Integrative Machine Learning Approaches for Identifying Loci Associated with Anthracnose Resistance in Strawberry
Abstract
1. Introduction
2. Results
2.1. Model Performance with Increasing Feature Count
2.2. Data Augmentation Drives Performance Gains in Non-Linear Genomic Prediction Algorithms
2.3. Impact of Data Augmentation on Model Performance Using a Minimal Set of in Formative Genotypes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Population Development
4.2. Genotyping Analysis
4.3. Pathogenicity Assay and Phenotypic Evaluation
4.4. Feature Selection
4.5. Data Preprocessing and Augmentation
4.6. Genomic Selection Model Training and Evaluation
4.7. Comparative Evaluation of Predictive Models Using Resistance-Associated SNP Subsets
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| CNN | convolutional neural network |
| FAOSTAT | Food and Agriculture Organization Corporate Statistical Database |
| G-BLUP | genomic best linear unbiased prediction |
| GBM | gradient boosting machine |
| GEBV | genomic estimated breeding value |
| GP | genomic prediction |
| GWAS | genome-wide association study |
| LASSO | least absolute shrinkage and selection operator |
| ML | machine learning |
| RF | random forest |
| RNN | recurrent neural network |
| SNP | single-nucleotide polymorphism |
| AUDPC | area under the disease progress curve |
| SVM | support vector machine |
References
- Charoenwoodhipong, P.; Zuelch, M.L.; Keen, C.L.; Hackman, R.M.; Holt, R.R. Strawberry (Fragaria × Ananassa) intake on human health and disease outcomes: A comprehensive literature review. Crit. Rev. Food Sci. Nutr. 2024, 65, 4884–4914. [Google Scholar] [CrossRef]
- Whitaker, V.M.; Knapp, S.J.; Hardigan, M.A.; Edger, P.P.; Slovin, J.P.; V Bassil, N.; Hytönen, T.; Mackenzie, K.K.; Lee, S.; Jung, S. A roadmap for research in octoploid strawberry. Hortic. Res. 2020, 7, 33. [Google Scholar] [CrossRef]
- Edger, P.P.; Poorten, T.J.; VanBuren, R.; Hardigan, M.A.; Colle, M.; McKain, M.R.; Smith, R.D.; Teresi, S.J.; Nelson, A.D.; Wai, C.M. Origin and evolution of the octoploid strawberry genome. Nat. Genet. 2019, 51, 541–547. [Google Scholar] [CrossRef]
- FAO. FAOSTAT Statistical Database; Food and Agriculture Organization of the United Nations: Rome, Italy, 2023. Available online: https://www.fao.org/faostat/en/ (accessed on 15 September 2025).
- Hu, S.; Zhang, Y.; Yu, H.; Zhou, J.; Hu, M.; Liu, A.; Wu, J.; Wang, H.; Zhang, C. Colletotrichum Spp. diversity between leaf anthracnose and crown rot from the same strawberry plant. Front. Microbiol. 2022, 13, 860694. [Google Scholar] [CrossRef]
- Peres, N.; Timmer, L.; Adaskaveg, J.; Correll, J. Lifestyles of Colletotrichum acutatum. Plant Dis. 2005, 89, 784–796. [Google Scholar] [CrossRef]
- Ozbudak, E.; Carrillo-Tarazona, Y.; Diaz, E.A.; Zambon, F.T.; Rossi, L.; Peres, N.A.; Raffaele, S.; Cano, L.M. Transcriptome analysis of Colletotrichum nymphaeae-Strawberry interaction reveals in planta expressed genes associated with virulence. Front. Plant Sci. 2025, 15, 1390926. [Google Scholar] [CrossRef]
- Morkeliūnė, A.; Rasiukevičiūtė, N.; Frercks, B.; Bendokas, V.; Antanynienė, R.; Mažeikienė, I.; Vaštakaitė-Kairienė, V.; Karklelienė, R.; Valiuškaitė, A. Evaluation of Strawberry Colletotrichum spp. Genetic Diversity in Lithuania. Agronomy 2025, 15, 720. [Google Scholar] [CrossRef]
- Howard, C.M.; Maas, J.; Chandler, C.; Albregts, E. Anthracnose of strawberry caused by the Colletotrichum complex in Florida. Plant Dis. 1992, 76, 976–981. [Google Scholar] [CrossRef]
- Forcelini, B.B.; Gonçalves, F.P.; Peres, N.A. Effect of inoculum concentration and interrupted wetness duration on the development of anthracnose fruit rot of strawberry. Plant Dis. 2017, 101, 372–377. [Google Scholar] [CrossRef]
- Zhang, L.; Huang, X.; He, C.; Zhang, Q.-Y.; Zou, X.; Duan, K.; Gao, Q. Novel fungal pathogenicity and leaf defense strategies are revealed by simultaneous transcriptome analysis of Colletotrichum fructicola and strawberry infected by this fungus. Front. Plant Sci. 2018, 9, 434. [Google Scholar] [CrossRef]
- Mertely, J.C.; Peres, N.A. Anthracnose Fruit Rot of Strawberry: PP-207/PP130, Rev. 9/2012; Edis, University of Florida, IFAS Extension: Gainesville, FL, USA, 2012; Available online: https://edis.ifas.ufl.edu/publication/PP130 (accessed on 30 July 2025).
- Salinas, N.; Verma, S.; Peres, N.; Whitaker, V.M. FaRCa1: A major subgenome-specific locus conferring resistance to Colletotrichum acutatum in strawberry. Theor. Appl. Genet. 2019, 132, 1109–1120. [Google Scholar] [CrossRef]
- Han, Y.; Zeng, X.; Xiang, F.; Ren, L.; Chen, F.; Gu, Y. Distribution and characteristics of Colletotrichum spp. associated with anthracnose of strawberry in Hubei, China. Plant Dis. 2016, 100, 996–1006. [Google Scholar] [CrossRef]
- Sleper, J.; Tapia, R.; Lee, S.; Whitaker, V. Within-family genomic selection in strawberry: Optimization of marker density, trial design, and training set composition. Plant Genome 2025, 18, e20550. [Google Scholar] [CrossRef]
- Meuwissen, T.H.; Hayes, B.J.; Goddard, M. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- Goddard, M.; Hayes, B. Genomic selection. J. Anim. Breed. Genet. 2007, 124, 323–330. [Google Scholar] [CrossRef]
- Bernardo, R. Molecular markers and selection for complex traits in plants: Learning from the last 20 years. Crop Sci. 2008, 48, 1649–1664. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, X.; Fu, J.; Wang, H.; Wang, J.; Huang, C.; Prasanna, B.M.; Olsen, M.S.; Wang, G.; Zhang, A. Enhancing genetic gain through genomic selection: From livestock to plants. Plant Commun. 2020, 1, 100005. [Google Scholar] [CrossRef]
- Farooq, M.; van Dijk, A.D.; Nijveen, H.; Mansoor, S.; de Ridder, D. Genomic prediction in plants: Opportunities for ensemble machine learning based approaches. F1000Research 2023, 11, 802. [Google Scholar] [CrossRef]
- Scheben, A.; Yuan, Y.; Edwards, D. Advances in genomics for adapting crops to climate change. Curr. Plant Biol. 2016, 6, 2–10. [Google Scholar] [CrossRef]
- Sandhu, K.S.; Shiv, A.; Kaur, G.; Meena, M.R.; Raja, A.K.; Vengavasi, K.; Mall, A.K.; Kumar, S.; Singh, P.K.; Singh, J. Integrated approach in genomic selection to accelerate genetic gain in sugarcane. Plants 2022, 11, 2139. [Google Scholar] [CrossRef]
- Gianola, D. Priors in whole-genome regression: The Bayesian alphabet returns. Genetics 2013, 194, 573–596. [Google Scholar] [CrossRef]
- Crossa, J.; Pérez-Rodríguez, P.; Cuevas, J.; Montesinos-López, O.; Jarquín, D.; De Los Campos, G.; Burgueño, J.; González-Camacho, J.M.; Pérez-Elizalde, S.; Beyene, Y. Genomic selection in plant breeding: Methods, models, and perspectives. Trends Plant Sci. 2017, 22, 961–975. [Google Scholar] [CrossRef]
- Habier, D.; Fernando, R.L.; Dekkers, J. The impact of genetic relationship information on genome-assisted breeding values. Genetics 2007, 177, 2389–2397. [Google Scholar] [CrossRef]
- Endelman, J.B. Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome 2011, 4, 250–255. [Google Scholar] [CrossRef]
- Pérez, P.; de Los Campos, G. Genome-wide regression and prediction with the BGLR statistical package. Genetics 2014, 198, 483–495. [Google Scholar] [CrossRef]
- González-Recio, O.; Forni, S. Genome-wide prediction of discrete traits using Bayesian regressions and machine learning. Genet. Sel. Evol. 2011, 43, 7. [Google Scholar] [CrossRef]
- Montesinos-López, A.; Montesinos-López, O.A.; Gianola, D.; Crossa, J.; Hernández-Suárez, C.M. Multi-environment genomic prediction of plant traits using deep learners with dense architecture. G3 Genes Genomes Genet. 2018, 8, 3813–3828. [Google Scholar] [CrossRef]
- Yoosefzadeh Najafabadi, M.; Hesami, M.; Eskandari, M. Machine learning-assisted approaches in modernized plant breeding programs. Genes 2023, 14, 777. [Google Scholar] [CrossRef]
- Montesinos-López, O.A.; Montesinos-López, A.; Pérez-Rodríguez, P.; Barrón-López, J.A.; Martini, J.W.; Fajardo-Flores, S.B.; Gaytan-Lugo, L.S.; Santana-Mancilla, P.C.; Crossa, J. A review of deep learning applications for genomic selection. BMC Genom. 2021, 22, 19. [Google Scholar] [CrossRef]
- Yamamoto, E.; Kataoka, S.; Shirasawa, K.; Noguchi, Y.; Isobe, S. Genomic selection for F1 hybrid breeding in strawberry (Fragaria × ananassa). Front. Plant Sci. 2021, 12, 645111. [Google Scholar] [CrossRef]
- Dalid, C.; Zheng, C.; Osorio, L.; Verma, S.; Abd-Elrahman, A.; Wang, X.; Whitaker, V.M. Genetic analysis of predicted vegetative biomass and biomass-related traits from digital phenotyping of strawberry. Plant Genome 2025, 18, e70018. [Google Scholar] [CrossRef]
- Chen, X.; Dai, D.; Zhao, S.; Shen, Y.; Wang, H.; Zhang, C. Genetic diversity of Colletotrichum spp. causing strawberry anthracnose in Zhejiang, China. Plant Dis. 2020, 104, 1351–1357. [Google Scholar] [CrossRef]
- Salinas, N.; Fan, Z.; Peres, N.; Lee, S.; Whitaker, V.M. FaRCa1 confers moderate resistance to the root necrosis form of strawberry anthracnose caused by Colletotrichum acutatum. HortScience 2020, 55, 693–698. [Google Scholar] [CrossRef]
- Azodi, C.B.; Bolger, E.; McCarren, A.; Roantree, M.; de Los Campos, G.; Shiu, S.-H. Benchmarking parametric and machine learning models for genomic prediction of complex traits. G3 Genes Genomes Genet. 2019, 9, 3691–3702. [Google Scholar] [CrossRef]
- Arruda, M.; Lipka, A.; Brown, P.; Krill, A.; Thurber, C.; Brown-Guedira, G.; Dong, Y.; Foresman, B.; Kolb, F. Comparing genomic selection and marker-assisted selection for Fusarium head blight resistance in wheat (Triticum aestivum L.). Mol. Breed. 2016, 36, 84. [Google Scholar] [CrossRef]
- Cerrudo, D.; Cao, S.; Yuan, Y.; Martinez, C.; Suarez, E.A.; Babu, R.; Zhang, X.; Trachsel, S. Genomic selection outperforms marker assisted selection for grain yield and physiological traits in a maize doubled haploid population across water treatments. Front. Plant Sci. 2018, 9, 366. [Google Scholar] [CrossRef]
- Buerstmayr, H.; Ban, T.; Anderson, J.A. QTL mapping and marker-assisted selection for Fusarium head blight resistance in wheat: A review. Plant Breed. 2009, 128, 1–26. [Google Scholar] [CrossRef]
- Steiner, B.; Michel, S.; Maccaferri, M.; Lemmens, M.; Tuberosa, R.; Buerstmayr, H. Exploring and exploiting the genetic variation of Fusarium head blight resistance for genomic-assisted breeding in the elite durum wheat gene pool. Theor. Appl. Genet. 2019, 132, 969–988. [Google Scholar] [CrossRef]
- Brauner, P.C.; Melchinger, A.E.; Schrag, T.A.; Utz, H.F.; Schipprack, W.; Kessel, B.; Ouzunova, M.; Miedaner, T. Low validation rate of quantitative trait loci for Gibberella ear rot resistance in European maize. Theor. Appl. Genet. 2017, 130, 175–186. [Google Scholar] [CrossRef]
- Thulasinathan, T.; Ayyenar, B.; Kambale, R.; Manickam, S.; Chellappan, G.; Shanmugavel, P.; Narayanan, M.B.; Swaminathan, M.; Muthurajan, R. Marker assisted introgression of resistance genes and phenotypic evaluation enabled identification of durable and broad-spectrum blast resistance in elite rice cultivar, CO 51. Genes 2023, 14, 719. [Google Scholar] [CrossRef]
- Thakur, S.; Singh, P.K.; Das, A.; Rathour, R.; Variar, M.; Prashanthi, S.; Singh, A.; Singh, U.; Chand, D.; Singh, N. Extensive sequence variation in rice blast resistance gene Pi54 makes it broad spectrum in nature. Front. Plant Sci. 2015, 6, 345. [Google Scholar] [CrossRef]
- Mir, Z.A.; Chandra, T.; Saharan, A.; Budhlakoti, N.; Mishra, D.; Saharan, M.; Mir, R.R.; Singh, A.K.; Sharma, S.; Vikas, V. Recent advances on genome-wide association studies (GWAS) and genomic selection (GS); prospects for Fusarium head blight research in Durum wheat. Mol. Biol. Rep. 2023, 50, 3885–3901. [Google Scholar] [CrossRef]
- Budhlakoti, N.; Kushwaha, A.K.; Rai, A.; Chaturvedi, K.; Kumar, A.; Pradhan, A.K.; Kumar, U.; Kumar, R.R.; Juliana, P.; Mishra, D. Genomic selection: A tool for accelerating the efficiency of molecular breeding for development of climate-resilient crops. Front. Genet. 2022, 13, 832153. [Google Scholar] [CrossRef]
- Shikha, K.; Shahi, J.; Vinayan, M.; Zaidi, P.; Singh, A.; Sinha, B. Genome-wide association mapping in maize: Status and prospects. 3 Biotech 2021, 11, 244. [Google Scholar] [CrossRef]
- Xu, Y.; Ma, K.; Zhao, Y.; Wang, X.; Zhou, K.; Yu, G.; Li, C.; Li, P.; Yang, Z.; Xu, C. Genomic selection: A breakthrough technology in rice breeding. Crop J. 2021, 9, 669–677. [Google Scholar] [CrossRef]
- Bernal Rubio, Y.L.; Gualdron Duarte, J.L.; Bates, R.; Ernst, C.; Nonneman, D.; Rohrer, G.; King, A.; Shackelford, S.; Wheeler, T.; Cantet, R.J.C. Meta-analysis of genome-wide association from genomic prediction models. Anim. Genet. 2016, 47, 36–48. [Google Scholar] [CrossRef]
- Upadhyaya, S.R.; Danilevicz, M.F.; Dolatabadian, A.; Neik, T.X.; Zhang, F.; Al-Mamun, H.A.; Bennamoun, M.; Batley, J.; Edwards, D. Genomics-based plant disease resistance prediction using machine learning. Plant Pathol. 2024, 73, 2298–2309. [Google Scholar] [CrossRef]
- Xu, Y.; Crouch, J.H. Marker-assisted selection in plant breeding: From publications to practice. Crop Sci. 2008, 48, 391–407. [Google Scholar] [CrossRef]
- Wang, X.; Xu, Y.; Hu, Z.; Xu, C. Genomic selection methods for crop improvement: Current status and prospects. Crop J. 2018, 6, 330–340. [Google Scholar] [CrossRef]
- Montesinos-López, O.A.; Martín-Vallejo, J.; Crossa, J.; Gianola, D.; Hernández-Suárez, C.M.; Montesinos-López, A.; Juliana, P.; Singh, R. New deep learning genomic-based prediction model for multiple traits with binary, ordinal, and continuous phenotypes. G3 Genes Genomes Genet. 2019, 9, 1545–1556. [Google Scholar] [CrossRef]
- Petrasch, S.; Mesquida-Pesci, S.D.; Pincot, D.D.; Feldmann, M.J.; Lopez, C.M.; Famula, R.; Hardigan, M.A.; Cole, G.S.; Knapp, S.J.; Blanco-Ulate, B. Genomic prediction of strawberry resistance to postharvest fruit decay caused by the fungal pathogen Botrytis cinerea. G3 2022, 12, jkab378. [Google Scholar] [CrossRef]
- Tapia, R.; Abd-Elrahman, A.; Osorio, L.; Whitaker, V.M.; Lee, S. Combining canopy reflectance spectrometry and genome-wide prediction to increase response to selection for powdery mildew resistance in cultivated strawberry. J. Exp. Bot. 2022, 73, 5322–5335. [Google Scholar] [CrossRef]
- Zingaretti, L.M.; Gezan, S.A.; Ferrão, L.F.V.; Osorio, L.F.; Monfort, A.; Muñoz, P.R.; Whitaker, V.M. Exploring Deep Learning for Complex Trait Genomic Prediction in Polyploid Outcrossing Species. Front. Plant Sci. 2020, 11, 25. [Google Scholar] [CrossRef]
- Sleper, J.A.; Azevedo, C.F.; Ferrão, L.F.; Whitaker, V.M. Using Genomic Selection to Examine Subgenome Dominance and Epistasis in Allopolyploid Strawberry. Res. Sq. Prepr. 2025. [Google Scholar] [CrossRef]
- Yeon, J.; Nguyen, T.T.P.; Kim, M.; Sim, S.-C. Prediction Accuracy of Genomic Estimated Breeding Values for Fruit Traits in Cultivated Tomato (Solanum lycopersicum L.). BMC Plant Biol. 2024, 24, 4934. [Google Scholar] [CrossRef]
- Han, K.; Ahn, H.-I.; Yang, H.-B.; Lee, Y.-R.; Lee, E.-S.; Lee, J.; Jang, C.-S.; Kim, D.-S. Identification of Genetic Loci Associated with Bolting Time in Radish (Raphanus sativus L.) by QTL Mapping and GWAS. Agronomy 2024, 14, 2700. [Google Scholar] [CrossRef]
- Hardigan, M.A.; Feldmann, M.J.; Lorant, A.; Bird, K.A.; Famula, R.; Acharya, C.; Cole, G.; Edger, P.P.; Knapp, S.J. Genome synteny has been conserved among the octoploid progenitors of cultivated strawberry over millions of years of evolution. Front. Plant Sci. 2020, 10, 1789. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Cox, D.R. Analysis of Binary Data; Routledge: Oxfordshire, UK, 2018. [Google Scholar]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Sirsat, M.S.; Oblessuc, P.R.; Ramiro, R.S. Genomic prediction of wheat grain yield using machine learning. Agriculture 2022, 12, 1406. [Google Scholar] [CrossRef]

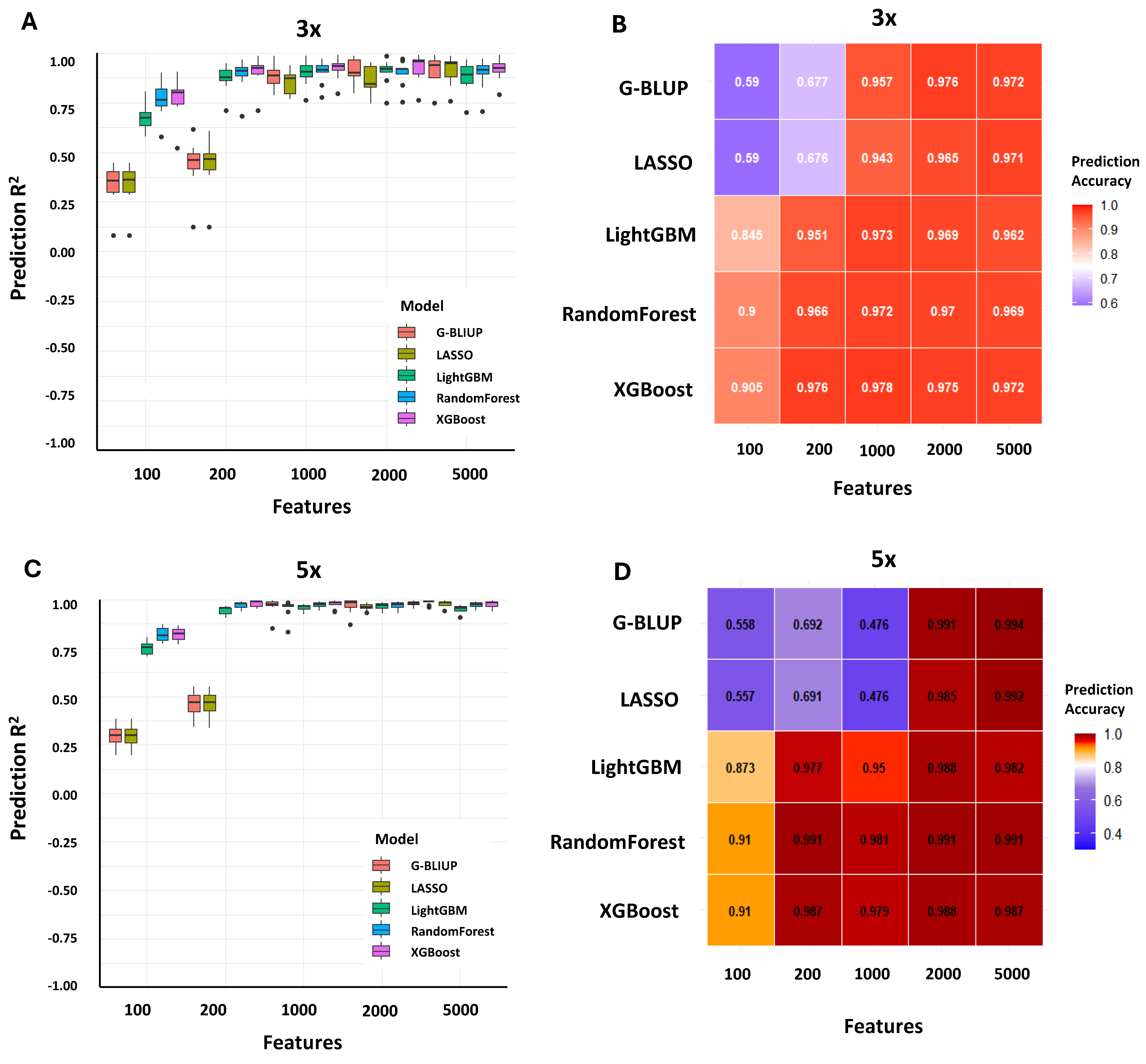

| Number of Features | Best Model | R2 a | RMSE b | MSE c | MAE d |
|---|---|---|---|---|---|
| 100 | XGBoost | 0.8724 | 2.937 | 8.626 | 1.29 |
| 200 | XGBoost | 0.9933 | 0.671 | 0.45 | 0.141 |
| 1000 | XGBoost | 0.9944 | 0.614 | 0.377 | 0.091 |
| 2000 | XGBoost | 0.9944 | 0.614 | 0.377 | 0.091 |
| 5000 | XGBoost | 0.9944 | 0.614 | 0.377 | 0.091 |
| Features | Augmentation | Best Model | R2 a | RMSE b | MSE c | MAE d |
|---|---|---|---|---|---|---|
| 100 | 3× | XGBoost | 0.8824 | 2.8403 | 8.0673 | 1.3259 |
| 100 | 5× | Random Forest | 0.8586 | 3.2169 | 10.3487 | 1.369 |
| 200 | 3× | XGBoost | 0.9887 | 0.8821 | 0.7781 | 0.2187 |
| 200 | 5× | XGBoost | 0.9918 | 0.774 | 0.599 | 0.1634 |
| 1000 | 3× | XGBoost | 0.9914 | 0.7664 | 0.5874 | 0.1313 |
| 1000 | 5× | XGBoost | 0.9936 | 0.6818 | 0.4649 | 0.102 |
| 2000 | 3× | XGBoost | 0.991 | 0.766 | 0.588 | 0.131 |
| 2000 | 5× | G-BLUP | 0.9937 | 0.6812 | 0.4641 | 0.1294 |
| 5000 | 3× | G-BLUP | 0.992 | 0.765 | 0.585 | 0.17 |
| 5000 | 5× | G-BLUP | 0.9937 | 0.6812 | 0.4641 | 0.1294 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jang, Y.J.; Yun, D.; Shin, W.; Goo, C.; Song, C.M.; Han, K.; Kim, S.; Kim, D.-S.; Lee, S.; Oh, Y. Integrative Machine Learning Approaches for Identifying Loci Associated with Anthracnose Resistance in Strawberry. Plants 2025, 14, 2889. https://doi.org/10.3390/plants14182889
Jang YJ, Yun D, Shin W, Goo C, Song CM, Han K, Kim S, Kim D-S, Lee S, Oh Y. Integrative Machine Learning Approaches for Identifying Loci Associated with Anthracnose Resistance in Strawberry. Plants. 2025; 14(18):2889. https://doi.org/10.3390/plants14182889
Chicago/Turabian StyleJang, Yoon Jeong, Dabin Yun, Wonyoung Shin, Changrim Goo, Chul Min Song, Koeun Han, Seolah Kim, Do-Sun Kim, Seonghee Lee, and Youngjae Oh. 2025. "Integrative Machine Learning Approaches for Identifying Loci Associated with Anthracnose Resistance in Strawberry" Plants 14, no. 18: 2889. https://doi.org/10.3390/plants14182889
APA StyleJang, Y. J., Yun, D., Shin, W., Goo, C., Song, C. M., Han, K., Kim, S., Kim, D.-S., Lee, S., & Oh, Y. (2025). Integrative Machine Learning Approaches for Identifying Loci Associated with Anthracnose Resistance in Strawberry. Plants, 14(18), 2889. https://doi.org/10.3390/plants14182889






