Changes in Cell Aggregation in Arabidopsis thaliana Suspension Culture Following Knockout of GAUT Gene Family Members
Abstract
1. Introduction
2. Results
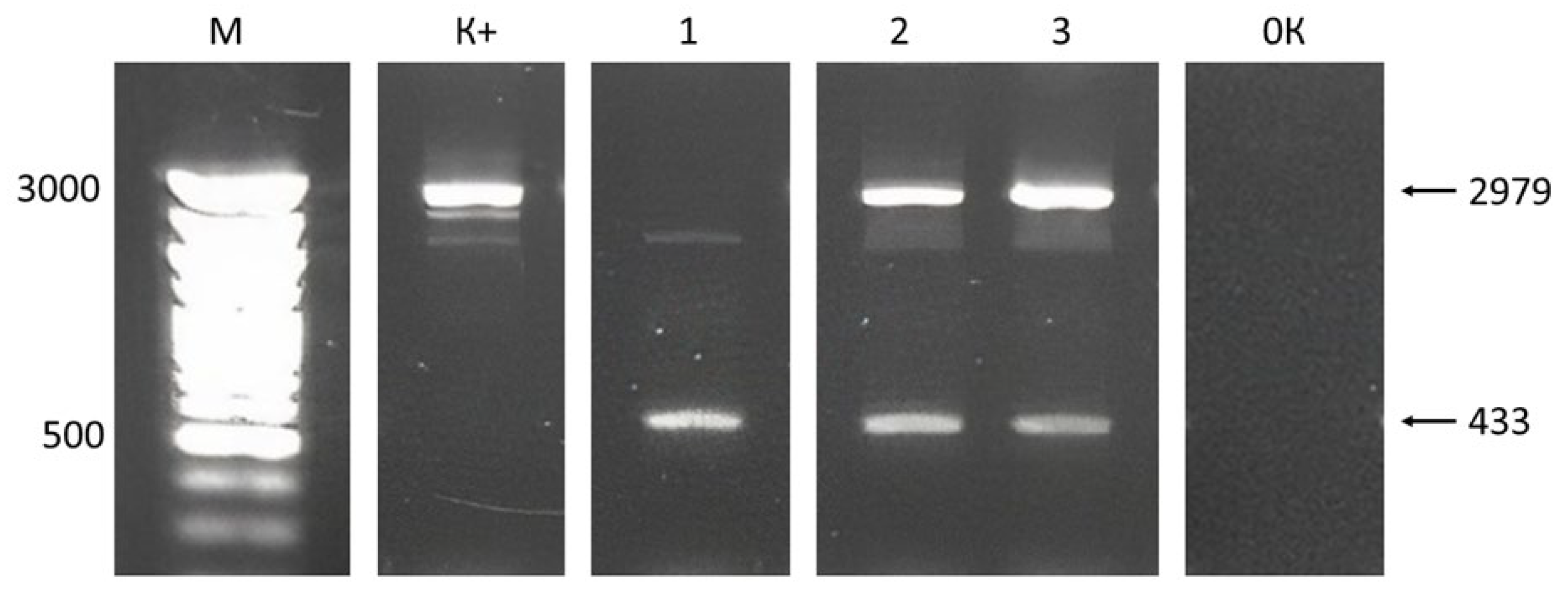
2.1. Plant Material Analysis
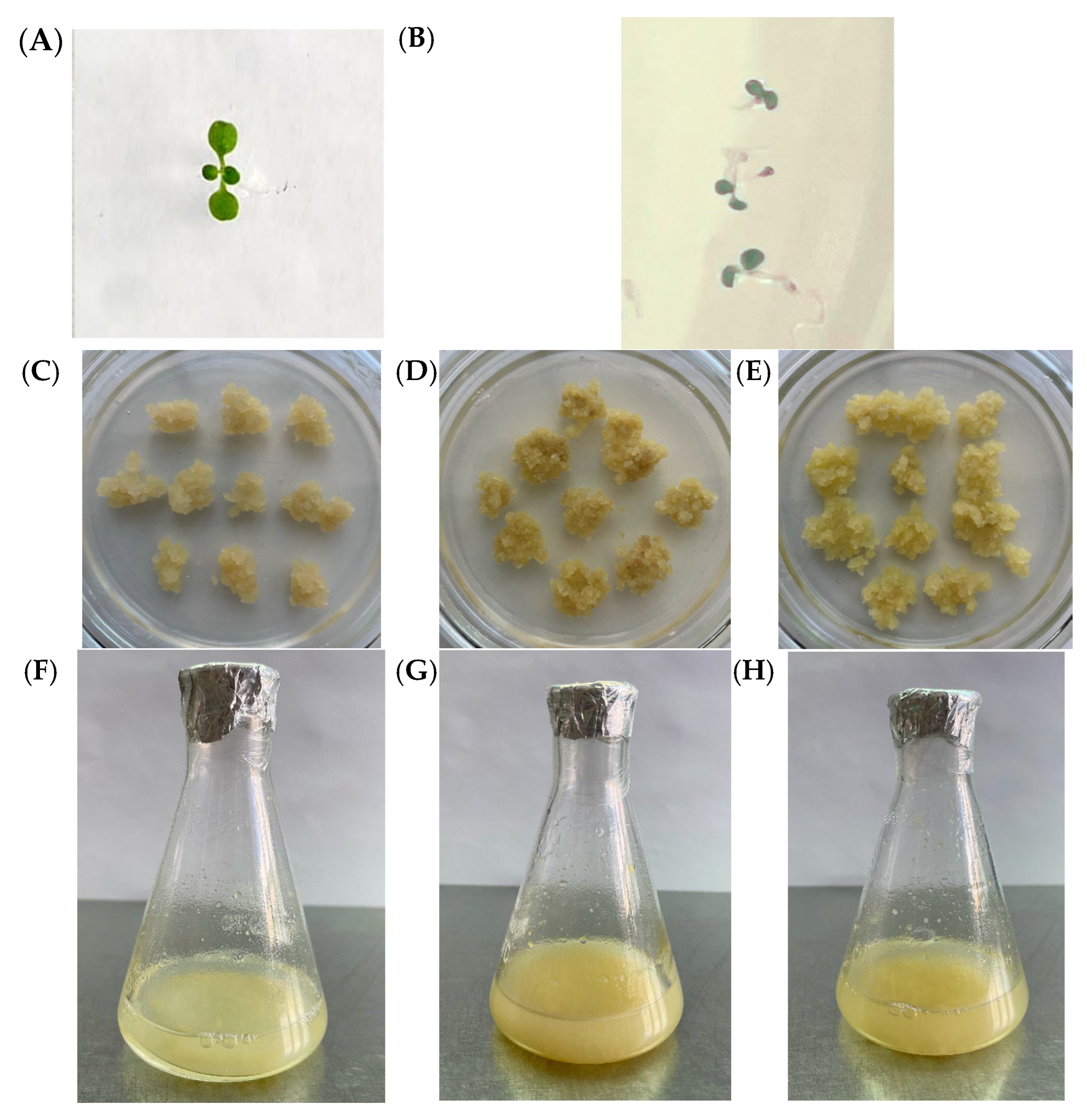
2.2. Phenotypic Characteristics of Callus and Suspension Cell Cultures with GAUT7 Knockout and the Qua1-1 Line
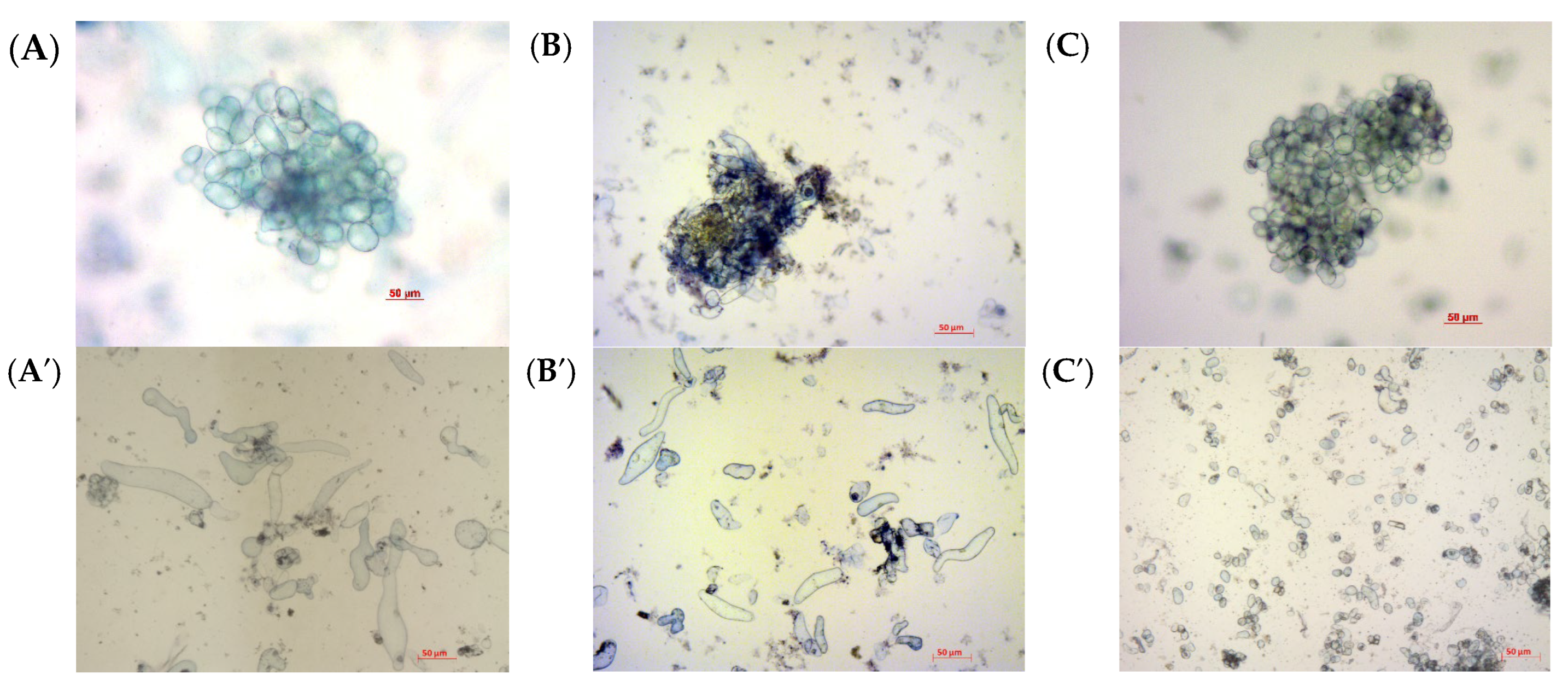
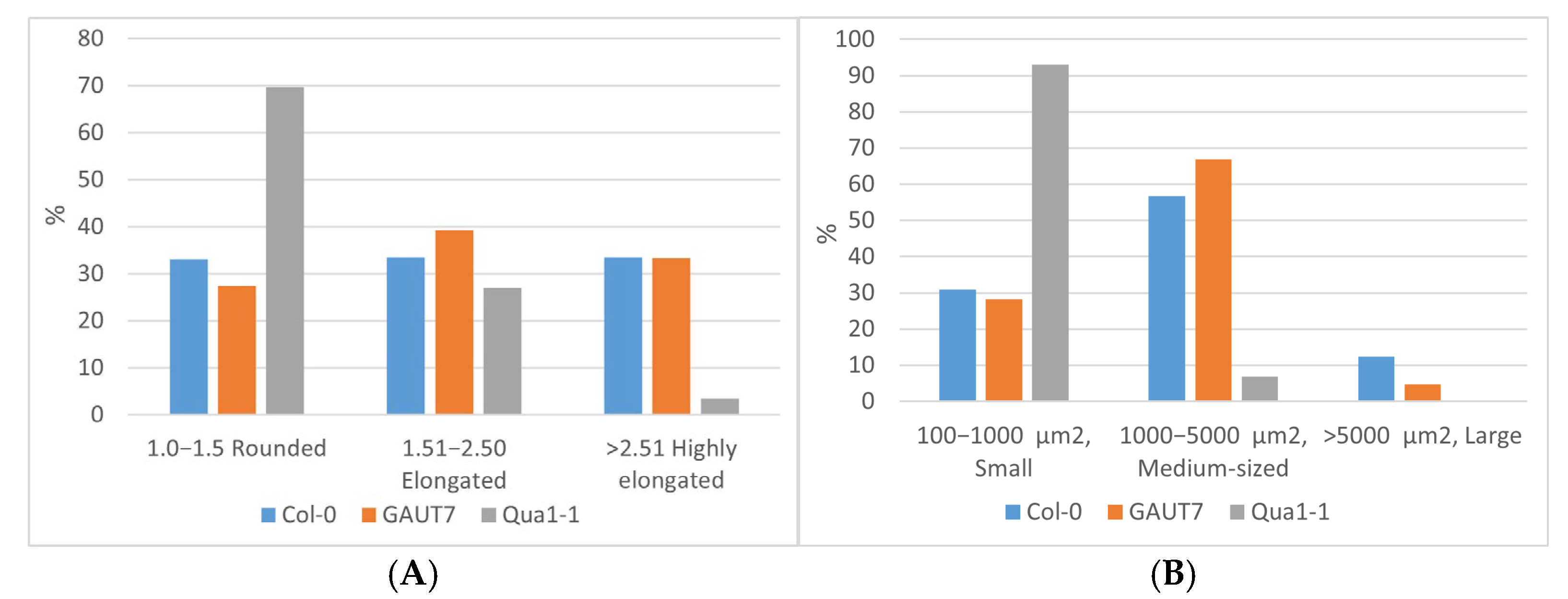
2.3. Biomass Accumulation and Aggregation of Suspension Cultures
2.4. Pectin Content Analysis in the Cell Wall
2.5. Analysis of Recombinant GFP Protein Accumulation
3. Discussion
4. Materials and Methods
4.1. Plant Materia
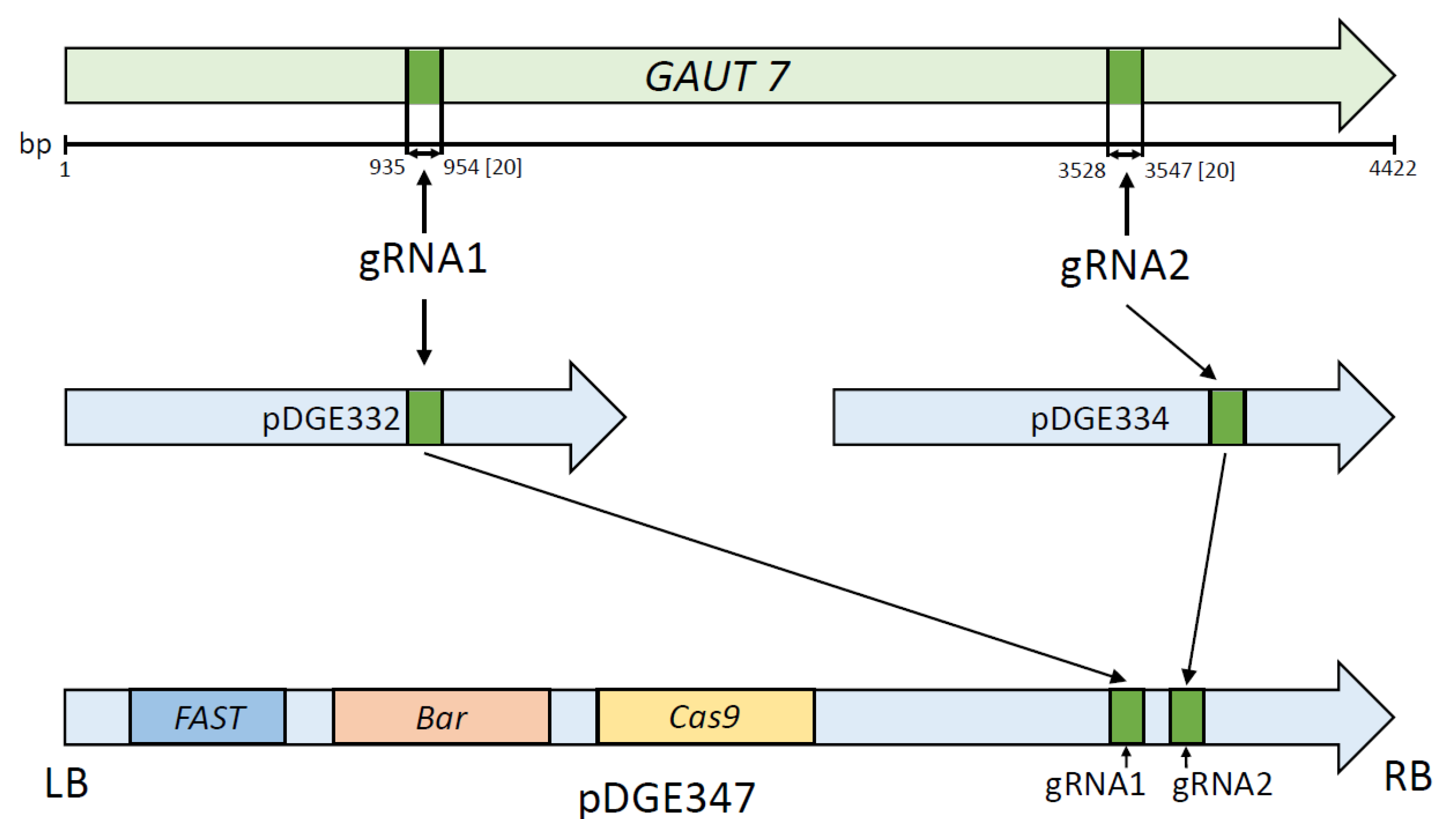
4.2. Plasmids Carrying Cas9 and Guide RNAs
4.3. Guide RNA Selection
4.4. Construction of the pDGE347_GAUT7 Genetic Construct
4.5. Delivery of the Genetic Construct into Plants
4.6. Analysis of Transformants for GAUT7 Deletion
4.7. Establishment of Suspension Cell Cultures
4.8. Analysis of Biomass Accumulation and Aggregation in Suspension Cultures
4.9. Analysis of Pectin Content in the Cell Wall
4.10. Light Microscopy
4.11. Quantification of Recombinant GFP Protein
4.12. Statistical Analysis
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cimini, S.; Ronci, M.B.; Barizza, E.; de Pinto, M.C.; Locato, V.; Lo Schiavo, F.; De Gara, L. Plant cell cultures as model systems to study programmed cell death. Plant Program. Cell Death Methods Protoc. 2018, 1743, 173–186. [Google Scholar]
- Tabata, H. Production of paclitaxel and the related taxanes by cell suspension cultures of Taxus species. Curr. Drug Targets 2006, 7, 453–461. [Google Scholar] [CrossRef]
- Chattopadhyay, S.; Datta, S.K.; Mahato, S.B. Production of L-DOPA from cell suspension culture of Mucuna pruriens f. pruriens. Plant Cell Rep. 1994, 13, 519–522. [Google Scholar] [CrossRef] [PubMed]
- Bapat, V.A.; Kavi Kishor, P.B.; Jalaja, N.; Jain, S.M.; Penna, S. Plant cell cultures: Biofactories for the production of bioactive compounds. Agronomy 2023, 13, 858. [Google Scholar] [CrossRef]
- Zagorskaya, A.A.; Deineko, E.V. Suspension-cultured plant cells as a platform for obtaining recombinant proteins. Russ. J. Plant Physiol. 2017, 64, 795–807. [Google Scholar] [CrossRef]
- Grabowski, G.A.; Golembo, M.; Shaaltiel, Y. Taliglucerase alfa: An enzyme replacement therapy using plant cell expression technology. Mol. Genet. Metab. 2014, 112, 1–8. [Google Scholar] [CrossRef]
- Huang, T.K.; Plesha, M.A.; Falk, B.W.; Dandekar, A.M.; McDonald, K.A. Bioreactor strategies for improving production yield and functionality of a recombinant human protein in transgenic tobacco cell cultures. Biotechnol. Bioeng. 2009, 102, 508–520. [Google Scholar] [CrossRef]
- Shin, Y.J.; Hong, S.Y.; Kwon, T.H.; Jang, Y.S.; Yang, M.S. High level of expression of recombinant human granulocyte-macrophage colony stimulating factor in transgenic rice cell suspension culture. Biotechnol. Bioeng. 2003, 82, 778–783. [Google Scholar] [CrossRef]
- Kolewe, M.E.; Henson, M.A.; Roberts, S.C. Analysis of aggregate size as a process variable affecting paclitaxel accumulation in Taxus suspension cultures. Biotechnol. Prog. 2011, 27, 1365–1372. [Google Scholar] [CrossRef]
- Daher, F.B.; Braybrook, S.A. How to let go: Pectin and plant cell adhesion. Front. Plant Sci. 2015, 6, 523. [Google Scholar] [CrossRef] [PubMed]
- Biswal, A.K.; Atmodjo, M.A.; Li, M.; Baxter, H.L.; Yoo, C.G.; Pu, Y.; Lee, Y.C.; Mazarei, M.; Black, I.M.; Zhang, J.Y.; et al. Sugar release and growth of biofuel crops are improved by downregulation of pectin biosynthesis. Nat. Biotechnol. 2018, 36, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Biswal, A.K.; Hao, Z.; Pattathil, S.; Yang, X.; Winkeler, K.; Collins, C.; Mohanty, S.S.; Richardson, E.A.; Gelineo-Albersheim, I.; Hunt, K.; et al. Downregulation of GAUT12 in Populus deltoides by RNA silencing results in reduced recalcitrance, increased growth and reduced xylan and pectin in a woody biofuel feedstock. Biotechnol. Biofuels 2015, 8, 41. [Google Scholar] [CrossRef]
- Atmodjo, M.A.; Sakuragi, Y.; Zhu, X.; Burrell, A.J.; Mohanty, S.S.; Atwood, J.A., 3rd; Orlando, R.; Scheller, H.V.; Mohnen, D. Galacturonosyltransferase (GAUT) 1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan: Galacturonosyltransferase complex. Proc. Natl. Acad. Sci. USA 2011, 108, 20225–20230. [Google Scholar] [CrossRef]
- Atmodjo, M.A.; Hao, Z.; Mohnen, D. Evolving Views of Pectin Biosynthesis. Annu. Rev. Plant Biol. 2013, 64, 747–779. [Google Scholar] [CrossRef]
- Sterling, J.D.; Atmodjo, M.A.; Inwood, S.E.; Kumar Kolli, V.S.; Quigley, H.F.; Hahn, M.G.; Mohnen, D. Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proc. Natl. Acad. Sci. USA 2006, 103, 5236–5241. [Google Scholar] [CrossRef]
- Frankevich, T.A.; Permyakova, N.V.; Sidorchuk, Y.V.; Deineko, E.V. Impact of GAUT1 Gene Knockout on Cell Aggregation in Arabidopsis thaliana Suspension Culture. BioTech 2025, 14, 2. [Google Scholar] [CrossRef]
- Leboeuf, E.; Guillon, F.; Thoiron, S.; Lahaye, M. Biochemical and immunohistochemical analysis of pectic polysaccharides in the cell walls of Arabidopsis mutant QUASIMODO 1 suspension-cultured cells: Implications for cell adhesion. J. Exp. Bot. 2005, 56, 3171–3182. [Google Scholar] [CrossRef]
- Jiang, F.; Doudna, J.A. CRISPR–Cas9 structures and mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef]
- Bortesi, L.; Zhu, C.; Zischewski, J.; Perez, L.; Bassié, L.; Nadi, R.; Forni, G.; Lade, S.B.; Soto, E.; Jin, X.; et al. Patterns of CRISPR/Cas9 activity in plants, animals and microbes. Plant Biotechnol. J. 2016, 14, 2203–2216. [Google Scholar] [CrossRef]
- Khatodia, S.; Bhatotia, K.; Passricha, N.; Khurana, S.M.; Tuteja, N. The CRISPR/Cas genome-editing tool: Application in improvement of crops. Front. Plant Sci. 2016, 7, 506. [Google Scholar] [CrossRef] [PubMed]
- Arora, L.; Narula, A. Gene editing and crop improvement using CRISPR-Cas9 system. Front. Plant Sci. 2017, 8, 1932. [Google Scholar] [CrossRef]
- Demirci, Y.; Zhang, B.; Unver, T. CRISPR/Cas9: An RNA-guided highly precise synthetic tool for plant genome editing. J. Cell. Physiol. 2018, 233, 1844–1859. [Google Scholar] [CrossRef]
- Caffall, K.H.; Pattathil, S.; Phillips, S.E.; Hahn, M.G.; Mohnen, D. Arabidopsis thaliana T-DNA mutants implicate GAUT genes in the biosynthesis of pectin and xylan in cell walls and seed testa. Mol. Plant 2009, 2, 1000–1014. [Google Scholar] [CrossRef] [PubMed]
- Durand, C.; Vicré-Gibouin, M.; Follet-Gueye, M.L.; Duponchel, L.; Moreau, M.; Lerouge, P.; Driouich, A. The organization pattern of root border-like cells of Arabidopsis is dependent on cell wall homogalacturonan. Plant Physiol. 2009, 150, 1411–1421. [Google Scholar] [CrossRef]
- Bouton, S.; Leboeuf, E.; Mouille, G.; Leydecker, M.T.; Talbotec, J.; Granier, F.; Lahaye, M.; Höfte, H.; Truong, H.N. QUASIMODO1 encodes a putative membrane-bound glycosyltransferase required for normal pectin synthesis and cell adhesion in Arabidopsis. Plant Cell 2002, 14, 2577–2590. [Google Scholar] [CrossRef] [PubMed]
- Lund, C.H.; Stenbæk, A.; Atmodjo, M.A.; Rasmussen, R.E.; Moller, I.E.; Erstad, S.M.; Biswal, A.K.; Mohnen, D.; Mravec, J.; Sakuragi, Y. Pectin synthesis and pollen tube growth in Arabidopsis involves three GAUT1 Golgi-anchoring proteins: GAUT5, GAUT6, and GAUT7. Front. Plant Sci. 2020, 11, 585774. [Google Scholar] [CrossRef]
- Xie, J.; Qi, B.; Mou, C.; Wang, L.; Jiao, Y.; Dou, Y.; Zheng, H. BREVIPEDICELLUS and ERECTA control the expression of AtPRX17 to prevent Arabidopsis callus browning. J. Exp. Bot. 2022, 73, 1516–1532. [Google Scholar] [CrossRef]
- Qiu, L.; Su, J.; Fu, Y.; Zhang, K. Genetic and Transcriptome Analyses of Callus Browning in Chaling Common Wild Rice (Oryza rufipogon Griff.). Genes 2023, 14, 2138. [Google Scholar] [CrossRef]
- Santos, R.B.; Abranches, R.; Fischer, R.; Sack, M.; Holland, T. Putting the spotlight back on plant suspension cultures. Front. Plant Sci. 2016, 7, 297. [Google Scholar] [CrossRef]
- Patil, R.A.; Kolewe, M.E.; Roberts, S.C. Cellular aggregation is a key parameter associated with long term variability in paclitaxel accumulation in Taxus suspension cultures. Plant Cell. Tissue Organ Cult. 2013, 112, 303–310. [Google Scholar] [CrossRef]
- Hepler, P.K.; Winship, L.J. Calcium at the cell wall-cytoplast interface. J. Integr. Plant Biol. 2010, 52, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Wdowiak, A.; Podgórska, A.; Szal, B. Calcium in plants: An important element of cell physiology and structure, signaling, and stress responses. Acta Physiol. Plant. 2024, 46, 108. [Google Scholar] [CrossRef]
- Verger, S.; Chabout, S.; Gineau, E.; Mouille, G. Cell adhesion in plants is under the control of putative O-fucosyltransferases. Development 2016, 143, 2536–2540. [Google Scholar] [CrossRef] [PubMed]
- Stuttmann, J.; Barthel, K.; Martin, P.; Ordon, J.; Erickson, J.L.; Herr, R.; Ferik, F.; Kretschmer, C.; Berner, T.; Keilwagen, J.; et al. Highly efficient multiplex editing: One-shot generation of 8× Nicotiana benthamiana and 12× Arabidopsis mutants. Plant J. 2021, 106, 8–22. [Google Scholar] [CrossRef] [PubMed]
- Concordet, J.P.; Haeussler, M. CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018, 46, 242–245. [Google Scholar] [CrossRef]
- Liu, H.; Ding, Y.; Zhou, Y.; Jin, W.; Xie, K.; Chen, L.L. CRISPR-P 2.0: An improved CRISPR-Cas9 tool for genome editing in plants. Mol. Plant 2017, 10, 530–532. [Google Scholar] [CrossRef]
- Zhang, X.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Kasajima, I.; Ide, Y.; Ohkama-Ohtsu, N.; Hayashi, H.; Yoneyama, T.; Fujiwara, T. A protocol for rapid DNA extraction from Arabidopsis thaliana for PCR analysis. Plant Mol. Biol. Report. 2004, 22, 49–52. [Google Scholar] [CrossRef]
- Schenk, R.U.; Hildebrandt, A.C. Medium and techniques for induction and growth of monocotyledonous and dicotyledonous plant cell cultures. Can. J. Bot. 1972, 50, 199–204. [Google Scholar] [CrossRef]
- Strober, W. Trypan blue exclusion test of cell viability. Curr. Protoc. Immunol. 2015, 111, A3-B. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.J.; Liu, Y.W.; Sun, M.X. Improved and high throughput quantitative measurements of weak GFP expression in transgenic plant materials. Plant Cell Rep. 2011, 30, 1253–1260. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]


| Suspension Cell Culture Line | Total Soluble Protein, mg/μL | GFP Protein, mg/μL | GFP as a Percentage of Total Soluble Protein, % |
|---|---|---|---|
| Col-0 GFP | 0.40 | 0.063 | 16 |
| GAUT7 | 0.96 | 0.053 | 5.6 * |
| Oligonucleotide Name | Nucleotide Sequence 5′–3′ |
|---|---|
| GAUT7_gRNA3_Forward | ATTGAATCAATCCAGTTCTTCCCA |
| GAUT7_gRNA3_Reverse | AAACTGGGAAGAACTGGATTGATT |
| GAUT7_gRNA4_Forward | ATTGCTTCCATATCAAGGTCCCAA |
| GAUT7_gRNA4_Reverse | AAACTTGGGACCTTGATATGGAAG |
| GAUT7_deltest_Up2 | TGTCACTGTTCAACCGGCTTCTT |
| GAUT7_deltest_Lo2 | CAATGCCCTCCATCTAGCAAGATC |
| pDGEtest up | ATAGCAATGACCAGTGCAAACAGTG |
| pDGEtest lo | CTCTTTTCTCTTAGGTTTACCCGCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frankevich, T.A.; Permyakova, N.V.; Sidorchuk, Y.V.; Deineko, E.V. Changes in Cell Aggregation in Arabidopsis thaliana Suspension Culture Following Knockout of GAUT Gene Family Members. Plants 2025, 14, 2816. https://doi.org/10.3390/plants14182816
Frankevich TA, Permyakova NV, Sidorchuk YV, Deineko EV. Changes in Cell Aggregation in Arabidopsis thaliana Suspension Culture Following Knockout of GAUT Gene Family Members. Plants. 2025; 14(18):2816. https://doi.org/10.3390/plants14182816
Chicago/Turabian StyleFrankevich, Tatyana A., Natalya V. Permyakova, Yury V. Sidorchuk, and Elena V. Deineko. 2025. "Changes in Cell Aggregation in Arabidopsis thaliana Suspension Culture Following Knockout of GAUT Gene Family Members" Plants 14, no. 18: 2816. https://doi.org/10.3390/plants14182816
APA StyleFrankevich, T. A., Permyakova, N. V., Sidorchuk, Y. V., & Deineko, E. V. (2025). Changes in Cell Aggregation in Arabidopsis thaliana Suspension Culture Following Knockout of GAUT Gene Family Members. Plants, 14(18), 2816. https://doi.org/10.3390/plants14182816







