The Genetic Loci Associated with Fiber Development in Upland Cotton (Gossypium hirsutum L.) Were Mapped by the BSA-Seq Technique
Abstract
1. Introduction
2. Results
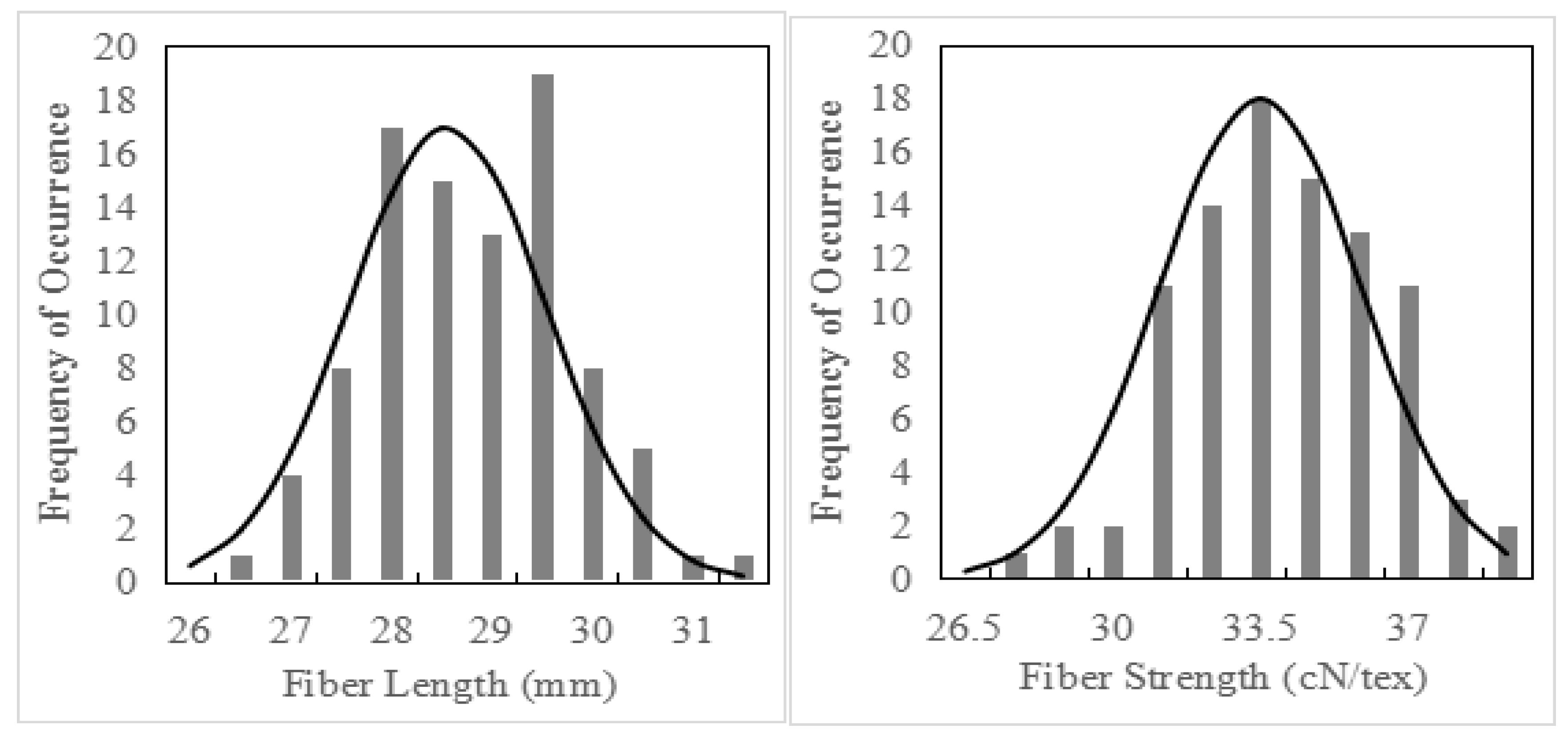
2.1. Statistical Analysis of Fiber Quality Traits
2.2. BSA-Seq Quality Assessment
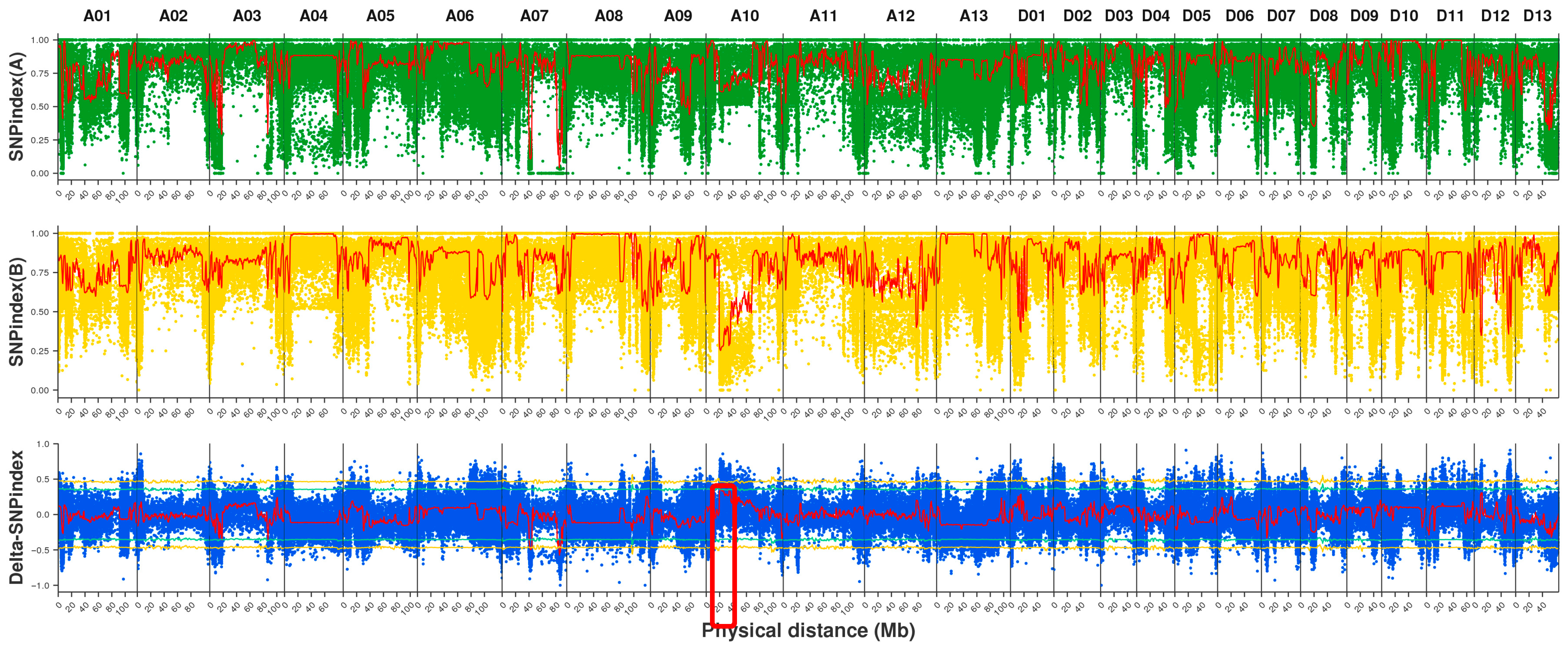
2.3. Mapping Analysis, SNP Detection, and Annotation
2.4. Candidate Region Localization and Gene Screening
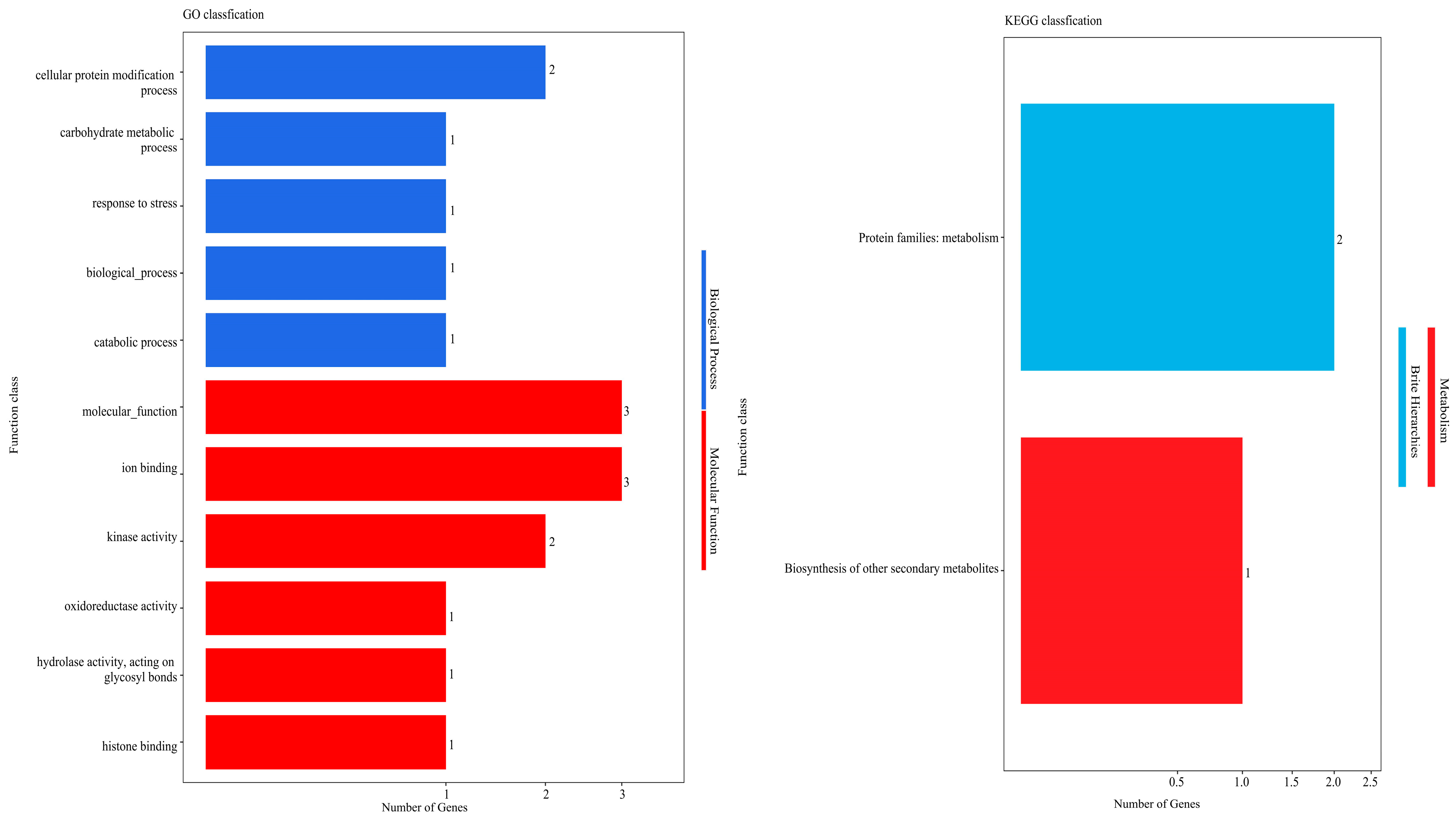
2.5. GO Classification and Enrichment Analysis
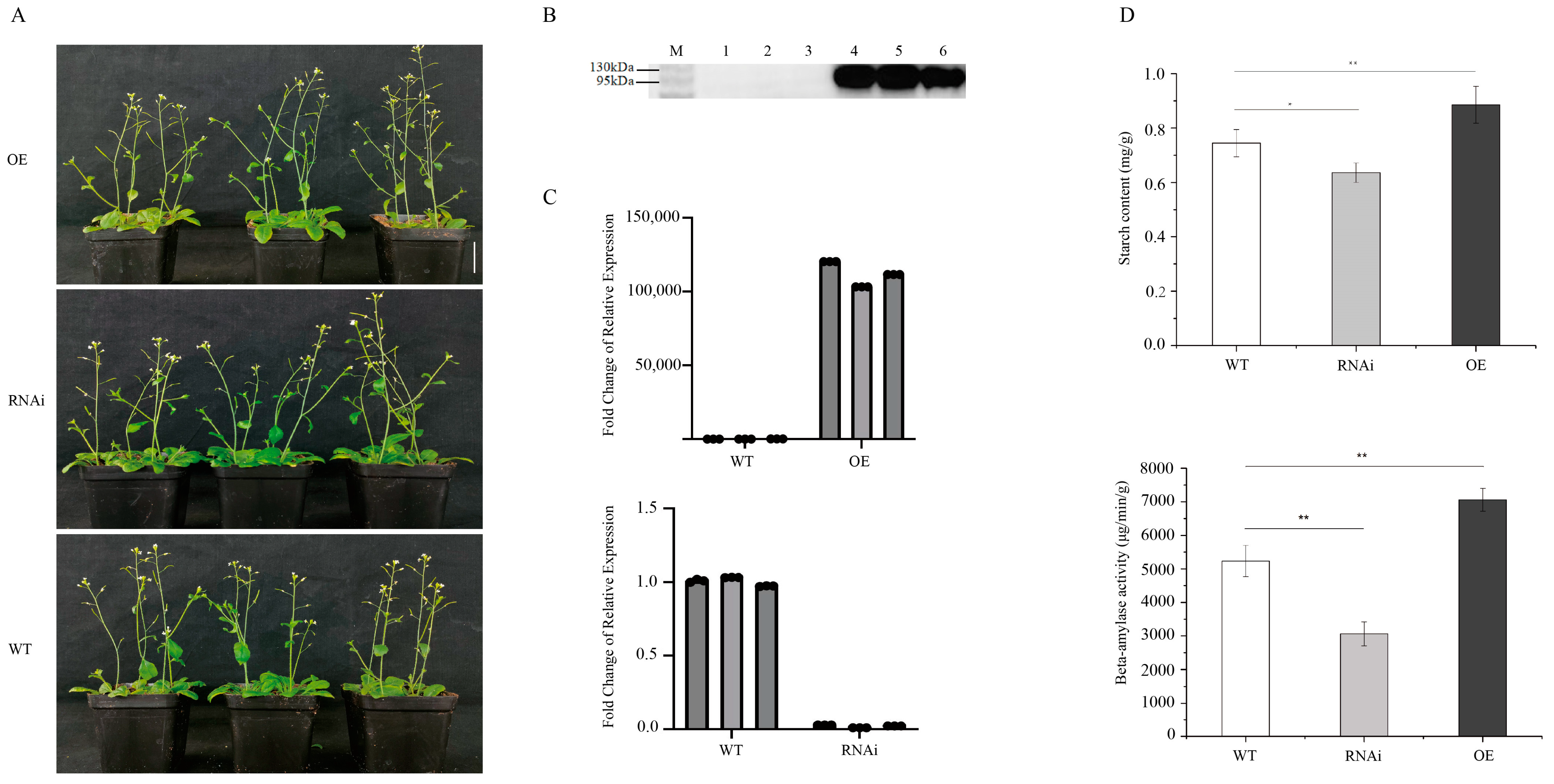
2.6. Functional Analysis of Candidate Genes GH_A10G1043 in Arabidopsis
3. Discussion
3.1. Parent Selection, Population Construction, and Fiber Quality Trait Analysis
3.2. Analysis of BSA-Seq Localization Results
3.3. GH_A10G1043 as a Candidate Gene Related to Cotton Fiber Strength
4. Materials and Methods
4.1. Plant Materials
4.2. Field Planting and Trait Investigation
4.3. BSA-Seq
4.3.1. DNA Extraction, Library Construction, and Sequencing
4.3.2. Reference Genome Alignment, SNP Detection, and Annotation
4.3.3. Candidate Region Analysis and Gene Identification
4.4. Gene Transformation and Identification
4.4.1. Arabidopsis Thaliana Transformation
4.4.2. Western Blot Analysis of Overexpression Plants
4.4.3. Quantitative RT-PCR (RT-qPCR) Validation
4.4.4. Determination of Starch Content and β-Amylase Activity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, J.; Fang, H.; Zhou, H.; Sanogo, S.; Ma, Z. Genetics, breeding, and marker-assisted selection for Verticillium wilt resistance in cotton. Crop Sci. 2014, 54, 1289–1303. [Google Scholar] [CrossRef]
- Wang, P.; He, S.; Sun, G.; Pan, Z.; Sun, J.; Geng, X.; Peng, Z.; Gong, W.; Wang, L.; Pang, B.; et al. Favorable pleiotropic loci for fiber yield and quality in upland cotton (Gossypium hirsutum). Sci. Rep. 2021, 11, 15935. [Google Scholar] [CrossRef]
- Shen, X.; Guo, W.; Zhu, X.; Yuan, Y.; Yu, J.Z.; Kohel, R.J.; Zhang, T. Molecular mapping of QTLs for fiber qualities in three diverse lines in Upland cotton using SSR markers. Mol. Breed. 2005, 15, 169–181. [Google Scholar] [CrossRef]
- Wang, M.; Tu, L.; Yuan, D.; Zhu, D.; Shen, C.; Li, J.; Liu, F.; Pei, L.; Wang, P.; Zhao, G.; et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 2019, 51, 224–229. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, J.; Fang, L.; Zhang, Z.; Ma, W.; Niu, Y.; Ju, L.; Deng, J.; Zhao, T.; Lian, J.; et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 2019, 51, 739–748. [Google Scholar] [CrossRef]
- Klein, H.; Xiao, Y.; Conklin, P.A.; Govindarajulu, R.; Kelly, J.A.; Scanlon, M.J.; Whipple, C.J.; Bartlett, M. Bulked-segregant analysis coupled to whole genome sequencing (BSA-Seq) for rapid gene cloning in maize. G3 Genes Genom. Genet. 2018, 8, 3583–3592. [Google Scholar] [CrossRef]
- Wei, T.; Zhang, L.; Zhu, R.; Jiang, X.; Yue, C.; Su, Y.; Ren, H.; Wang, M. A gain-of-function mutant of IAA7 inhibits stem elongation by transcriptional repression of EXPA5 genes in Brassica napus. Int. J. Mol. Sci. 2021, 22, 9018. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, X.; Pan, Q.; Li, P.; Liu, Y.; Lu, X.; Zhong, W.; Li, M.; Han, L.; Li, J.; et al. QTG-Seq accelerates QTL fine mapping through QTL partitioning and whole-genome sequencing of bulked segregant samples. Mol. Plant 2019, 12, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Michelmore, R.W.; Paran, I.; Kesseli, R. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 1991, 88, 9828–9832. [Google Scholar] [CrossRef]
- Zou, C.; Wang, P.; Xu, Y. Bulked sample analysis in genetics, genomics and crop improvement. Plant Biotechnol. J. 2016, 14, 1941–1955. [Google Scholar] [CrossRef] [PubMed]
- Abe, A.; Kosugi, S.; Yoshida, K.; Natsume, S.; Takagi, H.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Mitsuoka, C.; Tamiru, M.; et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 2012, 30, 174–178. [Google Scholar] [CrossRef]
- Gillmor, C.S.; Roeder, A.H.; Sieber, P.; Somerville, C.; Lukowitz, W. A genetic screen for mutations affecting cell division in the Arabidopsis thaliana embryo identifies seven loci required for cytokinesis. PLoS ONE 2016, 11, e0146492. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhu, J.; Chen, J.; Gao, F.; Xu, C.; Wu, H.; Chen, K.; Si, Z.; Yan, H.; Zhang, T. Rapid mapping and cloning of the virescent-1 gene in cotton by bulked segregant analysis–next generation sequencing and virus-induced gene silencing strategies. J. Exp. Bot. 2017, 68, 4125–4135. [Google Scholar] [CrossRef]
- Chen, Z.; Tang, D.; Hu, K.; Zhang, L.; Yin, Y.; Ni, J.; Li, P.; Wang, L.; Rong, T.; Liu, J. Combining QTL-seq and linkage mapping to uncover the genetic basis of single vs. paired spikelets in the advanced populations of two-ranked maize×teosinte. BMC Plant Biol. 2021, 21, 572. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Qi, F.; Hu, G.; Yang, Y.; Zhang, L.; Meng, J.; Han, Z.; Zhou, X.; Liu, H.; Ayaad, M.; et al. BSA-seq-based identification of a major additive plant height QTL with an effect equivalent to that of Semi-dwarf 1 in a large rice F2 population. Crop J. 2021, 9, 1428–1437. [Google Scholar] [CrossRef]
- Ochar, K.; Su, B.H.; Zhou, M.M.; Liu, Z.X.; Gao, H.W.; Lamlom, S.F.; Qiu, L.J. Identification of the genetic locus associated with the crinkled leaf phenotype in a soybean (Glycine max L.) mutant by BSA-Seq technology. J. Integr. Agr. 2022, 21, 3524–3539. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, L.; Mei, L.; Quampah, A.; He, Q.; Zhang, B.; Sun, W.; Zhang, X.; Shi, C.; Zhu, S. qOil-3, a major QTL identification for oil content in cottonseed across genomes and its candidate gene analysis. Ind. Crops Prod. 2020, 145, 112070. [Google Scholar] [CrossRef]
- Gao, J.; Shi, Y.; Wang, W.; Wang, Y.H.; Yang, H.; Shi, Q.H.; Chen, J.P.; Sun, Y.R.; Cai, L.W. Genome sequencing identified novel mechanisms underlying virescent mutation in upland cotton Gossypiuma hirsutum. BMC Genom. 2021, 22, 498. [Google Scholar] [CrossRef]
- Chen, W.; Yao, J.; Chu, L.; Yuan, Z.; Li, Y.; Zhang, Y. Genetic mapping of the nulliplex-branch gene (gb_nb1) in cotton using next-generation sequencing. Theor. Appl. Genet. 2015, 128, 539–547. [Google Scholar] [CrossRef]
- Wen, T.; Liu, C.; Wang, T.; Wang, M.; Tang, F.; He, L. Genomic mapping and identification of candidate genes encoding nulliplex-branch trait in sea-island cotton (Gossypium barbadense L.) by multi-omics analysis. Mol. Breed. 2021, 41, 34. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jia, X.; Guo, X.; Wei, H.; Zhang, M.; Wu, A.; Cheng, S.; Cheng, X.; Yu, S.; Wang, H. QTL and candidate gene identification of the node of the first fruiting branch (NFFB) by QTL-seq in upland cotton (Gossypium hirsutum L.). BMC Genom. 2021, 22, 882. [Google Scholar] [CrossRef] [PubMed]
- Chao, L.; Pan, Z.; Wang, J.; Wu, Y.; Shui, G.; Aini, N.; Tang, B.; Guo, C.; Han, P.; Shao, P.; et al. Genetic mapping and analysis of a compact plant architecture and precocious mutant in upland cotton. Plants 2022, 11, 1483. [Google Scholar] [CrossRef]
- Li, X.; Wang, W.; Wang, Z.; Li, K.; Lim, Y.P.; Piao, Z. Construction of chromosome segment substitution lines enables QTL mapping for flowering and morphological traits in Brassica rapa. Front. Plant Sci. 2015, 6, 432. [Google Scholar] [CrossRef]
- Wang, P.; Zhu, Y.; Song, X.; Cao, Z.; Ding, Y.; Liu, B.; Zhu, X.; Wang, S.; Guo, W.; Zhang, T. Inheritance of long staple fiber quality traits of Gossypium barbadense in G. hirsutum background using CSILs. Theor. Appl. Genet. 2012, 124, 1415–1428. [Google Scholar] [CrossRef]
- Yamamoto, T.; Yonemaru, J.; Yano, M. Towards the understanding of complex traits in rice: Substantially or superficially? DNA Res. 2009, 16, 141–154. [Google Scholar] [CrossRef]
- Bian, J.; He, H.; Shi, H.; Zhu, G.; Li, C.; Zhu, C.; Peng, X.; Yu, Q.; Fu, J.; He, X.; et al. Quantitative trait loci mapping for flag leaf traits in rice using a chromosome segment substitution line population. Plant Breed. 2014, 133, 203–209. [Google Scholar] [CrossRef]
- He, Q.; Yang, H.; Xiang, S.; Wang, W.; Xing, G.; Zhao, T.; Gai, J. QTL mapping for the number of branches and pods using wild chromosome segment substitution lines in soybean [Glycine max (L.) Merr.]. Plant Genet. Resour. 2014, 12 (Suppl. 1), S172–S177. [Google Scholar] [CrossRef]
- Si, Z.; Chen, H.; Zhu, X.; Cao, Z.; Zhang, T. Genetic dissection of lint yield and fiber quality traits of G. hirsutum in G. barbadense background. Mol. Breed. 2017, 37, 9. [Google Scholar] [CrossRef]
- Shi, Y.; Li, W.; Li, A.; Ge, R.; Zhang, B.; Li, J.; Liu, G.; Li, J.; Liu, A.; Shang, H.; et al. Constructing a high-density linkage map for Gossypium hirsutum × Gossypium barbadense and identifying QTLs for lint percentage. J. Integr. Plant Biol. 2015, 57, 450–467. [Google Scholar] [CrossRef]
- Stelly, D.M.; Saha, S.; Raska, D.A.; Jenkins, J.N. Registration of 17 upland (Gossypium hirsutum) cotton germplasm lines disomic for different G. barbadense chromosome or arm substitutions. Crop Sci. 2005, 45, 2663. [Google Scholar] [CrossRef]
- Luan, M.; Guo, X.; Zhang, Y.; Yao, J.; Chen, W. QTL mapping for agronomic and fibre traits using two interspecific chromosome substitution lines of Upland cotton. Plant Breed. 2009, 128, 671–679. [Google Scholar] [CrossRef]
- Shi, Y.; Zhang, B.; Liu, A.; Li, W.; Li, J.; Lu, Q.; Zhang, Z.; Li, S.; Gong, W.; Shang, H.; et al. Quantitative trait loci analysis of Verticillium wilt resistance in interspecific backcross populations of Gossypium hirsutum × Gossypium barbadense. BMC Genom. 2016, 17, 877. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Liu, A.; Li, J.; Zhang, J.; Zhang, B.; Ge, Q.; Jamshed, M.; Lu, Q.; Li, S.; Xiang, X.; et al. Dissecting the genetic basis of fiber quality and yield traits in interspecific backcross populations of Gossypium hirsutum × Gossypium barbadense. Mol. Genet. Genom. 2019, 294, 1385–1402. [Google Scholar] [CrossRef]
- Cao, Z.; Wang, P.; Zhu, X.; Chen, H.; Zhang, T. SSR marker-assisted improvement of fiber qualities in Gossypium hirsutum using G. barbadense introgression lines. Theor. Appl. Genet. 2014, 127, 587–594. [Google Scholar] [CrossRef]
- Guo, X.; Wang, H.; Wei, X.; Zhang, J.; Fu, X.; Ma, L.; Wei, H.; Yu, S. QTL mapping of fiber quality traits in two lower generation populations of upland cotton. Cotton Sci. 2021, 33, 33–41, (In Chinese with English Abstract). [Google Scholar]
- Zhu, P.; He, L.; Li, Y.; Huang, W.; Xi, F.; Lin, L.; Zhi, Q.; Zhang, W.; Tang, Y.T.; Geng, C.; et al. OTG-snpcaller: An optimized pipeline based on TMAP and GATK for SNP calling from ion torrent data. PLoS ONE 2014, 9, e97507, Erratum in PLoS ONE 2015, 10, e0139182. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from next-generation sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Yang, Y.; Sun, F.; Wang, P.; Yusuyin, M.; Kuerban, W.; Lai, C.; Li, C.; Ma, J.; Xiao, F. Genome-Wide Identification and Preliminary Functional Analysis of BAM (β-Amylase) Gene Family in Upland Cotton. Genes 2023, 14, 2077. [Google Scholar] [CrossRef]
- Shi, Y.; Liu, A.; Li, J.; Zhang, J.; Li, S.; Zhang, J.; Ma, L.; He, R.; Song, W.; Guo, L.; et al. Examining two sets of introgression lines across multiple environments reveals background-independent and stably expressed quantitative trait loci of fiber quality in cotton. Theor. Appl. Genet. 2020, 133, 2075–2093. [Google Scholar] [CrossRef]
- Zhang, K.; Yuan, M.; Xia, H.; He, L.; Ma, J.; Wang, M.; Zhao, H.; Hou, L.; Zhao, S.; Li, P.; et al. BSA-seq and genetic mapping reveals AhRt2 as a candidate gene responsible for red testa of peanut. Theor. Appl. Genet. 2022, 135, 1529–1540. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, J.; Liu, Z.; Qiu, L. Mapping of an incomplete dominant gene controlling multifoliolate leaf by BSA-seq in soybean (Glycine max L.). Acta Agron. Sin. 2020, 46, 1839–1849, (In Chinese with English Abstract). [Google Scholar]
- Takagi, H.; Tamiru, M.; Abe, A.; Yoshida, K.; Uemura, A.; Yaegashi, H.; Obara, T.; Oikawa, K.; Utsushi, H.; Kanzaki, E.; et al. MutMap accelerates breeding of a salt-tolerant rice cultivar. Nat. Biotechnol. 2015, 33, 445–449. [Google Scholar] [CrossRef]
- Lu, H.; Liu, T.; Joël, K.; Wang, S.; Qi, J.; Zhou, Q.; Sun, J.; Zhang, Z.; Weng, Y.; Huang, S. QTL-seq identifies an early flowering QTL located near flowering locus T in cucumber. Theor. Appl. Genet. 2014, 127, 1491–1499. [Google Scholar] [CrossRef]
- Illa-Berenguer, E.; Van Houten, J.; Huang, Z.; van der Knaap, E. Rapid and reliable identification of tomato fruit weight and locule number loci by QTL-seq. Theor. Appl. Genet. 2015, 128, 1329–1342. [Google Scholar] [CrossRef]
- Guo, G.; Wang, S.; Liu, J.; Pan, B.; Diao, W.; Ge, W.; Gao, C.; Snyder, J.C. Rapid identification of QTLs underlying resistance to Cucumber mosaic virus in pepper (Capsicum frutescens). Theor. Appl. Genet. 2017, 130, 41–52. [Google Scholar] [CrossRef]
- Ma, X.; Zheng, Z.; Lin, F.; Ge, T.; Sun, H. Genetic analysis and gene mapping of a low stigma exposed mutant gene by high-throughput sequencing. PLoS ONE 2018, 13, e0186942. [Google Scholar] [CrossRef]
- Zhao, C.; Zhao, G.; Geng, Z.; Wang, Z.; Wang, K.; Liu, S.; Zhang, H.; Guo, B.; Geng, J. Physical mapping and candidate gene prediction of fertility restorer gene of cytoplasmic male sterility in cotton. BMC Genom. 2018, 19, 6. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, D.; Yu, H.; Lin, B.; Hua, S.; Ding, H.; Fu, Y. Location and mapping of the determinate growth habit of Brassica napus by bulked segregant analysis (BSA) using whole genome re-sequencing. Sci. Agric. Sin. 2018, 51, 3029–3039, (In Chinese with English Abstract). [Google Scholar]
- Wang, B.; Liu, L.; Zhang, D.; Zhuang, Z.; Guo, H.; Qiao, X.; Wei, L.; Rong, J.; May, O.L.; Paterson, A.H.; et al. A genetic map between Gossypium hirsutum and the Brazilian endemic G. mustelinum and its application to QTL mapping. G3 Genes Genomes Genet. 2016, 6, 1673–1685. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Li, D.; Nie, Y.; Lin, Z. QTL mapping for yield and fiber quality traits using Gossypium mustelinum chromosome segment introgression lines. Acta Agron. Sin. 2017, 43, 1733–1745, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Miao, H.; Sun, P.; Miao, Y.; Liu, J.; Zhang, J.; Jia, C.; Wang, J.; Wang, Z.; Jin, Z.; Xu, B. Genome-wide identification and expression analysis of the β-amylase genes strongly associated with fruit development, ripening, and abiotic stress response in two banana cultivars. Front. Agric. Sci. Eng. 2016, 3, 346–356. [Google Scholar] [CrossRef]
- David, L.; Lee, S.; Bruderer, E.; Abt, M.; Fischer-Stettler, M.; Tschopp, M.; Solhaug, E.; Sanchez, K.; Zeeman, S. BETA-AMYLASE9 is a plastidial nonenzymatic regulator of leaf starch degradation. Plant Physiol. 2022, 188, 191–207. [Google Scholar] [CrossRef] [PubMed]
- Berndsen, C.; Storm, A.; Sardelli, A.; Hossain, S.; Clermont, K.; McFather, L.; Connor, M.; Monroe, J. The pseudoenzyme β-amylase 9 from Arabidopsis binds to and enhances the activity of α-amylase 3: A possible mechanism to promote stress-induced starch degradation. bioRxiv 2024. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, M.; Tian, J.; Xiao, F.; Xu, S.; Zuo, W.; Zhang, W. Improved photosynthetic capacity during the mid- and late reproductive stages contributed to increased cotton yield across four breeding eras in Xinjiang, China. Field Crops Res. 2019, 240, 177–184. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Deng, Y.; Li, J.; Wu, S.; Zhu, Y.; Chen, Y.; He, F. Integrated nr database in protein annotation system and its localization. Comput. Eng. 2006, 32, 71–72, (In Chinese with English Abstract). [Google Scholar]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32 (Suppl. 1), D277–D280. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef]
- Doitsidou, M.; Jarriault, S.; Poole, R.J. Next-generation sequencing-based approaches for mutation mapping and identification in Caenorhabditis elegans. Genetics 2016, 204, 451–474. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, T.; Yang, P.C. Western blot: Technique, theory, and trouble shooting. N. Am. J. Med. Sci. 2012, 4, 429–434. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
| Parents | Fiber Length/mm | Fiber Strength/cN·Tex−1 | Micronaire Value | Remark |
|---|---|---|---|---|
| Xinluzhong 60 | 29.10 | 33.60 | 4.80 | Gossypium hirsutum |
| Xinhai 36 | 38.70 | 46.70 | 4.50 | Gossypium barbadense |
| HC1 | 31.10 | 34.20 | 4.57 | higher fiber length and fiber strength |
| HC2 | 28.40 | 29.72 | 4.69 | lower fiber length and fiber strength |
| Mean Value | Standard Deviation | Coefficient of Variation | Skewness | Kurtosis | Maximum Value | Minimum Value | Range | |
|---|---|---|---|---|---|---|---|---|
| Fiber length (mm) | 29.56 | 1.18 | 3.98 | 0.07 | −0.38 | 32.71 | 26.73 | 5.98 |
| Fiber strength (cN/tex) | 33.69 | 2.46 | 7.32 | −0.20 | −0.19 | 38.70 | 26.70 | 12.00 |
| Sample | Mapped Reads | Total Reads | Mapping Rate (%) | Average Depth (×) | Coverage 1 (%) | Coverage 4 (%) |
|---|---|---|---|---|---|---|
| P36 | 321,236,430 | 322,838,918 | 99.50 | 16.19 | 93.44 | 88.63 |
| P60 | 324,778,375 | 325,993,176 | 99.63 | 16.75 | 99.07 | 97.62 |
| HC1 | 485,444,823 | 487,109,780 | 99.66 | 25.46 | 99.57 | 98.76 |
| HC2 | 487,003,898 | 488,691,023 | 99.65 | 25.14 | 99.60 | 98.75 |
| Reagent | 10% Separation Gel | 5% Concentrated Gel |
|---|---|---|
| H2O | 4.1 | 2.8 |
| 1.5 mol/L Tris-HCl (PH8.8) | 2.5 | - |
| 1.5 mol/L Tris-HCl (PH6.8) | - | 0.51 |
| 30% Acryamide | 3.3 | 0.67 |
| 10% SDS | 0.1 | 0.05 |
| 10% AP | 0.1 | 0.04 |
| TEMED | 0.005 | 0.003 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Sun, F.; Wei, X.; Wang, Z.; Ma, J.; Zhang, D.; Li, C.; Lai, C.; Fu, G.; Li, Y. The Genetic Loci Associated with Fiber Development in Upland Cotton (Gossypium hirsutum L.) Were Mapped by the BSA-Seq Technique. Plants 2025, 14, 2804. https://doi.org/10.3390/plants14172804
Yang Y, Sun F, Wei X, Wang Z, Ma J, Zhang D, Li C, Lai C, Fu G, Li Y. The Genetic Loci Associated with Fiber Development in Upland Cotton (Gossypium hirsutum L.) Were Mapped by the BSA-Seq Technique. Plants. 2025; 14(17):2804. https://doi.org/10.3390/plants14172804
Chicago/Turabian StyleYang, Yanlong, Fenglei Sun, Xin Wei, Zhengzheng Wang, Jun Ma, Dawei Zhang, Chunping Li, Chengxia Lai, Guoyong Fu, and Youzhong Li. 2025. "The Genetic Loci Associated with Fiber Development in Upland Cotton (Gossypium hirsutum L.) Were Mapped by the BSA-Seq Technique" Plants 14, no. 17: 2804. https://doi.org/10.3390/plants14172804
APA StyleYang, Y., Sun, F., Wei, X., Wang, Z., Ma, J., Zhang, D., Li, C., Lai, C., Fu, G., & Li, Y. (2025). The Genetic Loci Associated with Fiber Development in Upland Cotton (Gossypium hirsutum L.) Were Mapped by the BSA-Seq Technique. Plants, 14(17), 2804. https://doi.org/10.3390/plants14172804





