Genome-Wide Analysis of the Rice PcG Gene Family and Its Involvement in Salt Response and Development
Abstract
1. Introduction
2. Results
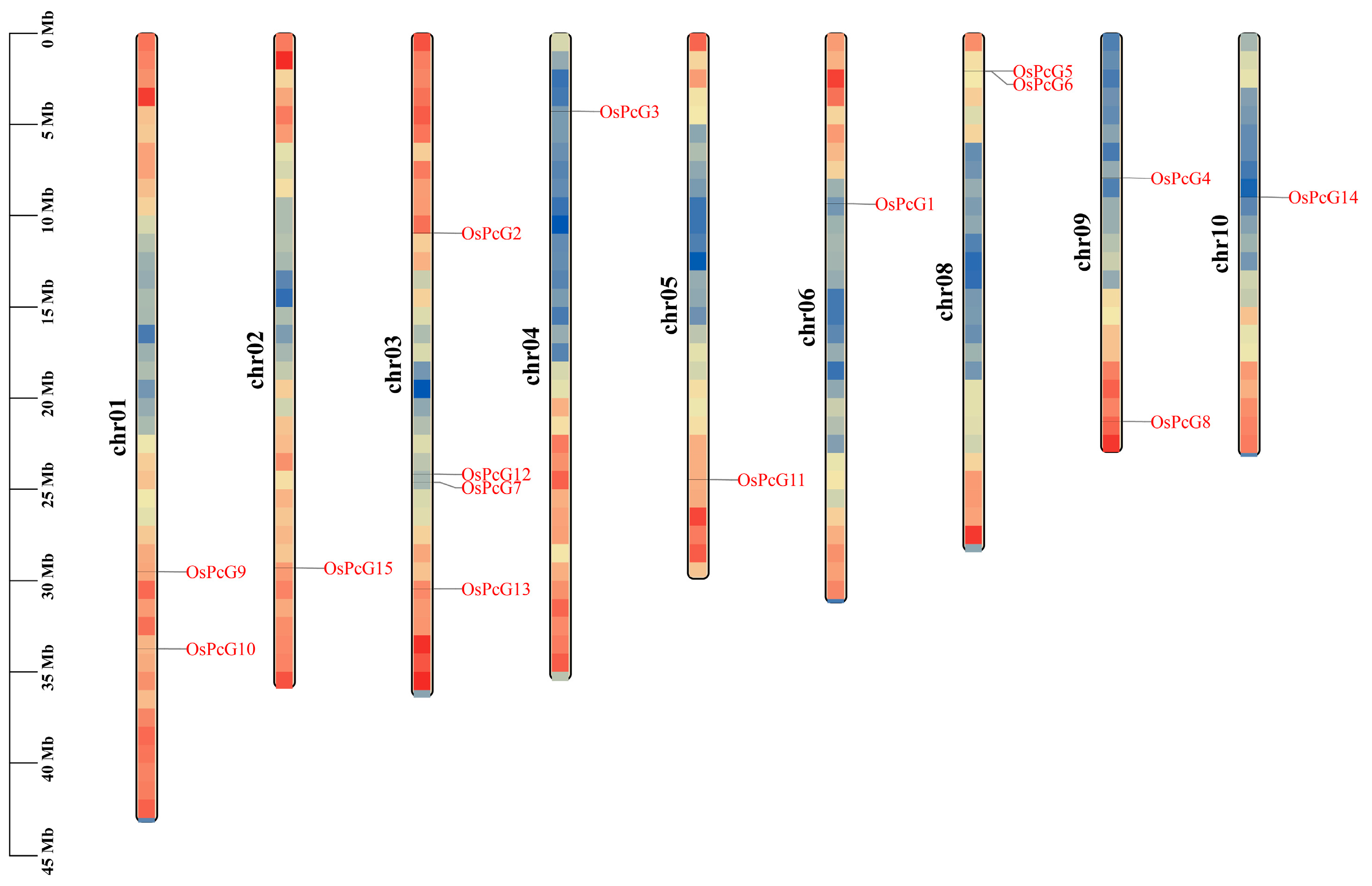
2.1. OsPcG Identification and Chromosome Localization
2.2. Physicochemical Properties and Predicted Subcellular Localization of OsPcG Proteins
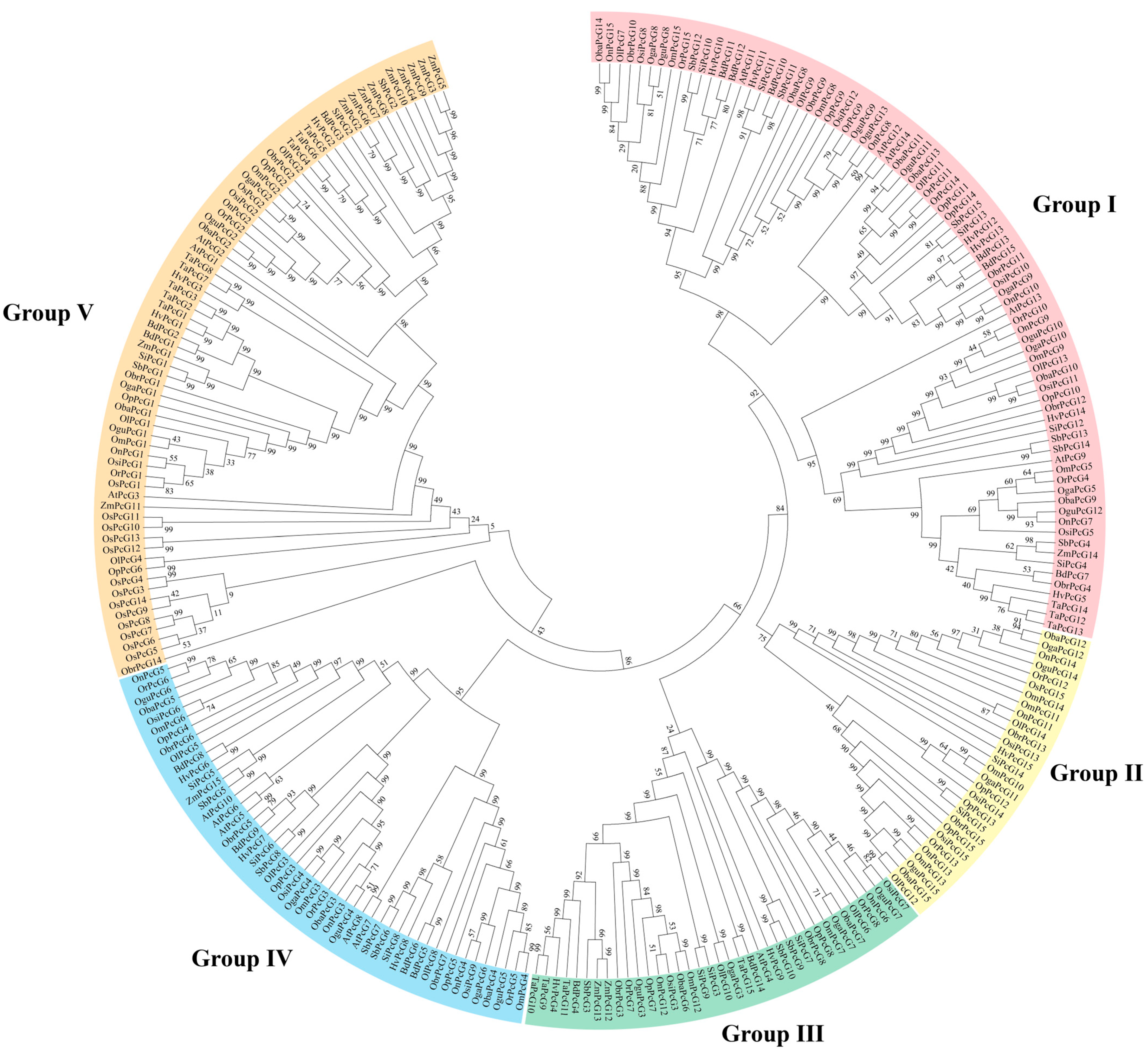
2.3. Classification of OsPcG and Phylogenetic Analysis of the Phylogenetic Tree
2.4. Structural Analysis of Rice OsPcG
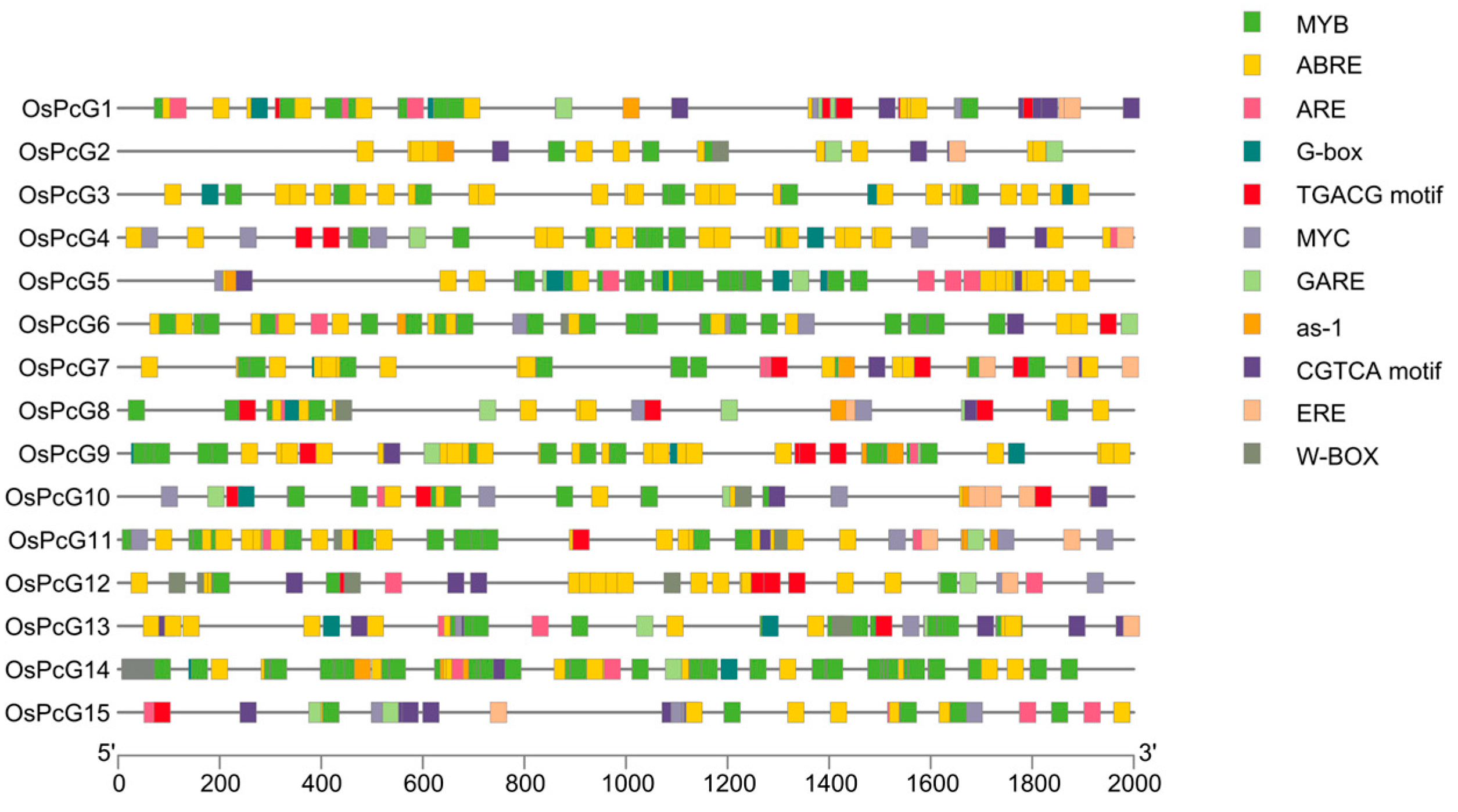
2.5. cis-Acting Elements in the OsPcG Promoter Region
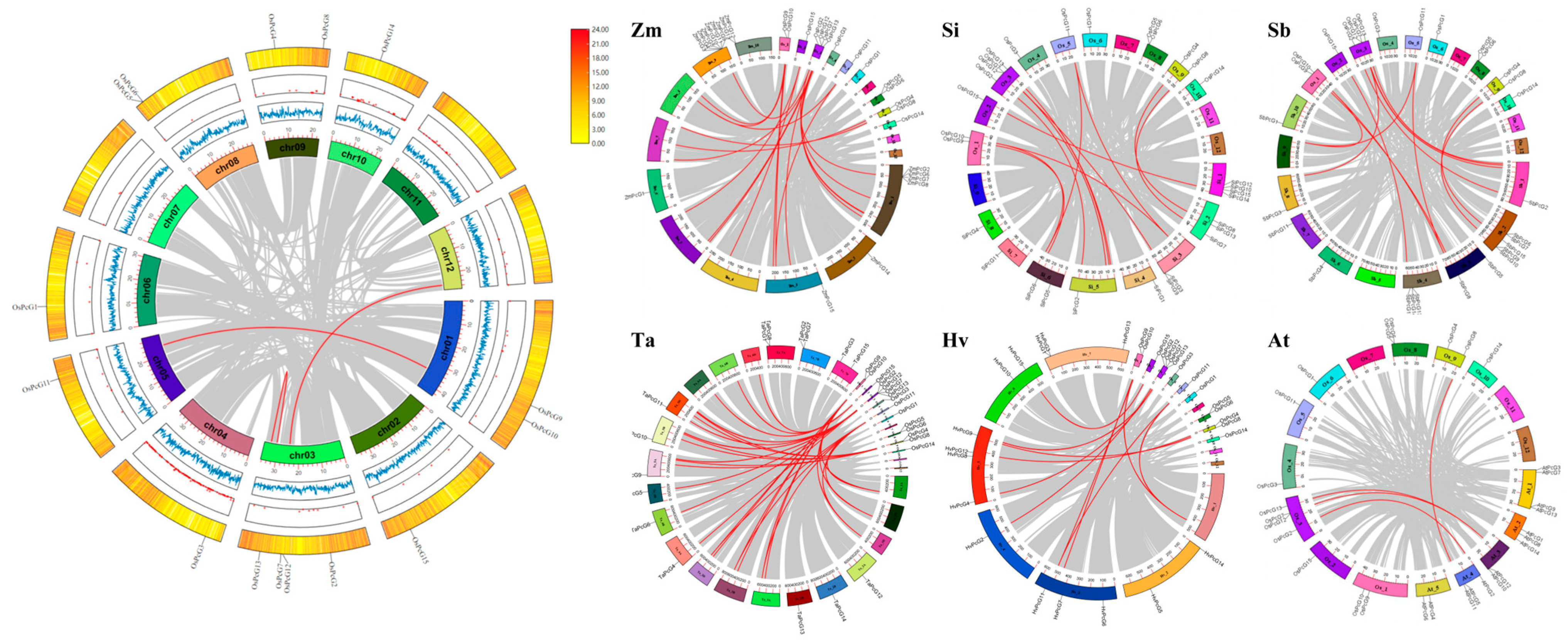
2.6. Analysis of Intra- and Inter-Species Covariance in the Rice PcG Family
2.7. Estimation of Covariance and Ka/Ks Ratio of OsPcG
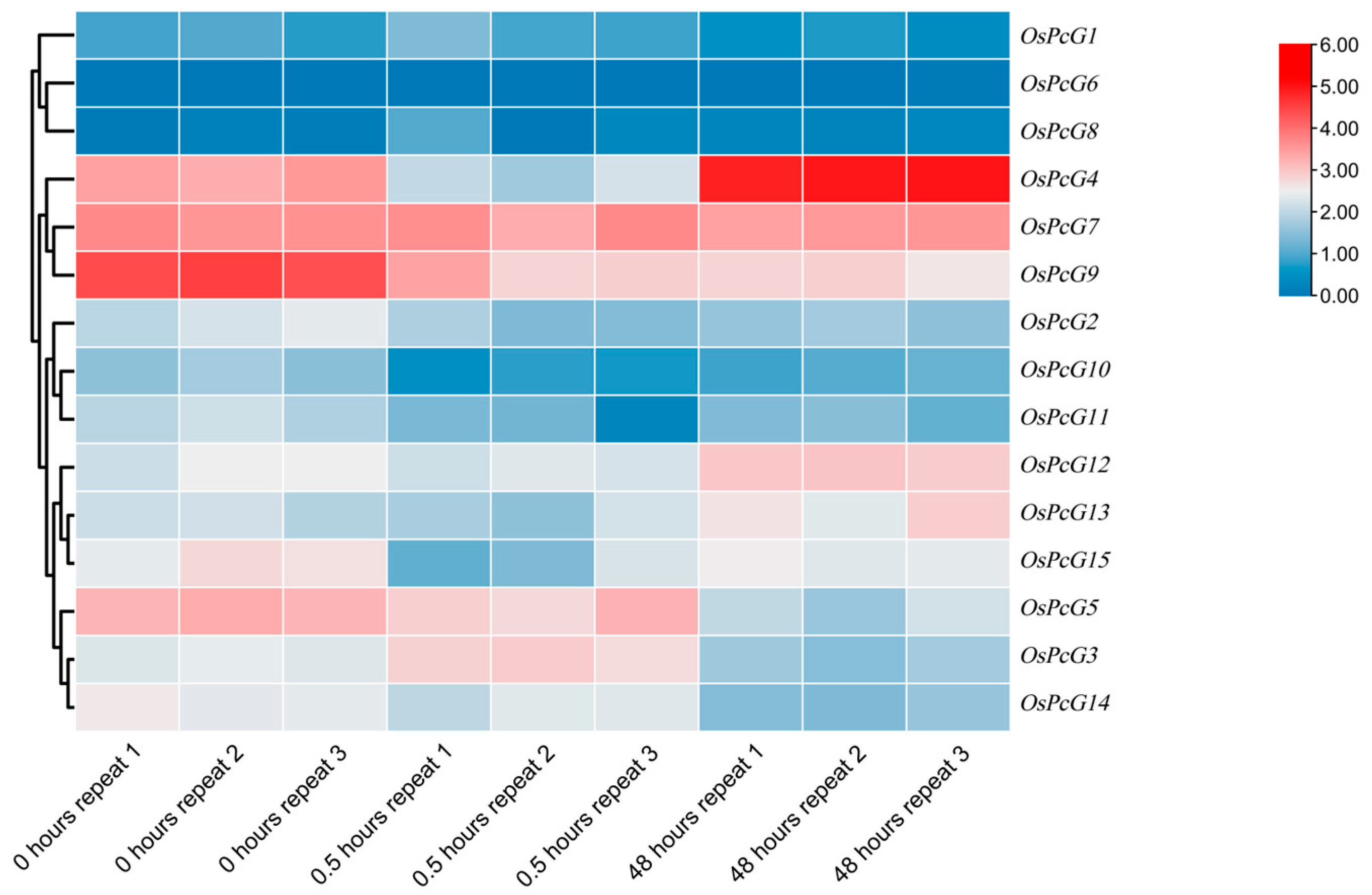
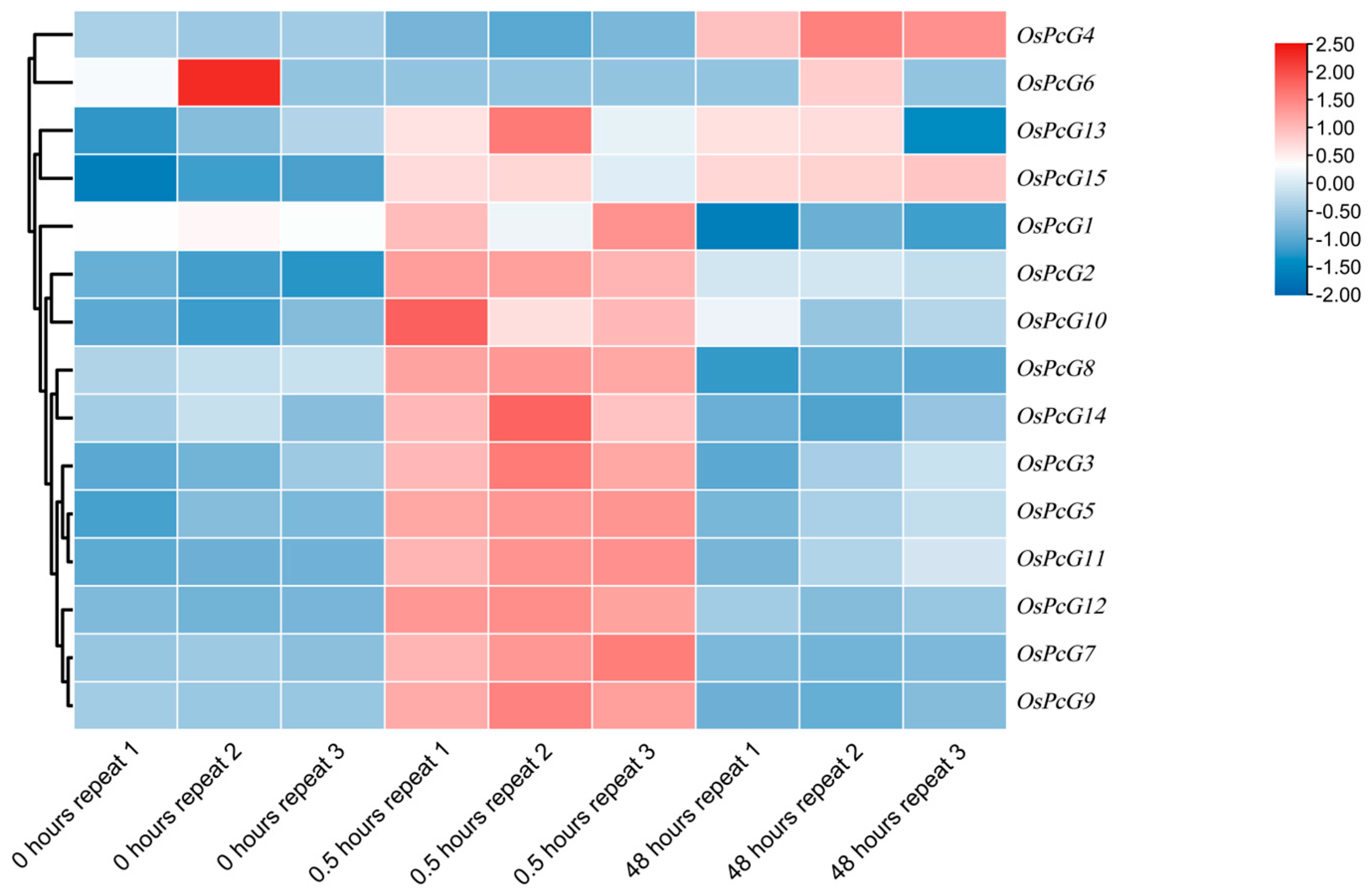
2.8. Analysis of the Expression Pattern of OsPcG Gene in Roots and Leaves
2.9. Subcellular Localization of OsFIE2
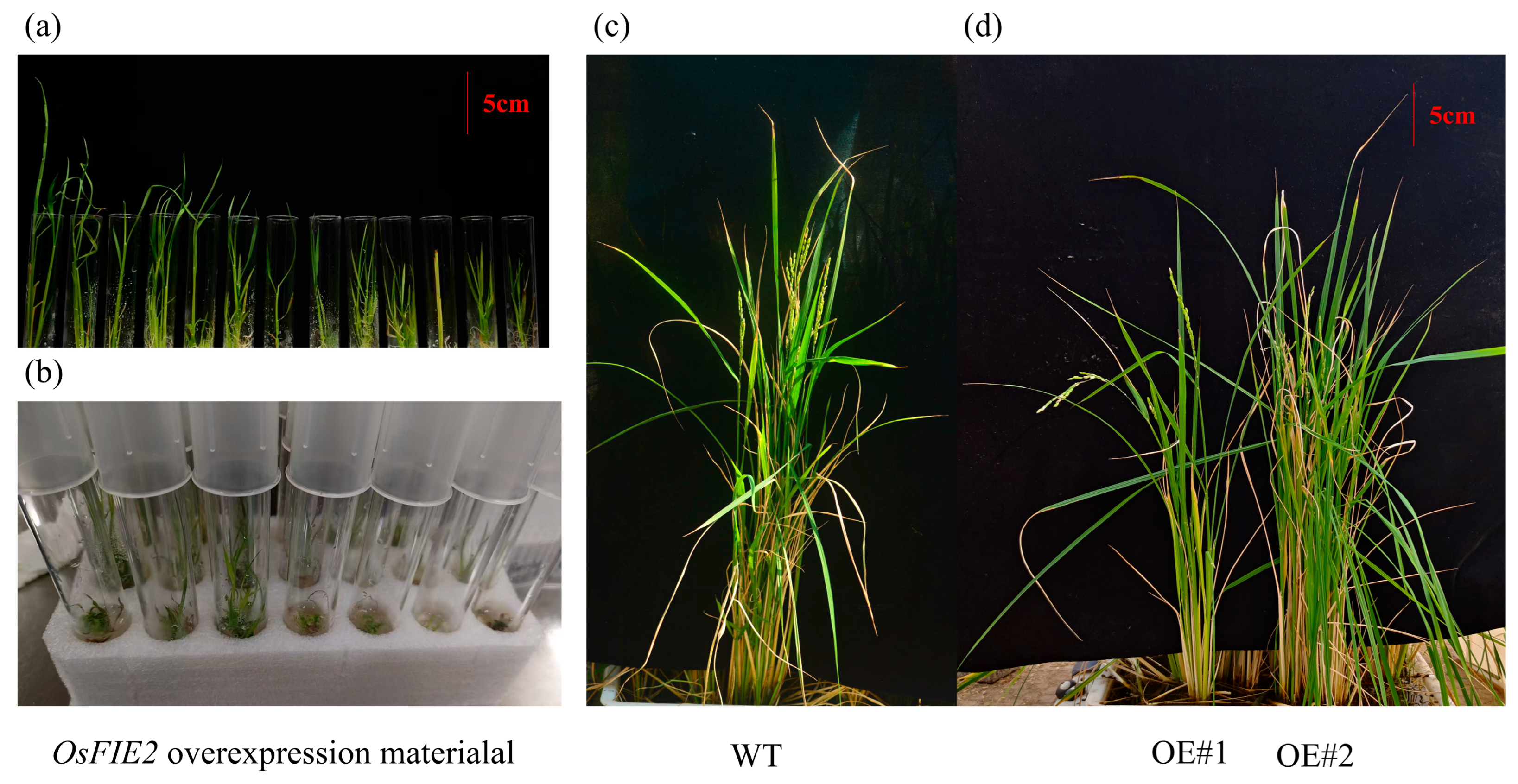
2.10. Role of FIE2 Gene in Regulating Plant Height in Transgenic Rice
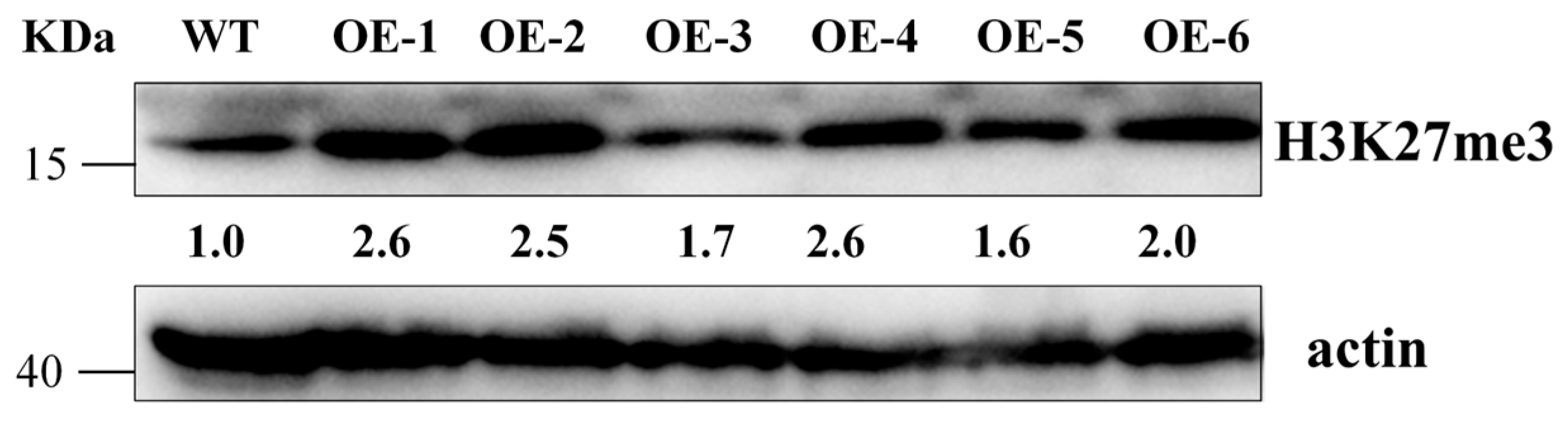
2.11. OsFIE2 Overexpression Enhances H3K27me3 Accumulation
3. Discussion
4. Materials and Methods
4.1. Identification of Members of the PcG Gene Family in Oryza sativa and Chromosome Localization
4.2. Predicting the Physicochemical Properties and Subcellular Localization of PcG Proteins
4.3. Gene Structure Analysis and Phylogenetic Development
4.4. Predicting cis-Acting Elements in Promoter Regions
4.5. Collinearity and Estimation of Ka/Ks Ratio Analysis
4.6. Plant Material
4.7. Transcript Expression Analysis During Root and Leaf Development in Rice
4.8. RNA Extraction and qRT-PCR Analysis
4.9. Subcellular Localization of the Rice PcG Family Member OsFIE2
4.10. Generation of OsFIE2 Overexpression Lines in Rice
4.11. Western Blot Analysis of H3K27me3 in OsFIE2 Overexpression Lines
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kagohara, L.T.; Stein-O’Brien, G.L.; Kelley, D.; Flam, E.; Wick, H.C.; Danilova, L.V.; Easwaran, H.; Favorov, A.V.; Qian, J.; Gaykalova, D.A.; et al. Epigenetic regulation of gene expression in cancer: Techniques, resources and analysis. Brief. Funct. Genomics 2018, 17, 49–63. [Google Scholar] [CrossRef]
- Mozgova, I.; Hennig, L. The polycomb group protein regulatory network. Annu. Rev. Plant Biol. 2015, 66, 269–296. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef]
- Jones, P.A.; Baylin, S.B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002, 3, 415–428. [Google Scholar] [CrossRef]
- Ojolo, S.P.; Cao, S.; Priyadarshani, S.V.; Li, W.; Yan, M.; Aslam, M.; Zhao, H.; Qin, Y. Regulation of Plant Growth and Development: A Review from a Chromatin Remodeling Perspective. Front. Plant Sci. 2018, 9, 1232. [Google Scholar] [CrossRef]
- Stillman, B. Histone Modifications: Insights into Their Influence on Gene Expression. Cell 2018, 175, 6–9. [Google Scholar] [CrossRef]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.N.; Zhu, C.; Jiang, J.; Zhang, H.; Zhu, J.K.; Duan, C.G. Epigenetic regulation in plant abiotic stress responses. J. Integr. Plant Biol. 2020, 62, 563–580. [Google Scholar] [CrossRef]
- Kim, J.M.; Sasaki, T.; Ueda, M.; Sako, K.; Seki, M. Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 2015, 6, 114. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Liu, X.; Singh, P.; Cui, Y.; Zimmerli, L.; Wu, K. Chromatin modifications and remodeling in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Lewis, E.B. A gene complex controlling segmentation in Drosophila. Nature 1978, 276, 565–570. [Google Scholar] [CrossRef]
- Jürgens, G. A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature 1985, 316, 153–155. [Google Scholar] [CrossRef]
- Simon, J.A.; Kingston, R.E. Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009, 10, 697–708. [Google Scholar] [CrossRef]
- Paro, R. Imprinting a determined state into the chromatin of Drosophila. Trends Genet. 1990, 6, 416–421. [Google Scholar] [CrossRef]
- Derkacheva, M.; Hennig, L. Variations on a theme: Polycomb group proteins in plants. J. Exp. Bot. 2014, 65, 2769–2784. [Google Scholar] [CrossRef]
- Czermin, B.; Melfi, R.; McCabe, D.; Seitz, V.; Imhof, A.; Pirrotta, V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 2002, 111, 185–196. [Google Scholar] [CrossRef]
- Shen, X.; Liu, Y.; Hsu, Y.J.; Fujiwara, Y.; Kim, J.; Mao, X.; Yuan, G.C.; Orkin, S.H. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell 2008, 32, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004, 431, 873–878. [Google Scholar] [CrossRef] [PubMed]
- Tavares, L.; Dimitrova, E.; Oxley, D.; Webster, J.; Poot, R.; Demmers, J.; Bezstarosti, K.; Taylor, S.; Ura, H.; Koide, H.; et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 2012, 148, 664–678. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.A.; Kingston, R.E. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell 2013, 49, 808–824. [Google Scholar] [CrossRef]
- Francis, N.J.; Saurin, A.J.; Shao, Z.; Kingston, R.E. Reconstitution of a functional core polycomb repressive complex. Mol. Cell 2001, 8, 545–556. [Google Scholar] [CrossRef] [PubMed]
- Mohd-Sarip, A.; Venturini, F.; Chalkley, G.E.; Verrijzer, C.P. Pleiohomeotic can link polycomb to DNA and mediate transcriptional repression. Mol. Cell Biol. 2002, 22, 7473–7483. [Google Scholar] [CrossRef]
- Yu, M.; Mazor, T.; Huang, H.; Huang, H.T.; Kathrein, K.L.; Woo, A.J.; Chouinard, C.R.; Labadorf, A.; Akie, T.E.; Moran, T.B.; et al. Direct recruitment of polycomb repressive complex 1 to chromatin by core binding transcription factors. Mol. Cell 2012, 45, 330–343. [Google Scholar] [CrossRef]
- Bu, S.; Lau, S.S.; Yong, W.L.; Zhang, H.; Thiagarajan, S.; Bashirullah, A.; Yu, F. Polycomb group genes are required for neuronal pruning in Drosophila. BMC Biol. 2023, 21, 33. [Google Scholar] [CrossRef] [PubMed]
- Angrand, P.O. Structure and Function of the Polycomb Repressive Complexes PRC1 and PRC2. Int. J. Mol. Sci. 2022, 23, 5971. [Google Scholar] [CrossRef]
- Turck, F.; Roudier, F.; Farrona, S.; Martin-Magniette, M.L.; Guillaume, E.; Buisine, N.; Gagnot, S.; Martienssen, R.A.; Coupland, G.; Colot, V. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 2007, 3, e86. [Google Scholar] [CrossRef]
- Zhang, X.; Germann, S.; Blus, B.J.; Khorasanizadeh, S.; Gaudin, V.; Jacobsen, S.E. The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat. Struct. Mol. Biol. 2007, 14, 869–871. [Google Scholar] [CrossRef]
- Sanchez-Pulido, L.; Devos, D.; Sung, Z.R.; Calonje, M. RAWUL: A new ubiquitin-like domain in PRC1 ring finger proteins that unveils putative plant and worm PRC1 orthologs. BMC Genom. 2008, 9, 308. [Google Scholar] [CrossRef]
- Cao, R.; Wang, L.; Wang, H.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002, 298, 1039–1043. [Google Scholar] [CrossRef] [PubMed]
- Barbour, H.; Daou, S.; Hendzel, M.; Affar, E.B. Polycomb group-mediated histone H2A monoubiquitination in epigenome regulation and nuclear processes. Nat. Commun. 2020, 11, 5947. [Google Scholar] [CrossRef]
- Bemer, M.; Grossniklaus, U. Dynamic regulation of Polycomb group activity during plant development. Curr. Opin. Plant Biol. 2012, 15, 523–529. [Google Scholar] [CrossRef]
- Shen, Q.; Lin, Y.; Li, Y.; Wang, G. Dynamics of H3K27me3 Modification on Plant Adaptation to Environmental Cues. Plants 2021, 10, 1165. [Google Scholar] [CrossRef]
- Müller, J.; Hart, C.M.; Francis, N.J.; Vargas, M.L.; Sengupta, A.; Wild, B.; Miller, E.L.; O’Connor, M.B.; Kingston, R.E.; Simon, J.A. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 2002, 111, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Spillane, C.; MacDougall, C.; Stock, C.; Köhler, C.; Vielle-Calzada, J.P.; Nunes, S.M.; Grossniklaus, U.; Goodrich, J. Interaction of the Arabidopsis polycomb group proteins FIE and MEA mediates their common phenotypes. Curr. Biol. 2000, 10, 1535–1538. [Google Scholar] [CrossRef]
- Poza-Viejo, L.; Payá-Milans, M.; Wilkinson, M.D.; Piñeiro, M.; Jarillo, J.A.; Crevillén, P. Brassica rapa CURLY LEAF is a major H3K27 methyltransferase regulating flowering time. Planta 2024, 260, 27. [Google Scholar] [CrossRef]
- Luo, M.; Bilodeau, P.; Dennis, E.S.; Peacock, W.J.; Chaudhury, A. Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 2000, 97, 10637–10642. [Google Scholar] [CrossRef]
- Gendall, A.R.; Levy, Y.Y.; Wilson, A.; Dean, C. The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 2001, 107, 525–535. [Google Scholar] [CrossRef]
- Alexandre, C.; Möller-Steinbach, Y.; Schönrock, N.; Gruissem, W.; Hennig, L. Arabidopsis MSI1 is required for negative regulation of the response to drought stress. Mol. Plant 2009, 2, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Bowers, J.E.; Wang, X.; Ming, R.; Alam, M.; Paterson, A.H. Synteny and collinearity in plant genomes. Science 2008, 320, 486–488. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Zhang, Y.; Deng, X.; Li, S.; Zhang, C.; Li, Y. Comparative genomic and transcriptomic analysis suggests the evolutionary dynamic of GH3 genes in Gramineae crops. Front. Plant Sci. 2019, 10, 1297. [Google Scholar] [CrossRef]
- Guruprasad, K.; Reddy, B.V.; Pandit, M.W. Correlation between stability of a protein and its dipeptide composition: A novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. 1990, 4, 155–161. [Google Scholar] [CrossRef]
- Hu, H.; Jia, Y.; Hao, Z.; Ma, G.; Xie, Y.; Wang, C.; Ma, D. Lipidomics-based insights into the physiological mechanism of wheat in response to heat stress. Plant Physiol. Biochem. 2023, 205, 108190. [Google Scholar] [CrossRef]
- Knetsch, T.G.J.; Ubbink, M. The effect of lipid composition on the thermal stability of nanodiscs. Biochim. Biophys. Acta Biomembr. 2024, 1866, 184239. [Google Scholar] [CrossRef]
- Paterson, A.H.; Bowers, J.E.; Chapman, B.A. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. USA 2004, 101, 9903–9908. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Huang, H.; Lin, X.; Liu, P.; Zhao, L.; Nie, W.F.; Zhu, J.K.; Lang, Z. MSI4/FVE is required for accumulation of 24-nt siRNAs and DNA methylation at a subset of target regions of RNA-directed DNA methylation. Plant J. 2021, 108, 347–357. [Google Scholar] [CrossRef]
- Niu, Q.; Song, Z.; Tang, K.; Chen, L.; Wang, L.; Ban, T.; Guo, Z.; Kim, C.; Zhang, H.; Duan, C.G.; et al. A histone H3K4me1-specific binding protein is required for siRNA accumulation and DNA methylation at a subset of loci targeted by RNA-directed DNA methylation. Nat. Commun. 2021, 12, 3367. [Google Scholar] [CrossRef]
- Ye, R.; Wang, M.; Du, H.; Chhajed, S.; Koh, J.; Liu, K.H.; Shin, J.; Wu, Y.; Shi, L.; Xu, L.; et al. Glucose-driven TOR-FIE-PRC2 signalling controls plant development. Nature 2022, 609, 986–993. [Google Scholar] [CrossRef]
- Li, S.; Zhou, B.; Peng, X.; Kuang, Q.; Huang, X.; Yao, J.; Du, B.; Sun, M.X. OsFIE2 plays an essential role in the regulation of rice vegetative and reproductive development. New Phytol. 2014, 201, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Pan, M.; E, Z.; Zhou, Y.; Niu, B.; Chen, C. Functional divergence of two duplicated Fertilization Independent Endosperm genes in rice with respect to seed development. Plant J. 2020, 104, 124–137. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Sung, S. Polycomb-mediated gene silencing in Arabidopsis thaliana. Mol. Cells 2014, 37, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Schuettengruber, B.; Chourrout, D.; Vervoort, M.; Leblanc, B.; Cavalli, G. Genome regulation by polycomb and trithorax proteins. Cell 2007, 128, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Merini, W.; Romero-Campero, F.J.; Gomez-Zambrano, A.; Zhou, Y.; Turck, F.; Calonje, M. The Arabidopsis Polycomb Repressive Complex 1 (PRC1) Components AtBMI1A, B, and C Impact Gene Networks throughout All Stages of Plant Development. Plant Physiol. 2017, 173, 627–641. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef]
- Yu, C.S.; Cheng, C.W.; Su, W.C.; Chang, K.C.; Huang, S.W.; Hwang, J.K.; Lu, C.H. CELLO2GO: A web server for protein subCELlular LOcalization prediction with functional gene ontology annotation. PLoS ONE 2014, 9, e99368. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]



| Sequence ID | Number of Amino Acid | Molecular Weight | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Prediction of Subcellular Localization |
|---|---|---|---|---|---|---|---|
| OsPcG1 | 896 | 100,295.81 | 8.02 | 51.46 | 63.98 | −0.747 | Nuclear |
| OsPcG2 | 895 | 99,862.93 | 8 | 51.02 | 65.74 | −0.704 | Nuclear |
| OsPcG3 | 624 | 71,546.95 | 5.87 | 49.48 | 78.48 | −0.502 | Nuclear |
| OsPcG4 | 604 | 68,615.51 | 6.52 | 51.82 | 76.69 | −0.434 | Nuclear |
| OsPcG5 | 376 | 42,035.94 | 5.68 | 44.98 | 86.33 | −0.107 | Plasma Membrane/Extracellular |
| OsPcG6 | 466 | 51,848.95 | 7.55 | 48.48 | 79.27 | −0.295 | Chloroplast |
| OsPcG7 | 428 | 48,359.93 | 4.77 | 47.79 | 80.21 | −0.49 | Cytoplasmic/Nuclear |
| OsPcG8 | 410 | 44,741.85 | 4.98 | 44.39 | 79.76 | −0.363 | Cytoplasmic |
| OsPcG9 | 525 | 57,532.24 | 6.24 | 45.7 | 70.61 | −0.546 | Cytoplasmic |
| OsPcG10 | 490 | 53,639.04 | 8.22 | 59.71 | 65.18 | −0.754 | Nuclear |
| OsPcG11 | 488 | 53,639.50 | 5.5 | 67.1 | 63.22 | −0.867 | Nuclear |
| OsPcG12 | 510 | 56,251.41 | 8.86 | 69.16 | 65.43 | −0.856 | Nuclear |
| OsPcG13 | 439 | 48,637.46 | 9.37 | 54.32 | 75.49 | −0.726 | Nuclear |
| OsPcG14 | 415 | 45,675.33 | 4.89 | 64.84 | 63.23 | −0.954 | Nuclear |
| OsPcG15 | 1607 | 181,498.29 | 6.16 | 47.39 | 70.83 | −0.571 | Nuclear |
| Seq_1 | Seq_2 | EffectiveLen | AverageS-Sites | AverageN-Sites | Ka | Ks | Ka/Ks | cN | cS | pN | pS |
|---|---|---|---|---|---|---|---|---|---|---|---|
| OsPcG10 | XP_015640607.1 | 1410 | 331.58 | 1078.42 | 0.22 | 0.99 | 0.23 | 208.5 | 182.5 | 0.19 | 0.55 |
| OsPcG12 | XP_015620739.1 | 1377 | 321.92 | 1055.08 | 0.22 | 0.63 | 0.34 | 200.2 | 137.8 | 0.19 | 0.43 |
| OsPcG13 | XP_015628300.1 | 1065 | 251.75 | 813.25 | 0.54 | 1.90 | 0.28 | 311.2 | 173.8 | 0.38 | 0.69 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, Z.; Cao, J.; Li, C.; Liu, J.; Yang, X.; Cheng, X. Genome-Wide Analysis of the Rice PcG Gene Family and Its Involvement in Salt Response and Development. Plants 2025, 14, 2805. https://doi.org/10.3390/plants14172805
Shi Z, Cao J, Li C, Liu J, Yang X, Cheng X. Genome-Wide Analysis of the Rice PcG Gene Family and Its Involvement in Salt Response and Development. Plants. 2025; 14(17):2805. https://doi.org/10.3390/plants14172805
Chicago/Turabian StyleShi, Ziang, Jun Cao, Chuheng Li, Jun Liu, Xinlei Yang, and Xiliu Cheng. 2025. "Genome-Wide Analysis of the Rice PcG Gene Family and Its Involvement in Salt Response and Development" Plants 14, no. 17: 2805. https://doi.org/10.3390/plants14172805
APA StyleShi, Z., Cao, J., Li, C., Liu, J., Yang, X., & Cheng, X. (2025). Genome-Wide Analysis of the Rice PcG Gene Family and Its Involvement in Salt Response and Development. Plants, 14(17), 2805. https://doi.org/10.3390/plants14172805






