, L.; Li, Z.; Jiang, D. Epigenetic control of plant abiotic stress responses. J. Genet. Genom. 2024, 52, 129–144. [Google Scholar] [CrossRef]
- Tao, X.; Feng, S.; Zhao, T.; Guan, X. Efficient chromatin profiling of H3K4me3 modification in cotton using CUT&Tag. Plant Methods 2020, 16, 120. [Google Scholar] [CrossRef]
- Lu, Z.F.; Hofmeister, B.T.; Vollmers, C.; DuBois, R.M.; Schmitz, R.J. Combining ATAC-seq with nuclei sorting for discovery of cis-regulatory regions in plant genomes. Nucleic Acids Res. 2017, 45, e41. [Google Scholar] [CrossRef]
- Seale, K.; Horvath, S.; Teschendorff, A.; Eynon, N.; Voisin, S. Making sense of the ageing methylome. Nat. Rev. Genet. 2022, 23, 585–605. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.K. Epigenetic gene regulation in plants and its potential applications in crop improvement. Nat. Rev. Mol. Cell Biol. 2024, 26, 51–67. [Google Scholar] [CrossRef]
- Liu, R.; Lang, Z.B. The mechanism and function of active DNA demethylation in plants. J. Integr. Plant Biol. 2020, 62, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Tong, W.; Li, R.P.; Huang, J.; Zhao, H.J.; Ge, R.H.; Wu, Q.; Mallano, A.I.; Wang, Y.L.; Li, F.D.; Deng, W.W.; et al. Divergent DNA methylation contributes to duplicated gene evolution and chilling response in tea plants. Plant J. 2021, 106, 1312–1327. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, H.; Ma, Y.; Gao, B.; Guan, P.; Huang, X.; Ouyang, W.; Guo, M.; Chen, G.; Li, G.; et al. Domains Rearranged Methylase 2 maintains DNA methylation at large DNA hypomethylated shores and long-range chromatin interactions in rice. Plant Biotechnol. J. 2023, 21, 2333–2347. [Google Scholar] [CrossRef] [PubMed]
- Zemach, A.; Kim, M.Y.; Hsieh, P.H.; Coleman-Derr, D.; Eshed-Williams, L.; Thao, K.; Harmer, S.L.; Zilberman, D. The Nucleosome Remodeler DDM1 Allows DNA Methyltransferases to Access H1-Containing Heterochromatin. Cell 2013, 153, 193–205. [Google Scholar] [CrossRef]
- Lindroth, A.M.; Cao, X.F.; Jackson, J.P.; Zilberman, D.; McCallum, C.M.; Henikoff, S.; Jacobsen, S.E. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 2001, 292, 2077–2080. [Google Scholar] [CrossRef]
- Jackson, J.P.; Lindroth, A.M.; Cao, X.F.; Jacobsen, S.E. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 2002, 416, 556–560. [Google Scholar] [CrossRef]
- Zhang, H.M.; Lang, Z.B.; Zhu, J.K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef]
- Zhang, H.M.; Deng, X.Y.; Miki, D.; Cutler, S.; La, H.G.; Hou, Y.J.; Oh, J.; Zhu, J.K. Sulfamethazine Suppresses Epigenetic Silencing in by Impairing Folate Synthesis. Plant Cell 2012, 24, 1230–1241. [Google Scholar] [CrossRef]
- Ouyang, W.Z.; Xiong, D.; Li, G.L.; Li, X.W. Unraveling the 3D Genome Architecture in Plants: Present and Future. Mol. Plant 2020, 13, 1676–1693. [Google Scholar] [CrossRef]
- Pontvianne, F.; Carpentier, M.C.; Durut, N.; Pavlistová, V.; Jaske, K.; Schorová, S.; Parrinello, H.; Rohmer, M.; Pikaard, C.S.; Fojtová, M.; et al. Identification of Nucleolus-Associated Chromatin Domains Reveals a Role for the Nucleolus in 3D Organization of the A. thaliana Genome. Cell Rep. 2016, 16, 1574–1587. [Google Scholar] [CrossRef] [PubMed]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Wang, L.; Pan, W. Comparison of Hi-C-Based Scaffolding Tools on Plant Genomes. Genes 2023, 14, 2147. [Google Scholar] [CrossRef] [PubMed]
- Hinnant, T.; Ning, W.; Lechler, T. Compartment specific responses to contractility in the small intestinal epithelium. PLoS Genet. 2024, 20, e1010899. [Google Scholar] [CrossRef]
- Li, Z.; Sun, L.H.; Xu, X.; Liu, Y.T.; He, H.; Deng, X.W. Light control of three-dimensional chromatin organization in soybean. Plant Biotechnol. J. 2024, 22, 2596–2611. [Google Scholar] [CrossRef]
- Crevillen, P.; Sonmez, C.; Wu, Z.; Dean, C. A gene loop containing the floral repressor FLC is disrupted in the early phase of vernalization. EMBO J. 2013, 32, 140–148. [Google Scholar] [CrossRef]
- Li, M.; Wang, R.; Wu, X.; Wang, J. Homoeolog expression bias and expression level dominance (ELD) in four tissues of natural allotetraploid Brassica napus. BMC Genom. 2020, 21, 330. [Google Scholar] [CrossRef]
- Hu, Y.; Xiong, J.; Shalby, N.; Zhuo, C.; Jia, Y.; Yang, Q.Y.; Tu, J. Comparison of dynamic 3D chromatin architecture uncovers heterosis for leaf size in Brassica napus. J. Adv. Res. 2022, 42, 289–301. [Google Scholar] [CrossRef]
- Okhovat, M.; VanCampen, J.; Nevonen, K.A.; Harshman, L.; Li, W.; Layman, C.E.; Ward, S.; Herrera, J.; Wells, J.; Sheng, R.R.; et al. TAD evolutionary and functional characterization reveals diversity in mammalian TAD boundary properties and function. Nat. Commun. 2023, 14, 8111. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Romero-Campero, F.J.; Yang, M.; Baile, F.; Cao, Y.; Shu, J.; Luo, L.; Wang, D.; Sun, S.; Yan, P.; et al. Binding by the Polycomb complex component BMI1 and H2A monoubiquitination shape local and long-range interactions in the Arabidopsis genome. Plant Cell 2023, 35, 2484–2503. [Google Scholar] [CrossRef]
- Ma, M.; Wang, Y.D.; Sun, Z.F.; Zhao, R.Z.; Li, H.H.; Li, X.X.; Zhu, H.F.; Yang, X.D.; Zhang, C.W.; Fang, Y.D. Regulation of transcriptional homeostasis by DNA methylation upon genome duplication in pak choi. Mol. Hortic. 2025, 5, 22. [Google Scholar] [CrossRef]
- Wang, L.; Mai, C.; He, S.; Niu, B.; Jia, G.; Yang, T.; Xu, Y.; Ren, M.; Zhao, X.; Liu, X.; et al. Dynamic 3D chromatin organization and epigenetic regulation of gene expression in peanut nodules. J. Integr. Plant Biol. 2025, 1–19. [Google Scholar] [CrossRef]
- Ma, N.; Li, X.; Ci, D.; Zeng, H.Y.; Zhang, C.; Xie, X.; Zhong, C.; Deng, X.W.; Li, D.; He, H. Chromatin Topological Domains Associate with the Rapid Formation of Tandem Duplicates in Plants. Adv. Sci. 2025, 12, e2408861. [Google Scholar] [CrossRef] [PubMed]
- Soltis, P.S.; Marchant, D.B.; Van de Peer, Y.; Soltis, D.E. Polyploidy and genome evolution in plants. Curr. Opin. Genet. Dev. 2015, 35, 119–125. [Google Scholar] [CrossRef]
- Xiang, D.Q.; Quilichini, T.D.; Liu, Z.Y.; Gao, P.; Pan, Y.L.; Li, Q.; Nilsen, K.T.; Venglat, P.; Esteban, E.; Pasha, A.; et al. The Transcriptional Landscape of Polyploid Wheats and Their Diploid Ancestors during Embryogenesis and Grain Development. Plant Cell 2019, 31, 2888–2911. [Google Scholar] [CrossRef] [PubMed]
- Li, X.D.; Yu, E.; Fan, C.C.; Zhang, C.Y.; Fu, T.D.; Zhou, Y.M. Developmental, cytological and transcriptional analysis of autotetraploid. Planta 2012, 236, 579–596. [Google Scholar] [CrossRef]
- Miao, L.F.; Xu, W.Y.; Liu, Y.H.; Huang, X.Y.; Chen, Z.; Wang, H.F.; Wang, Z.H.; Chen, Y.M.; Song, Q.X.; Zhang, J.; et al. Reshaped DNA methylation cooperating with homoeolog-divergent expression promotes improved root traits in synthesized tetraploid wheat. New Phytol. 2024, 242, 507–523. [Google Scholar] [CrossRef]
- Zhao, T.; Guan, X.Y.; Hu, Y.; Zhang, Z.Q.; Yang, H.; Shi, X.W.; Han, J.; Mei, H.; Wang, L.Y.; Shao, L.; et al. Population-wide DNA methylation polymorphisms at single-nucleotide resolution in 207 cotton accessions reveal epigenomic contributions to complex traits. Cell Res. 2024, 34, 859–872. [Google Scholar] [CrossRef] [PubMed]
- He, X.Y.; Chen, Y.W.; Xia, Y.T.; Hong, X.Y.; You, H.Q.; Zhang, R.; Liang, Z.S.; Cui, Q.; Zhang, S.C.; Zhou, M.; et al. DNA methylation regulates biosynthesis of tanshinones and phenolic acids during growth of Salvia miltiorrhiza. Plant Physiol. 2024, 194, 2086–2100. [Google Scholar] [CrossRef]
- Xie, H.H.; Zheng, Y.D.; Xue, M.Y.; Huang, Y.L.; Qian, D.W.; Zhao, M.L.; Li, J.G. DNA methylation-mediated ROS production contributes to seed abortion in litchi. Mol. Hortic. 2024, 4, 12. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.L.; Wan, Y.T.; Liu, H.N.; Pei, M.S.; He, G.Q.; Guo, D.L. CHH hypermethylation contributes to the early ripening of grapes revealed by DNA methylome landscape of ‘Kyoho’ and its bud mutant. Hortic. Res. 2025, 12, uhae285. [Google Scholar] [CrossRef]
- Yu, H.; Gao, M.; Guo, C.C.; Wang, H.F. Reduced CHH methylation levels reveal a critical role of aging pathway genes in Moso bamboo flowering. Hortic. Plant J. 2025, 11, 1341–1352. [Google Scholar] [CrossRef]
- Lee, J.; Lee, S.; Park, K.; Shin, S.Y.; Frost, J.M.; Hsieh, P.H.; Shin, C.; Fischer, R.L.; Hsieh, T.F.; Choi, Y. Distinct regulatory pathways contribute to dynamic CHH methylation patterns in transposable elements throughout embryogenesis. Front. Plant Sci. 2023, 14, 1204279. [Google Scholar] [CrossRef]
- He, L.; Zhao, C.; Zhang, Q.; Zinta, G.; Wang, D.; Lozano-Duran, R.; Zhu, J.K. Pathway conversion enables a double-lock mechanism to maintain DNA methylation and genome stability. Proc. Natl. Acad. Sci. USA 2021, 118, e2107320118. [Google Scholar] [CrossRef]
- Stevens, T.J.; Lando, D.; Basu, S.; Atkinson, L.P.; Cao, Y.; Lee, S.F.; Leeb, M.; Wohlfahrt, K.J.; Boucher, W.; O’Shaughnessy-Kirwan, A.; et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 2017, 544, 59–64. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, R.; Liang, J.; Cai, X.; Zhang, L.; Guo, H.; Zhang, Z.; Wu, J.; Wang, X. BL-Hi-C reveals the 3D genome structure of Brassica crops with high sensitivity. Hortic. Res. 2024, 11, uhae017. [Google Scholar] [CrossRef]
- Wang, M.; Wang, P.; Lin, M.; Ye, Z.; Li, G.; Tu, L.; Shen, C.; Li, J.; Yang, Q.; Zhang, X. Evolutionary dynamics of 3D genome architecture following polyploidization in cotton. Nat. Plants 2018, 4, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.F.; Jia, G.H.; Jiang, X.Y.; Cao, S.; Chen, Z.J.; Song, Q.X. Altered chromatin architecture and gene expression during polyploidization and domestication of soybean. Plant Cell 2021, 33, 1430–1446. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Feng, S.; Duttke, S.H.; Potok, M.E.; Zhang, Y.; Gallego-Bartolome, J.; Liu, W.; Jacobsen, S.E. DNA methylation-linked chromatin accessibility affects genomic architecture in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118, e2023347118. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Hou, X.; Wang, D.; Cheng, Z.; Wang, Y.; Jiao, Y. A near-complete assembly of an Arabidopsis thaliana genome. Mol. Plant 2022, 15, 1247–1250. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Kumar, L.; Futschik, M.E. Mfuzz: A software package for soft clustering of microarray data. Bioinformation 2007, 2, 5–7. [Google Scholar] [CrossRef]
- Krueger, F.; Andrews, S.R. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011, 27, 1571–1572. [Google Scholar] [CrossRef]
- Akalin, A.; Kormaksson, M.; Li, S.; Garrett-Bakelman, F.E.; Figueroa, M.E.; Melnick, A.; Mason, C.E. methylKit: A comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012, 13, R87. [Google Scholar] [CrossRef] [PubMed]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Wolff, J.; Bhardwaj, V.; Nothjunge, S.; Richard, G.; Renschler, G.; Gilsbach, R.; Manke, T.; Backofen, R.; Ramirez, F.; Gruning, B.A. Galaxy HiCExplorer: A web server for reproducible Hi-C data analysis, quality control and visualization. Nucleic Acids Res. 2018, 46, W11–W16. [Google Scholar] [CrossRef]
- van der Weide, R.H.; van den Brand, T.; Haarhuis, J.H.I.; Teunissen, H.; Rowland, B.D.; de Wit, E. Hi-C analyses with GENOVA: A case study with cohesin variants. Nar. Genom. Bioinform. 2021, 3, lqab040. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Cresswell, K.G.; Dozmorov, M.G. TADCompare: An R Package for Differential and Temporal Analysis of Topologically Associated Domains. Front. Genet. 2020, 11, 158. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Su, Y.; Chang, L.; Jiao, G.; Ou, Y.; Yang, M.; Xu, C.; Liu, P.; Wang, Z.; Qi, Z.; et al. Increased long-distance and homo-trans interactions related to H3K27me3 in Arabidopsis hybrids. J. Integr. Plant Biol. 2024, 66, 208–227. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, R.; Ni, Z.; Zhang, D.; Fang, Y. The Association Between DNA Methylation and Three-Dimensional Genome During Whole Genome Doubling in Arabidopsis thaliana. Plants 2025, 14, 2959. https://doi.org/10.3390/plants14192959
Zhao R, Ni Z, Zhang D, Fang Y. The Association Between DNA Methylation and Three-Dimensional Genome During Whole Genome Doubling in Arabidopsis thaliana. Plants. 2025; 14(19):2959. https://doi.org/10.3390/plants14192959
Chicago/Turabian StyleZhao, Ranze, Zhongqiu Ni, Dingyu Zhang, and Yuda Fang. 2025. "The Association Between DNA Methylation and Three-Dimensional Genome During Whole Genome Doubling in Arabidopsis thaliana" Plants 14, no. 19: 2959. https://doi.org/10.3390/plants14192959
APA StyleZhao, R., Ni, Z., Zhang, D., & Fang, Y. (2025). The Association Between DNA Methylation and Three-Dimensional Genome During Whole Genome Doubling in Arabidopsis thaliana. Plants, 14(19), 2959. https://doi.org/10.3390/plants14192959





