Genome-Wide Identification and Expression Analysis of the Melon B-BOX (BBX) Gene Family in Response to Abiotic and Biotic Stresses
Abstract
1. Introduction
2. Results
2.1. Identification of BBX Genes in Melon
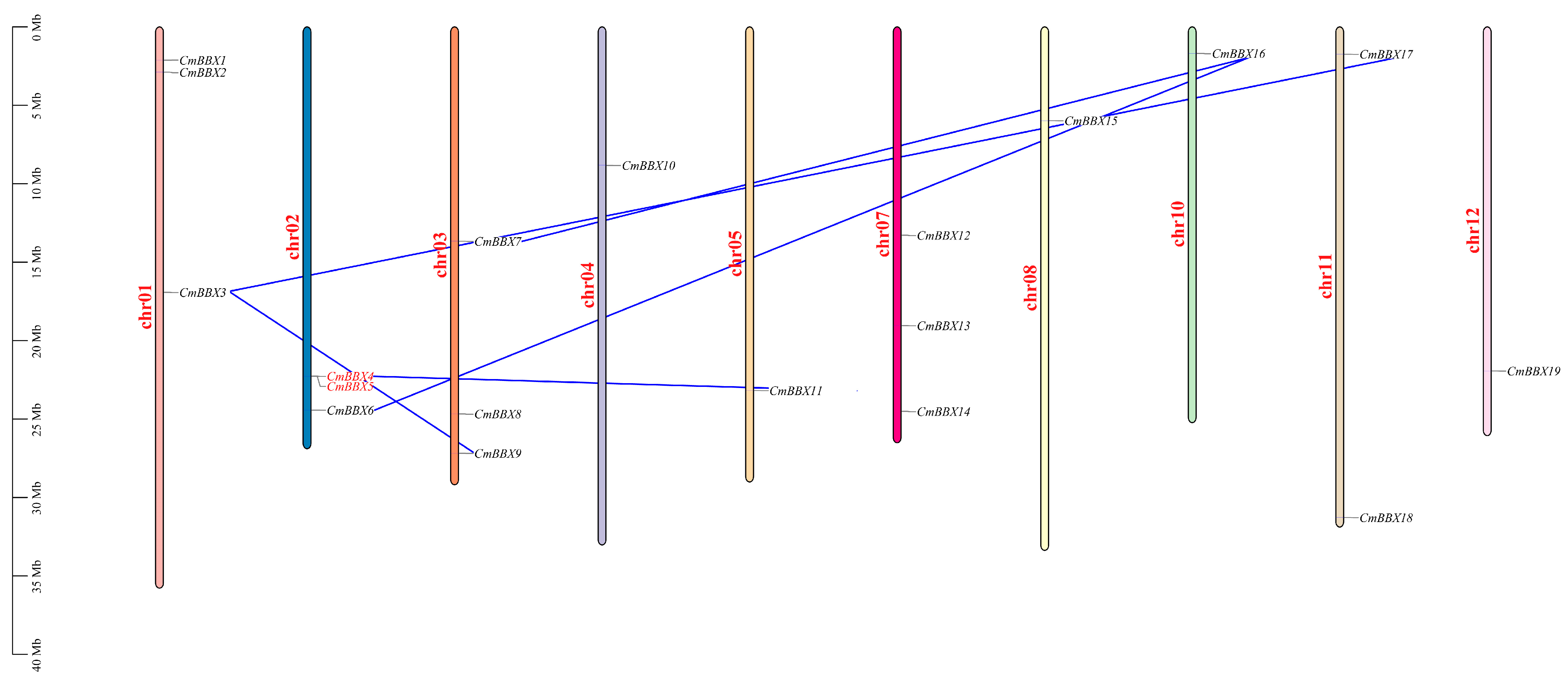
2.2. Chromosomal Location of CmBBXs
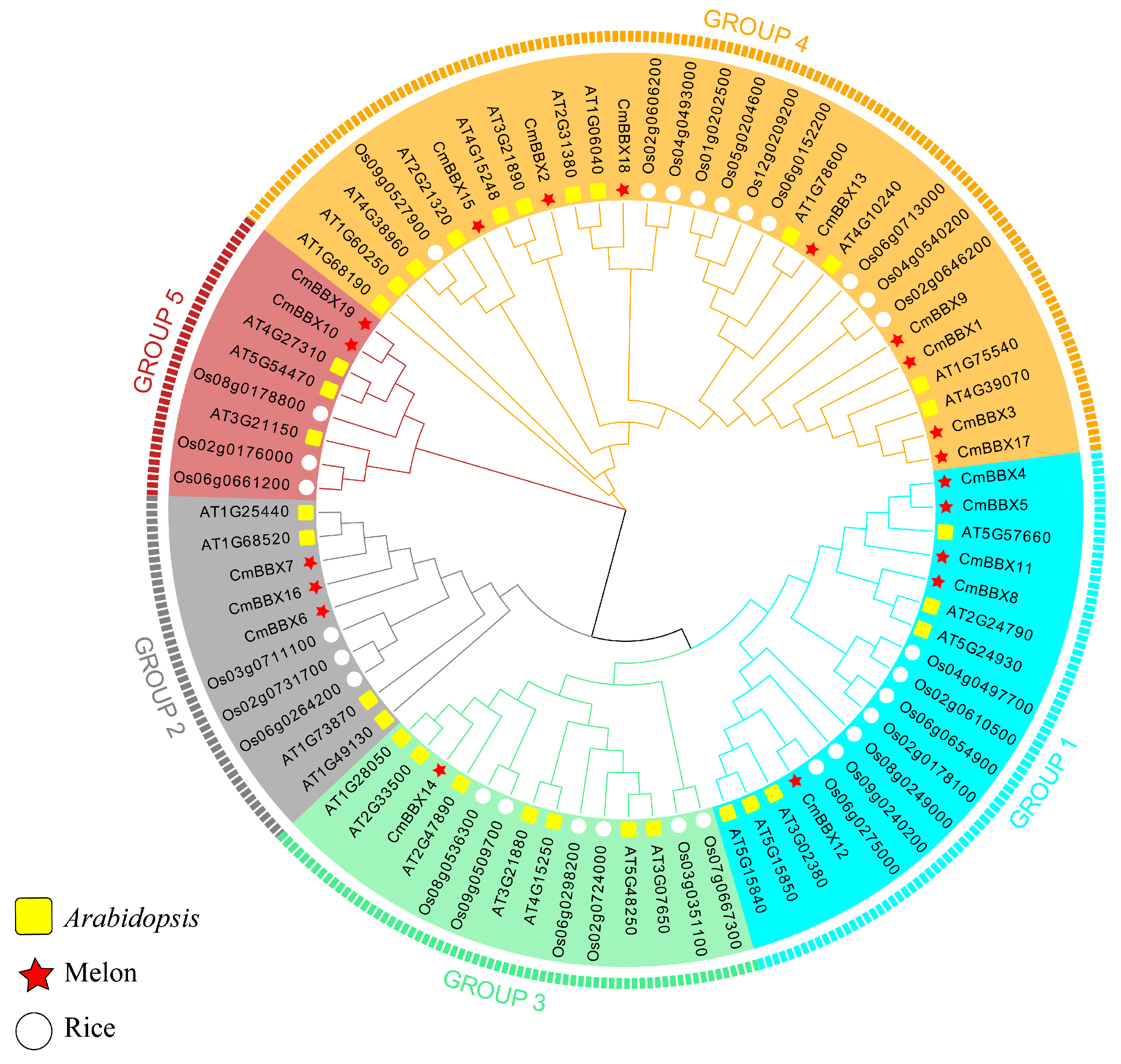
2.3. Phylogenetic Tree Analysis of BBX Family Genes
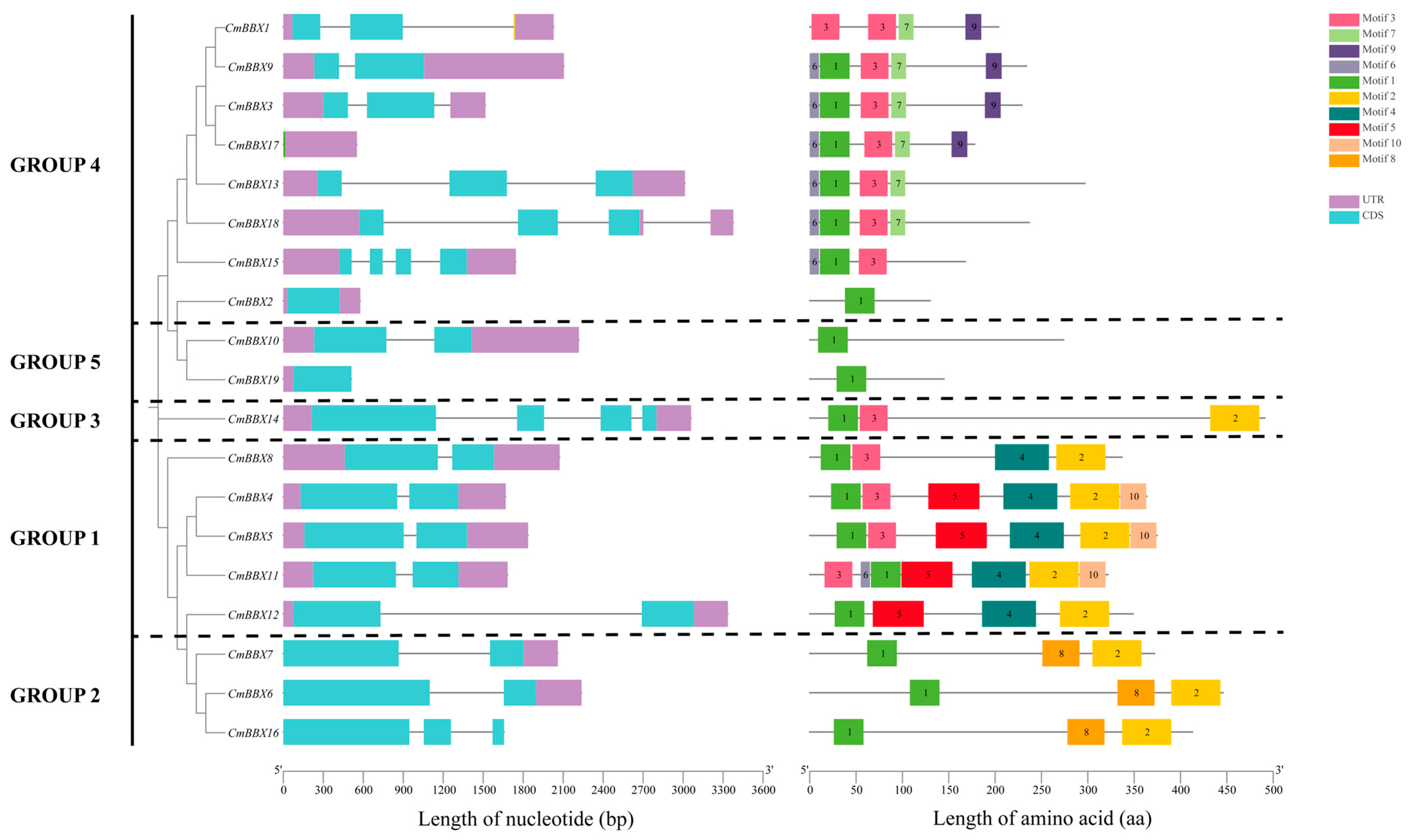
2.4. Gene Structure and Conserved Motif Analysis of CmBBXs
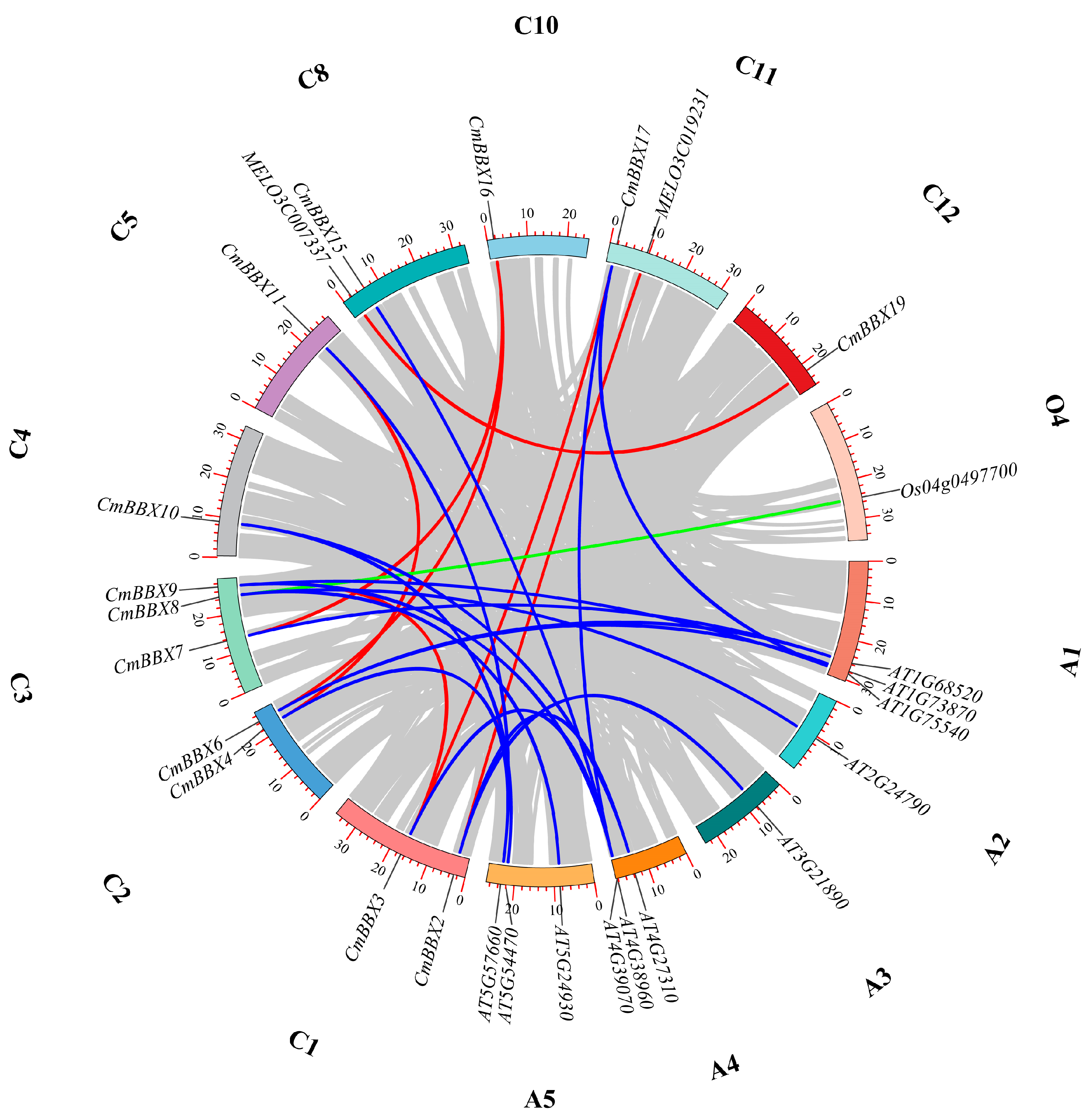
2.5. Gene Duplication and Synteny Analysis
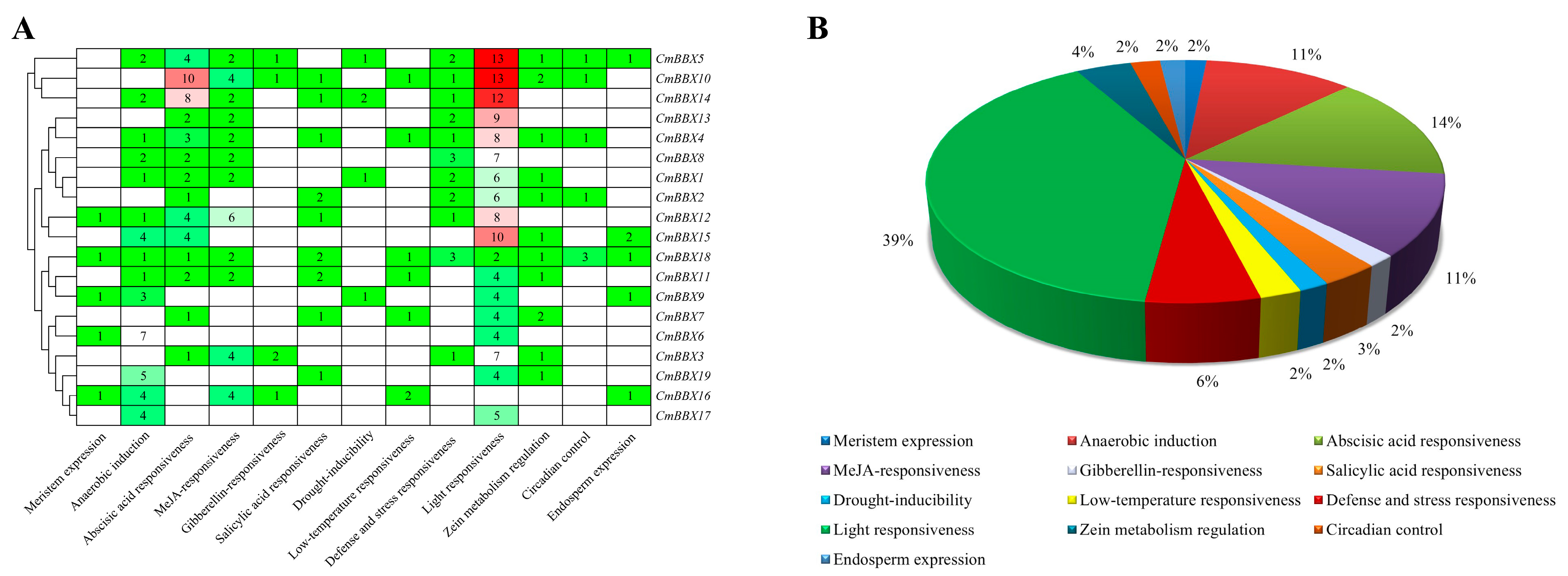
2.6. Analysis of the Cis-Acting Elements in CmBBXs
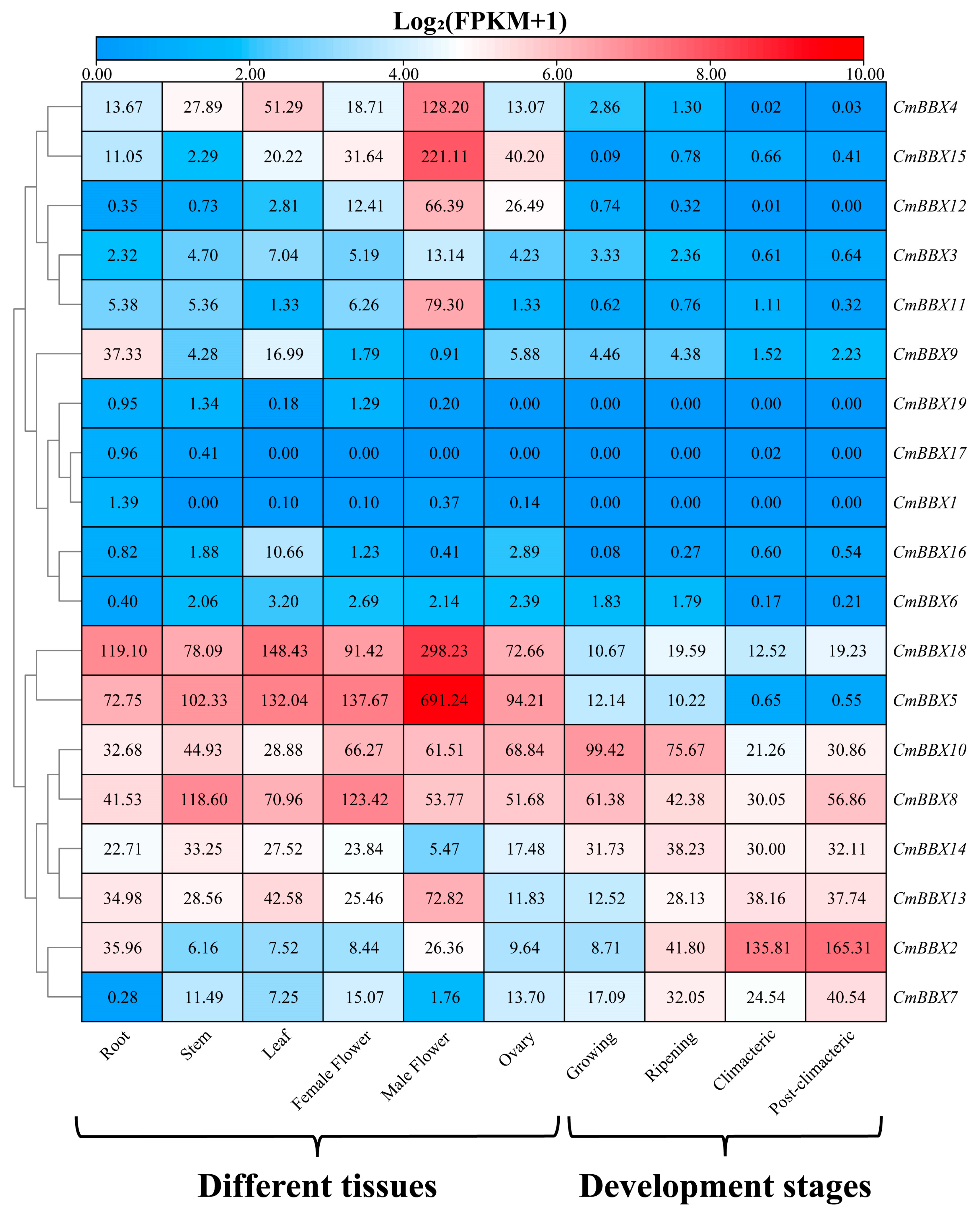
2.7. Tissue-Specific Expression Analysis of CmBBXs
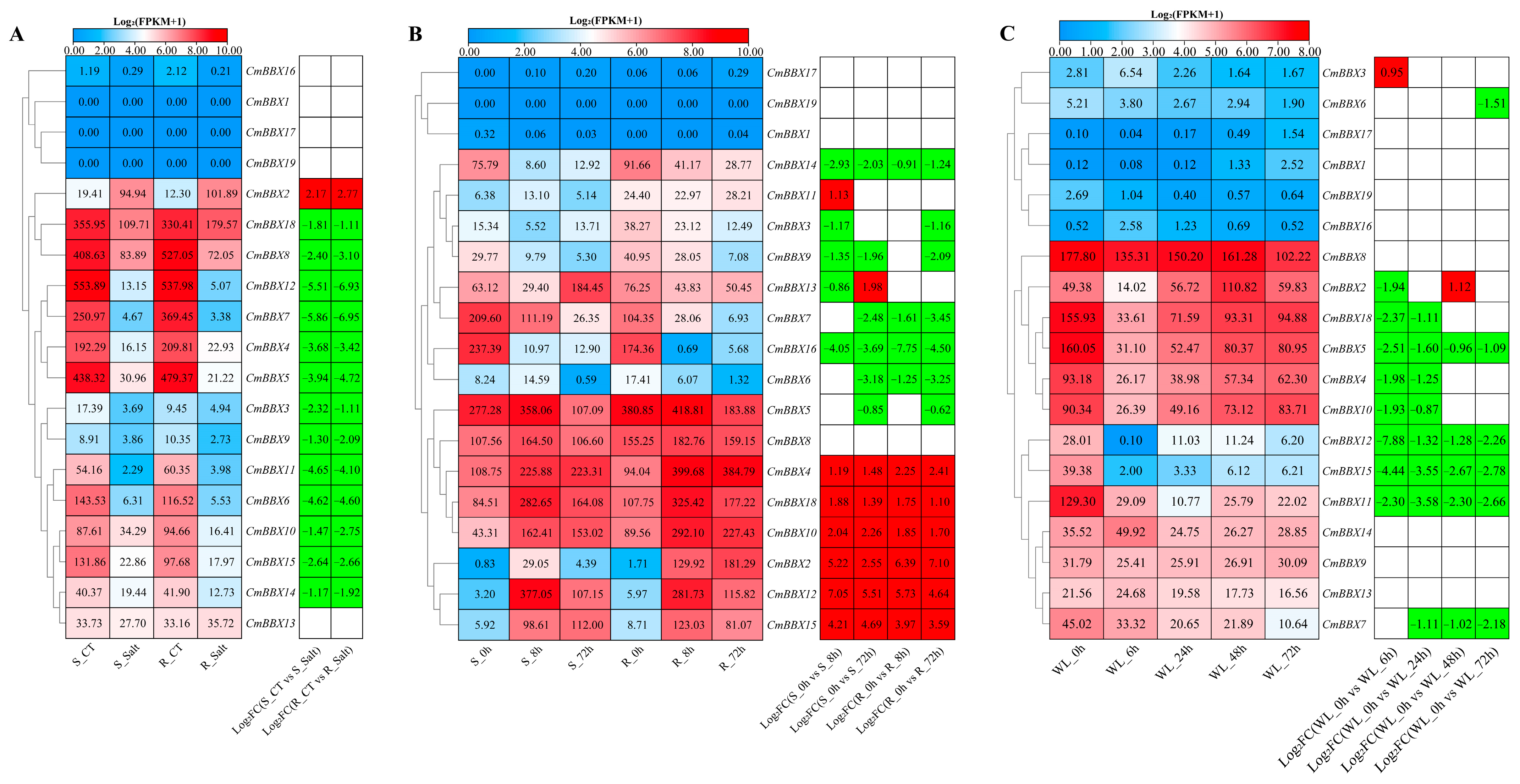
2.8. Expression Patterns Analysis of CmBBXs Under Abiotic Stresses
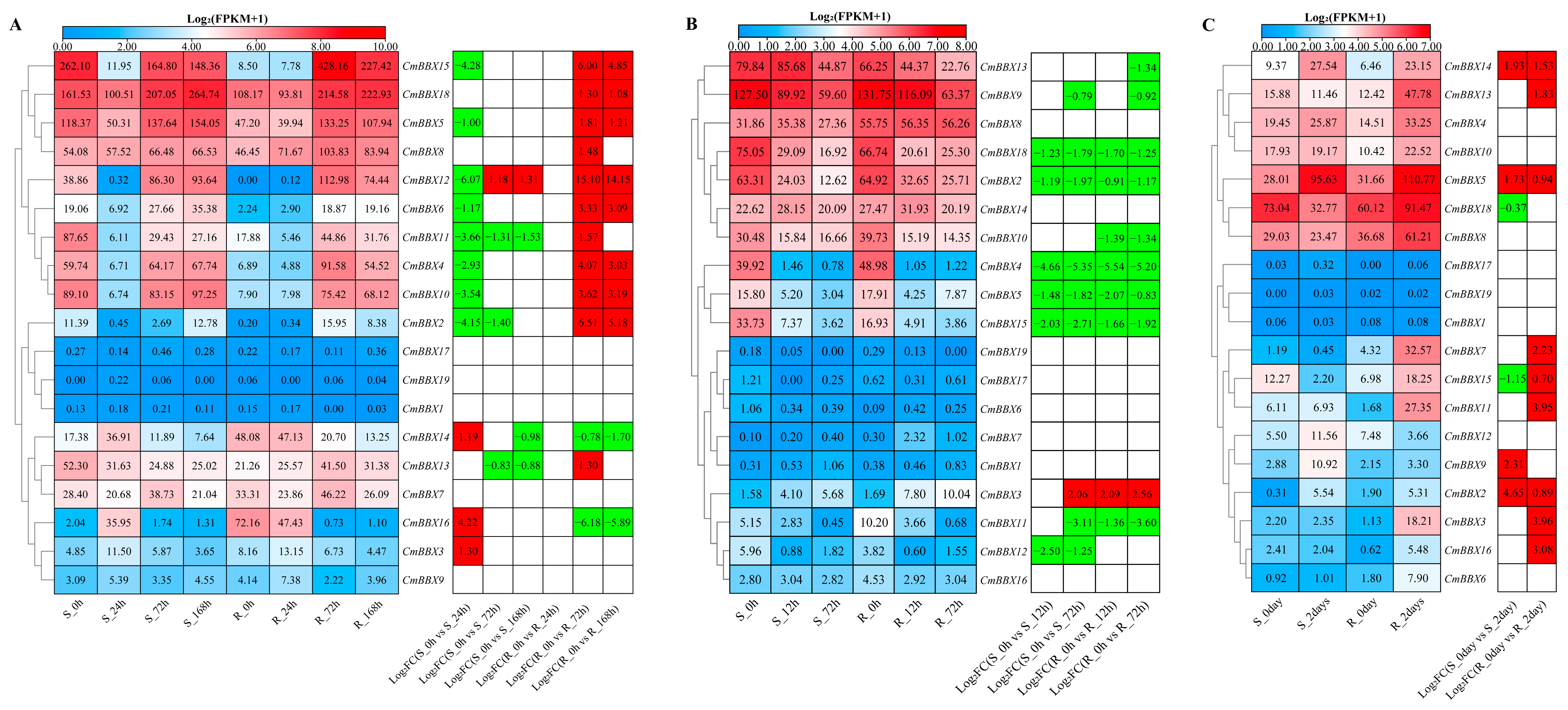
2.9. Expression Patterns Analysis of CmBBXs Under Biotic Stresses
2.10. Regulation Patterns of CmBBXs Under Stresses
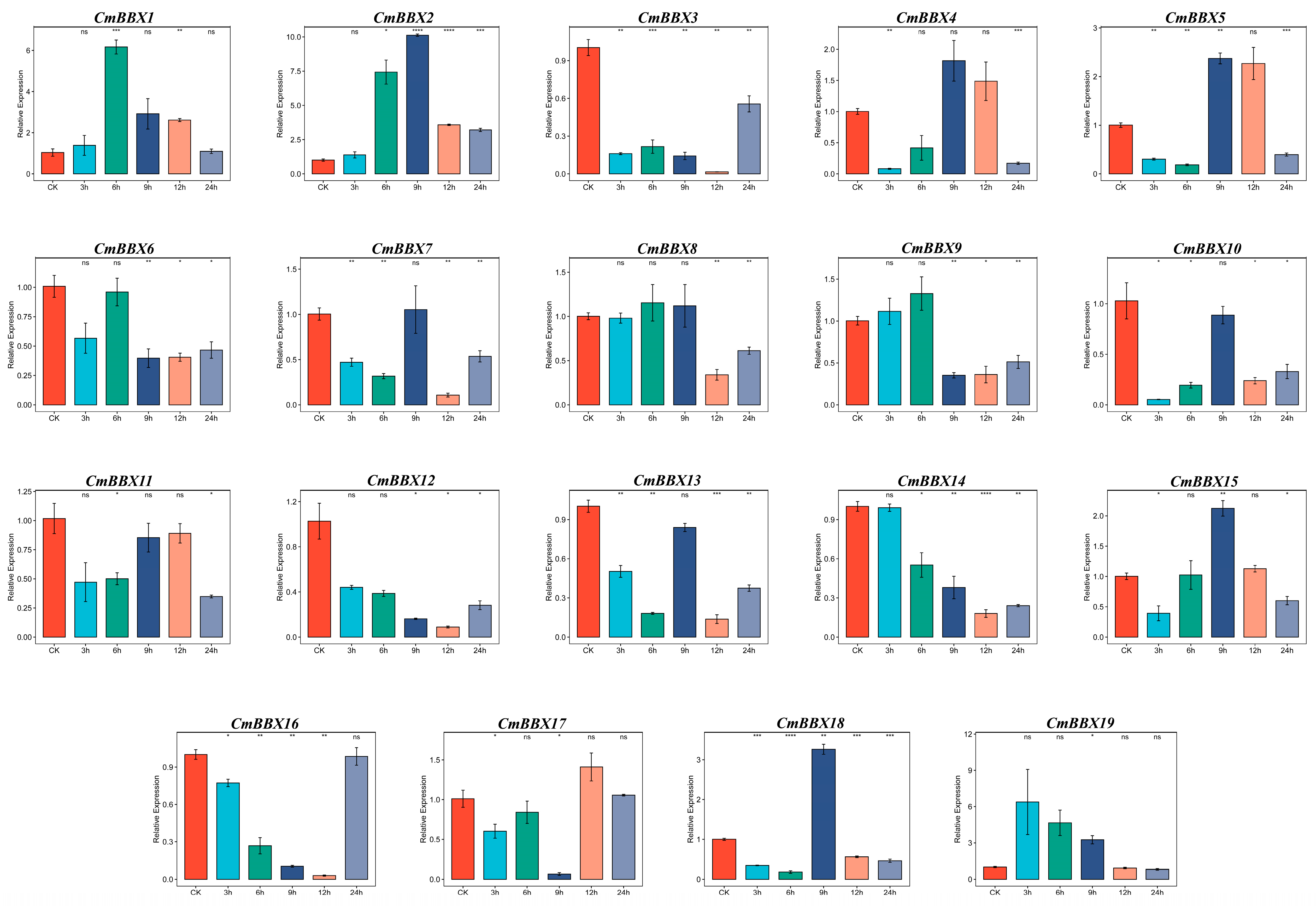
2.11. RT-qPCR Analysis of the CmBBXs
3. Discussion
4. Materials and Methods
4.1. Identification and Chromosomal Distribution of Melon BBX Gene Family Members
4.2. Analysis of the BBX Gene Family Characteristics and Phylogenetic Evolution in Melons
4.3. Synteny Analysis of BBX Family Genes from Melon, A. thaliana, and Rice
4.4. Tissue-Specific Expression of Melon BBX Family Genes
4.5. Analysis of Expression Patterns of Melon BBX Gene Family Under Various Stresses
4.6. RT-qPCR Analysis of Melon Under Salt Stress
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Strader, L.; Weijers, D.; Wagner, D. Plant transcription factors—Being in the right place with the right company. Curr. Opin. Plant Biol. 2022, 65, 102136. [Google Scholar] [CrossRef]
- Tian, F.; Yang, D.C.; Meng, Y.Q.; Jin, J.; Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2020, 48, D1104–D1113. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Noman, A.; Aqeel, M.; Khalid, N.; Islam, W.; Sanaullah, T.; Anwar, M.; Khan, S.; Ye, W.; Lou, Y. Zinc finger protein transcription factors: Integrated line of action for plant antimicrobial activity. Microb. Pathog. 2019, 132, 141–149. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Liao, J.; Chen, Q.; Jin, W.; Li, S.; Zhu, T.; Li, S. Identification and Characterization of the BBX Gene Family in Bambusa pervariabilis x Dendrocalamopsis grandis and Their Potential Role under Adverse Environmental Stresses. Int. J. Mol. Sci. 2023, 24, 13465. [Google Scholar] [CrossRef] [PubMed]
- Khanna, R.; Kronmiller, B.; Maszle, D.R.; Coupland, G.; Holm, M.; Mizuno, T.; Wu, S.H. The Arabidopsis B-box zinc finger family. Plant Cell 2009, 21, 3416–3420. [Google Scholar] [CrossRef] [PubMed]
- Putterill, J.; Robson, F.; Lee, K.; Simon, R.; Coupland, G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 1995, 80, 847–857. [Google Scholar] [CrossRef]
- Lyu, G.; Li, D.; Li, S. Bioinformatics analysis of BBX family genes and its response to UV-B in Arabidopsis thaliana. Plant Signal. Behav. 2020, 15, 1782647. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, X.; Weng, X.; Wang, L.; Xie, W. The rice B-box zinc finger gene family: Genomic identification, characterization, expression profiling and diurnal analysis. PLoS ONE 2012, 7, e48242. [Google Scholar] [CrossRef]
- Obel, H.O.; Cheng, C.; Li, Y.; Tian, Z.; Njogu, M.K.; Li, J.; Lou, Q.; Yu, X.; Yang, Z.; Ogweno, J.O.; et al. Genome-Wide Identification of the B-Box Gene Family and Expression Analysis Suggests Their Potential Role in Photoperiod-Mediated beta-Carotene Accumulation in the Endocarp of Cucumber (Cucumis sativus L.) Fruit. Genes 2022, 13, 658. [Google Scholar] [CrossRef]
- Shan, B.; Bao, G.; Shi, T.; Zhai, L.; Bian, S.; Li, X. Genome-wide identification of BBX gene family and their expression patterns under salt stress in soybean. BMC Genom. 2022, 23, 820. [Google Scholar] [CrossRef]
- Wang, X.; Guo, H.; Jin, Z.; Ding, Y.; Guo, M. Comprehensive Characterization of B-Box Zinc Finger Genes in Citrullus lanatus and Their Response to Hormone and Abiotic Stresses. Plants 2023, 12, 2634. [Google Scholar] [CrossRef] [PubMed]
- Chu, Z.; Wang, X.; Li, Y.; Yu, H.; Li, J.; Lu, Y.; Li, H.; Ouyang, B. Genomic Organization, Phylogenetic and Expression Analysis of the B-BOX Gene Family in Tomato. Front. Plant Sci. 2016, 7, 1552. [Google Scholar] [CrossRef]
- Ma, R.; Chen, J.; Huang, B.; Huang, Z.; Zhang, Z. The BBX gene family in Moso bamboo (Phyllostachys edulis): Identification, characterization and expression profiles. BMC Genom. 2021, 22, 533. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, H.; Huang, J.; Shi, T.; Meng, Z.; Chen, Q.; Deng, J. Genome-wide analysis of BBX gene family in Tartary buckwheat (Fagopyrum tataricum). PeerJ 2021, 9, e11939. [Google Scholar] [CrossRef]
- Bai, B.; Lu, N.; Li, Y.; Guo, S.; Yin, H.; He, Y.; Sun, W.; Li, W.; Xie, X. OsBBX14 promotes photomorphogenesis in rice by activating OsHY5L1 expression under blue light conditions. Plant Sci. 2019, 284, 192–202. [Google Scholar] [CrossRef]
- Liu, Y.; Cheng, H.; Cheng, P.; Wang, C.; Li, J.; Liu, Y.; Song, A.; Chen, S.; Chen, F.; Wang, L.; et al. The BBX gene CmBBX22 negatively regulates drought stress tolerance in chrysanthemum. Hortic. Res. 2022, 9, uhac181. [Google Scholar] [CrossRef]
- Yan, H.; Marquardt, K.; Indorf, M.; Jutt, D.; Kircher, S.; Neuhaus, G.; Rodriguez-Franco, M. Nuclear localization and interaction with COP1 are required for STO/BBX24 function during photomorphogenesis. Plant Physiol. 2011, 156, 1772–1782. [Google Scholar] [CrossRef]
- Tian, Z.; Han, J.; Che, G.; Hasi, A. Genome-wide characterization and expression analysis of SAUR gene family in Melon (Cucumis melo L.). Planta 2022, 255, 123. [Google Scholar] [CrossRef]
- Castanera, R.; Ruggieri, V.; Pujol, M.; Garcia-Mas, J.; Casacuberta, J.M. An Improved Melon Reference Genome with Single-Molecule Sequencing Uncovers a Recent Burst of Transposable Elements with Potential Impact on Genes. Front. Plant Sci. 2019, 10, 1815. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chen, J.; Zhang, F.; Song, Y. Transcriptome analysis of callus from melon. Gene 2019, 684, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Slovin, S.; Carissimo, A.; Panariello, F.; Grimaldi, A.; Bouche, V.; Gambardella, G.; Cacchiarelli, D. Single-Cell RNA Sequencing Analysis: A Step-by-Step Overview. Methods Mol. Biol. 2021, 2284, 343–365. [Google Scholar] [CrossRef]
- Imadi, S.R.; Kazi, A.G.; Ahanger, M.A.; Gucel, S.; Ahmad, P. Plant transcriptomics and responses to environmental stress: An overview. J. Genet. 2015, 94, 525–537. [Google Scholar] [CrossRef]
- Ling, Y.; Xiong, X.; Yang, W.; Liu, B.; Shen, Y.; Xu, L.; Lu, F.; Li, M.; Guo, Y.; Zhang, X. Comparative Analysis of Transcriptomics and Metabolomics Reveals Defense Mechanisms in Melon Cultivars against Pseudoperonospora cubensis Infection. Int. J. Mol. Sci. 2023, 24, 17552. [Google Scholar] [CrossRef]
- Yang, D.; Chen, H.; Zhang, Y.; Wang, Y.; Zhai, Y.; Xu, G.; Ding, Q.; Wang, M.; Zhang, Q.; Lu, X.; et al. Genome-Wide Identification and Expression Analysis of the Melon Aldehyde Dehydrogenase (ALDH) Gene Family in Response to Abiotic and Biotic Stresses. Plants 2024, 13, 2939. [Google Scholar] [CrossRef]
- Wu, B.; Wang, L.; Pan, G.; Li, T.; Li, X.; Hao, J. Genome-wide characterization and expression analysis of the auxin response factor (ARF) gene family during melon (Cucumis melo L.) fruit development. Protoplasma 2020, 257, 979–992. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yadav, V.; Yan, X.; Cheng, D.; Wei, C.; Zhang, X. Systematic genome-wide analysis of the ethylene-responsive ACS gene family: Contributions to sex form differentiation and development in melon and watermelon. Gene 2021, 805, 145910. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Dai, Y.; Noman, M.; Li, R.; Li, X.; Wu, X.; Wang, H.; Song, F.; Li, D. Genome-wide characterization and functional analysis of the melon TGA gene family in disease resistance through ectopic overexpression in Arabidopsis thaliana. Plant Physiol. Biochem. 2024, 212, 108784. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Botto, J.F. The BBX family of plant transcription factors. Trends Plant Sci. 2014, 19, 460–470. [Google Scholar] [CrossRef]
- Wu, Z.; Fu, D.; Gao, X.; Zeng, Q.; Chen, X.; Wu, J.; Zhang, N. Characterization and expression profiles of the B-box gene family during plant growth and under low-nitrogen stress in Saccharum. BMC Genom. 2023, 24, 79. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, Y.; Yu, T.; Li, J.; Qiu, X.; Zhu, C.; Liu, J.; Dang, F.; Yang, Y. Characterization of the B-BOX gene family in pepper and the role of CaBBX14 in defense response against Phytophthora capsici infection. Int. J. Biol. Macromol. 2023, 237, 124071. [Google Scholar] [CrossRef]
- Zhang, S.; Chen, C.; Li, L.; Meng, L.; Singh, J.; Jiang, N.; Deng, X.; He, Z.; Lemaux, P.G. Evolutionary expansion, gene structure, and expression of the rice wall-associated kinase gene family. Plant Physiol. 2005, 139, 1107–1124. [Google Scholar] [CrossRef]
- Qu, R.; Cao, Y.; Tang, X.; Sun, L.; Wei, L.; Wang, K. Identification and expression analysis of the WRKY gene family in Isatis indigotica. Gene 2021, 783, 145561. [Google Scholar] [CrossRef]
- Lee, J. The Principles and Applications of High-Throughput Sequencing Technologies. Dev. Reprod. 2023, 27, 9–24. [Google Scholar] [CrossRef]
- Chen, T.Y.; You, L.; Hardillo, J.A.U.; Chien, M.P. Spatial Transcriptomic Technologies. Cells 2023, 12, 2042. [Google Scholar] [CrossRef]
- Tholl, D. Biosynthesis and biological functions of terpenoids in plants. Adv. Biochem. Eng. Biotechnol. 2015, 148, 63–106. [Google Scholar] [CrossRef] [PubMed]
- Hurgobin, B.; Lewsey, M.G. Applications of cell- and tissue-specific ‘omics to improve plant productivity. Emerg. Top. Life Sci. 2022, 6, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Coruh, C.; Xu, G.; Martins, L.M.; Bourbousse, C.; Lambolez, A.; Law, J.A. The CLASSY family controls tissue-specific DNA methylation patterns in Arabidopsis. Nat. Commun. 2022, 13, 244. [Google Scholar] [CrossRef] [PubMed]
- Walsh, J.R.; Woodhouse, M.R.; Andorf, C.M.; Sen, T.Z. Tissue-specific gene expression and protein abundance patterns are associated with fractionation bias in maize. BMC Plant Biol. 2020, 20, 4. [Google Scholar] [CrossRef]
- Li, W.; Xiong, R.; Chu, Z.; Peng, X.; Cui, G.; Dong, L. Transcript-Wide Identification and Characterization of the BBX Gene Family in Trichosanthes kirilowii and Its Potential Roles in Development and Abiotic Stress. Plants 2025, 14, 975. [Google Scholar] [CrossRef]
- Ouyang, Y.; Pan, X.; Wei, Y.; Wang, J.; Xu, X.; He, Y.; Zhang, X.; Li, Z.; Zhang, H. Genome-wide identification and characterization of the BBX gene family in pineapple reveals that candidate genes are involved in floral induction and flowering. Genomics 2022, 114, 110397. [Google Scholar] [CrossRef]
- Koellhoffer, J.P.; Xing, A.; Moon, B.P.; Li, Z. Tissue-specific expression of a soybean hypersensitive-induced response (HIR) protein gene promoter. Plant Mol. Biol. 2015, 87, 261–271. [Google Scholar] [CrossRef]
- Baba-Kasai, A.; Hara, N.; Takano, M. Tissue-specific and light-dependent regulation of phytochrome gene expression in rice. Plant Cell Environ. 2014, 37, 2654–2666. [Google Scholar] [CrossRef]
- Li, Y.; Tong, Y.; Ye, J.; Zhang, C.; Li, B.; Hu, S.; Xue, X.; Tian, Q.; Wang, Y.; Li, L.; et al. Genome-Wide Characterization of B-Box Gene Family in Salvia miltiorrhiza. Int. J. Mol. Sci. 2023, 24, 2146. [Google Scholar] [CrossRef]
- Song, K.; Li, B.; Wu, H.; Sha, Y.; Qin, L.; Chen, X.; Liu, Y.; Tang, H.; Yang, L. The Function of BBX Gene Family under Multiple Stresses in Nicotiana tabacum. Genes 2022, 13, 1841. [Google Scholar] [CrossRef]
- Nakaminami, K.; Seki, M. RNA Regulation in Plant Cold Stress Response. Adv. Exp. Med. Biol. 2018, 1081, 23–44. [Google Scholar] [CrossRef]
- Hu, J.; Cai, J.; Xu, T.; Kang, H. Epitranscriptomic mRNA modifications governing plant stress responses: Underlying mechanism and potential application. Plant Biotechnol. J. 2022, 20, 2245–2257. [Google Scholar] [CrossRef]
- Kwon, C.; Lee, J.H.; Yun, H.S. SNAREs in Plant Biotic and Abiotic Stress Responses. Mol. Cells 2020, 43, 501–508. [Google Scholar] [CrossRef] [PubMed]
- Strzemski, M.; Dresler, S. Impact of Biotic/Abiotic Stress Factors on Plant Specialized Metabolites. Int. J. Mol. Sci. 2024, 25, 5742. [Google Scholar] [CrossRef] [PubMed]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef]
- Yu, J.; Wu, S.; Sun, H.; Wang, X.; Tang, X.; Guo, S.; Zhang, Z.; Huang, S.; Xu, Y.; Weng, Y.; et al. CuGenDBv2: An updated database for cucurbit genomics. Nucleic Acids Res. 2023, 51, D1457–D1464. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, W327–W331. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, J.; Paterson, A.H. MCScanX-transposed: Detecting transposed gene duplications based on multiple colinearity scans. Bioinformatics 2013, 29, 1458–1460. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Tian, Y.; Bai, S.; Dang, Z.; Hao, J.; Zhang, J.; Hasi, A. Genome-wide identification and characterization of long non-coding RNAs involved in fruit ripening and the climacteric in Cucumis melo. BMC Plant Biol. 2019, 19, 369. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, G.; Yan, C.; Cao, N.; Yang, H.; Le, M.; Zhu, F. Depicting the molecular responses of adventitious rooting to waterlogging in melon hypocotyls by transcriptome profiling. 3 Biotech 2021, 11, 351. [Google Scholar] [CrossRef]
- Zhu, Q.; Gao, P.; Wan, Y.; Cui, H.; Fan, C.; Liu, S.; Luan, F. Comparative transcriptome profiling of genes and pathways related to resistance against powdery mildew in two contrasting melon genotypes. Sci. Hortic. 2018, 227, 169–180. [Google Scholar] [CrossRef]
- Yang, T.; Liu, J.; Li, X.; Amanullah, S.; Lu, X.; Zhang, M.; Zhang, Y.; Luan, F.; Liu, H.; Wang, X. Transcriptomic Analysis of Fusarium oxysporum Stress-Induced Pathosystem and Screening of Fom-2 Interaction Factors in Contrasted Melon Plants. Front. Plant Sci. 2022, 13, 961586. [Google Scholar] [CrossRef]
- Intana, W.; Wonglom, P.; Suwannarach, N.; Sunpapao, A. Trichoderma asperelloides PSU-P1 Induced Expression of Pathogenesis-Related Protein Genes against Gummy Stem Blight of Muskmelon (Cucumis melo) in Field Evaluation. J. Fungi 2022, 8, 156. [Google Scholar] [CrossRef] [PubMed]

| Gene ID | CDS Size (bp) | Number of Amino Acid (aa) | Molecular Weight (kDa) | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Prediction of Subcellular Location |
|---|---|---|---|---|---|---|---|---|
| MELO3C018684/CmBBX1 | 615 | 204 | 22.70 | 5.96 | 43.51 | 66.57 | −0.419 | Nuclear |
| MELO3C018801/CmBBX2 | 393 | 130 | 14.57 | 7.48 | 34.68 | 76.46 | −0.285 | Extracellular |
| MELO3C013430/CmBBX3 | 690 | 229 | 24.92 | 6.12 | 50.08 | 65.33 | −0.399 | Nuclear |
| MELO3C017501/CmBBX4 | 1095 | 364 | 40.13 | 6.01 | 43.97 | 71.48 | −0.302 | Nuclear |
| MELO3C017500/CmBBX5 | 1128 | 375 | 41.10 | 5.96 | 44.18 | 67.04 | −0.351 | Nuclear |
| MELO3C017304/CmBBX6 | 1341 | 446 | 51.38 | 5.86 | 51.61 | 69.08 | −0.708 | Nuclear |
| MELO3C011576/CmBBX7 | 1119 | 372 | 42.68 | 8.35 | 55.04 | 66.80 | −0.8 | Nuclear |
| MELO3C011317/CmBBX8 | 1014 | 337 | 36.88 | 6.08 | 43.97 | 69.23 | −0.494 | Nuclear |
| MELO3C010985/CmBBX9 | 705 | 234 | 24.95 | 6.69 | 48.46 | 85.85 | −0.051 | Extracellular |
| MELO3C018174/CmBBX10 | 825 | 274 | 30.04 | 4.35 | 61.29 | 52.66 | −0.839 | Nuclear |
| MELO3C004050/CmBBX11 | 969 | 322 | 35.47 | 8.22 | 51.92 | 65.47 | −0.397 | Nuclear |
| MELO3C018930/CmBBX12 | 1050 | 349 | 39.05 | 5.23 | 43.96 | 61.26 | −0.805 | Nuclear |
| MELO3C016115/CmBBX13 | 894 | 297 | 32.36 | 5.22 | 55.88 | 62.42 | −0.452 | Nuclear |
| MELO3C017755/CmBBX14 | 1476 | 491 | 54.32 | 5.58 | 42.34 | 67.54 | −0.56 | Nuclear |
| MELO3C007886/CmBBX15 | 507 | 168 | 18.85 | 6.59 | 55.32 | 72.50 | −0.576 | Extracellular |
| MELO3C012248/CmBBX16 | 1242 | 413 | 46.28 | 5.51 | 41.04 | 63.34 | −0.708 | Nuclear |
| MELO3C023341/CmBBX17 | 537 | 178 | 19.44 | 7.01 | 62.73 | 75.11 | −0.25 | Nuclear |
| MELO3C022445/CmBBX18 | 714 | 237 | 26.05 | 4.95 | 43.37 | 74.14 | −0.29 | Extracellular |
| MELO3C002548/CmBBX19 | 438 | 145 | 16.14 | 5.58 | 55.01 | 65.86 | −0.559 | Nuclear |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Li, Y.; Wang, Y.; Yan, C.; Yang, D.; Xing, Y.; Lu, X. Genome-Wide Identification and Expression Analysis of the Melon B-BOX (BBX) Gene Family in Response to Abiotic and Biotic Stresses. Plants 2025, 14, 2715. https://doi.org/10.3390/plants14172715
Zhang Y, Li Y, Wang Y, Yan C, Yang D, Xing Y, Lu X. Genome-Wide Identification and Expression Analysis of the Melon B-BOX (BBX) Gene Family in Response to Abiotic and Biotic Stresses. Plants. 2025; 14(17):2715. https://doi.org/10.3390/plants14172715
Chicago/Turabian StyleZhang, Yu, Yin Li, Yan Wang, Congsheng Yan, Dekun Yang, Yujie Xing, and Xiaomin Lu. 2025. "Genome-Wide Identification and Expression Analysis of the Melon B-BOX (BBX) Gene Family in Response to Abiotic and Biotic Stresses" Plants 14, no. 17: 2715. https://doi.org/10.3390/plants14172715
APA StyleZhang, Y., Li, Y., Wang, Y., Yan, C., Yang, D., Xing, Y., & Lu, X. (2025). Genome-Wide Identification and Expression Analysis of the Melon B-BOX (BBX) Gene Family in Response to Abiotic and Biotic Stresses. Plants, 14(17), 2715. https://doi.org/10.3390/plants14172715





