Group 1 LEA Proteins in Durum Wheat: Evolution, Expression, and Roles in Abiotic Stress Tolerance
Abstract
1. Introduction
2. Results
2.1. Identification and Characterization of TtEM (LEA_5) Genes in Durum Wheat
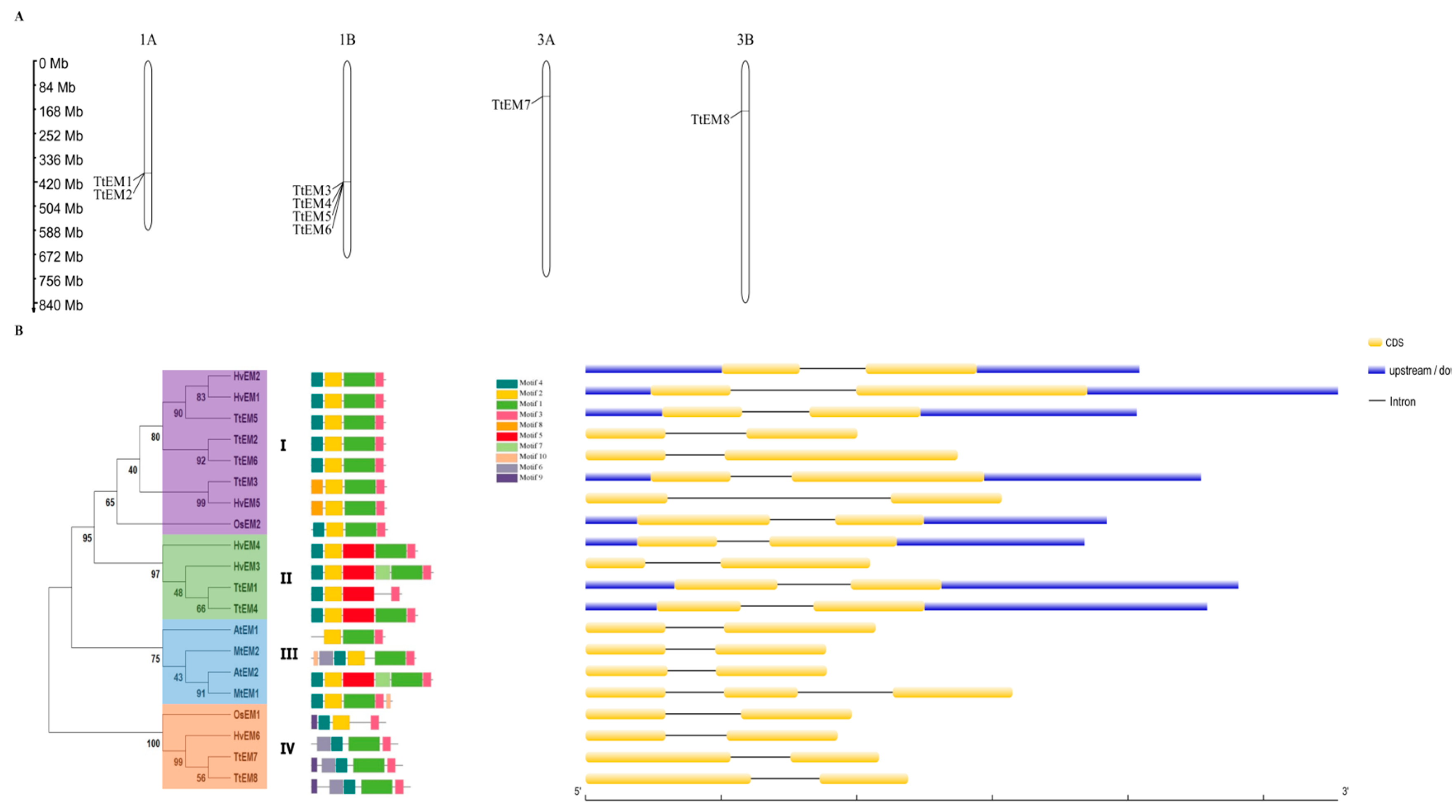
2.2. Chromosome Distribution and Phylogenetic Analysis
2.3. Gene Structure and Conserved Motif Analyses of EM Genes
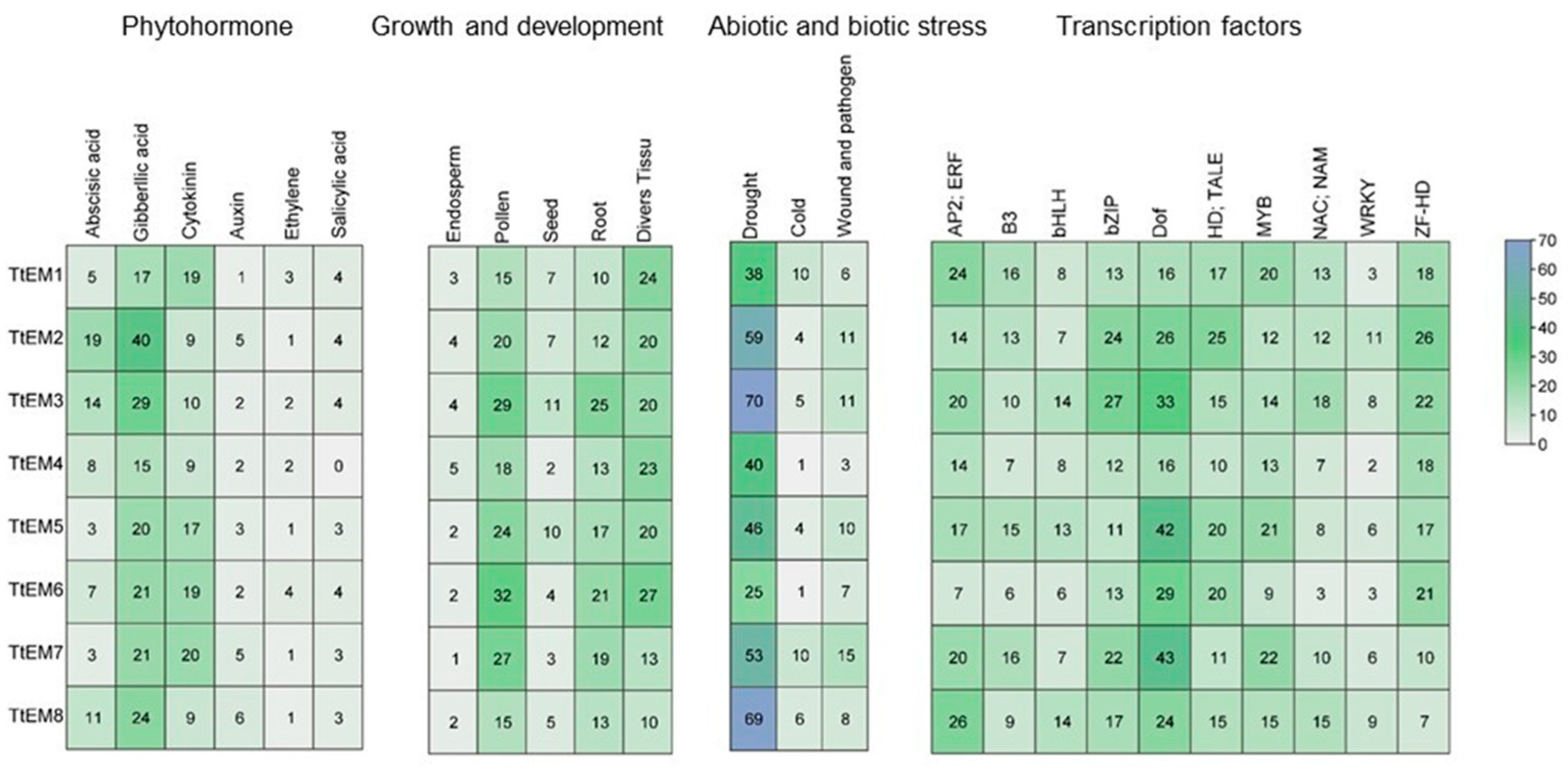
2.4. Analysis of Cis-Acting Elements in Promoters of TtEM Genes
2.5. Expression Patterns of TtEM1 and TtEM4 Genes in Different Tissues and Under Stress Treatments
2.6. Expression, Purification, and Structural Characterization of TtEM1 and TtEM4 Proteins
2.7. Preservation of LDH Activity Under Different Stress Treatments
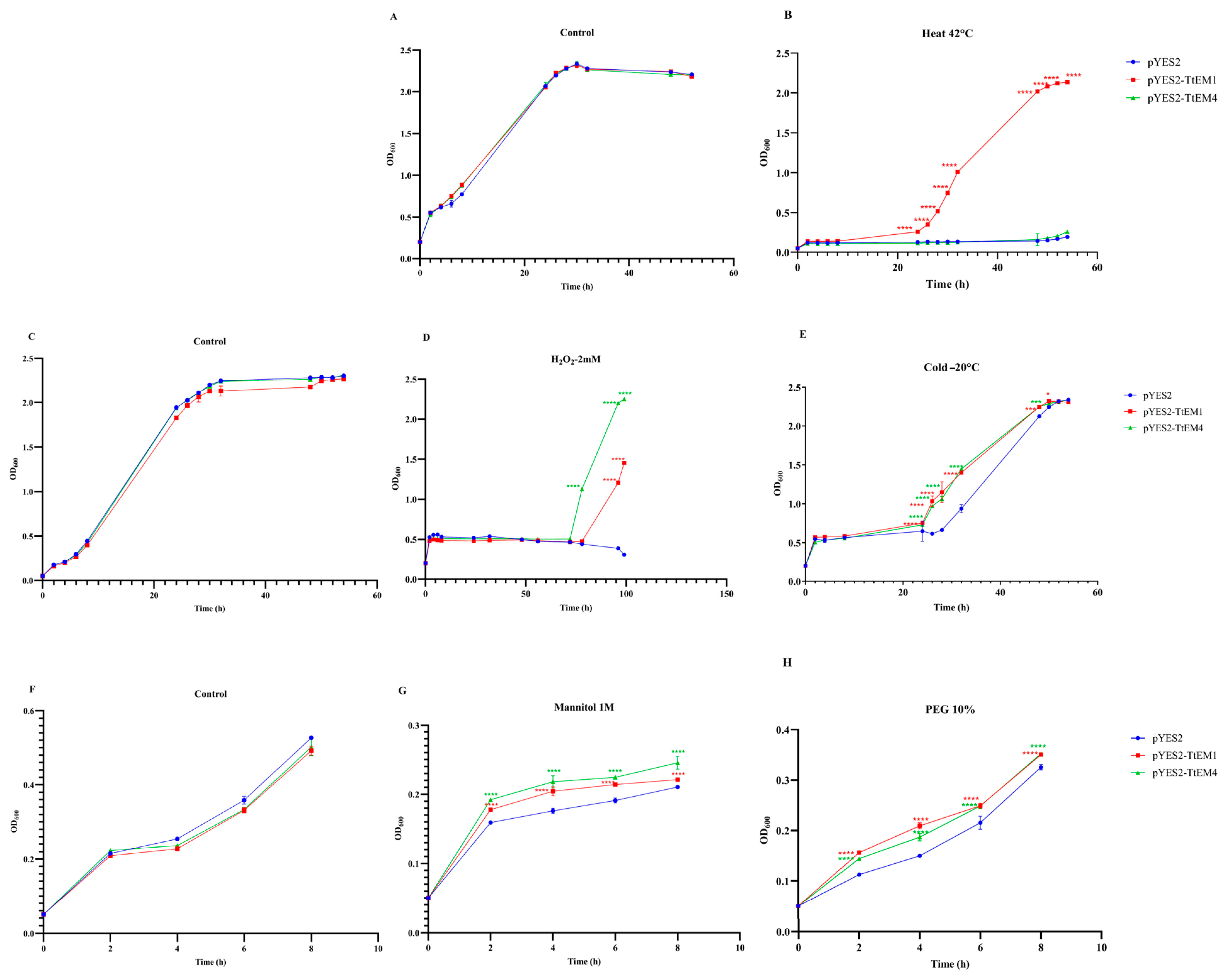
2.8. Overexpression of TtEM1 and TtEM4 Enhanced Yeast Cell Tolerance to Abiotic Stress
3. Discussion
4. Materials and Methods
4.1. Identification and Analysis of LEA_5 Family Members in Durum Wheat
4.2. Analysis of Phylogeny, Genes Structures, and Conserved Motifs
4.3. Cis-Acting Elements and Transcription Factors Analysis
4.4. Plant Material, Growth Conditions, and Stress Treatments
4.5. RNA Isolation, cDNA Synthesis, and qRT-PCR Analysis
4.6. Expression and Purification of TtEM1 and TtEM4 Proteins
4.7. Evaluation of Heat Stability and Protease Sensitivity of TtEM1 and TtEM4 Proteins
4.8. Lactate Dehydrogenase (LDH) Protection Assays
4.9. Evaluation of Stress Tolerance in Yeast Cells Expressing TtEM1 or TtEM4
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Martínez-Moreno, F.; Solís, I.; Noguero, D.; Blanco, A.; Özberk, İ.; Nsarellah, N.; Elias, E.; Mylonas, I.; Soriano, J.M. Durum Wheat in the Mediterranean Rim: Historical Evolution and Genetic Resources. Genet. Resour. Crop Evol. 2020, 67, 1415–1436. [Google Scholar] [CrossRef]
- Saharan, B.S.; Brar, B.; Duhan, J.S.; Kumar, R.; Marwaha, S.; Rajput, V.D.; Minkina, T. Molecular and Physiological Mechanisms to Mitigate Abiotic Stress Conditions in Plants. Life 2022, 12, 1634. [Google Scholar] [CrossRef]
- Li, S.; Meng, H.; Yang, Y.; Zhao, J.; Xia, Y.; Wang, S.; Wang, F.; Zheng, G.; Li, J. Overexpression of AtruLEA1 from Acer Truncatum Bunge Enhanced Arabidopsis Drought and Salt Tolerance by Improving ROS-Scavenging Capability. Plants 2025, 14, 117. [Google Scholar] [CrossRef]
- Dure, L.; Greenway, S.C.; Galau, G.A. Developmental Biochemistry of Cottonseed Embryogenesis and Germination: Changing Messenger Ribonucleic Acid Populations As Shown by in Vitro and in Vivo Protein Synthesis. Biochemistry 1981, 20, 4162–4168. [Google Scholar] [CrossRef]
- LeBlanc, B.M.; Hand, S.C. Target Enzymes Are Stabilized by AfrLEA6 and a Gain of α-Helix Coincides with Protection by a Group 3 LEA Protein during Incremental Drying. Biochim. Biophys. Acta—Proteins Proteom. 2021, 1869, 140642. [Google Scholar] [CrossRef]
- Amara, I.; Zaidi, I.; Masmoudi, K.; Ludevid, M.D.; Pagès, M.; Goday, A.; Brini, F. Insights into Late Embryogenesis Abundant (LEA) Proteins in Plants: From Structure to the Functions. Am. J. Plant Sci. 2014, 05, 3440–3455. [Google Scholar] [CrossRef]
- Celik Altunoglu, Y.; Baloglu, M.C.; Baloglu, P.; Yer, E.N.; Kara, S. Genome-Wide Identification and Comparative Expression Analysis of LEA Genes in Watermelon and Melon Genomes. Physiol. Mol. Biol. Plants 2017, 23, 5–21. [Google Scholar] [CrossRef]
- Wang, R.; Yan, S.J.; Liu, C.; Guo, H.; Cui, Y.N. Comparative Physiological and Gene Expression Analyses Reveal Mechanisms Involved in Maintaining Photosynthesis Capacity, Alleviating Ion Toxicity and Oxidative Stress of Kentucky Bluegrass under NaCl Treatment. Plants 2024, 13, 2107. [Google Scholar] [CrossRef]
- Janis, B.; Belott, C.; Brockman, T.; Menze, M.A. Functional and Conformational Plasticity of an Animal Group 1 LEA Protein. Biomolecules 2022, 12, 425. [Google Scholar] [CrossRef]
- Hunault, G.; Jaspard, E. LEAPdb: A Database for the Late Embryogenesis Abundant Proteins. BMC Genom. 2010, 11, 221. [Google Scholar] [CrossRef]
- Soulages, J.L.; Kim, K.; Walters, C.; Cushman, J.C. Temperature-Induced Extended Helix/Random Coil Transitions in a Group 1 Late Embryogenesis-Abundant Protein from Soybean. Plant Physiol. 2002, 128, 822–832. [Google Scholar] [CrossRef]
- Raga-Carbajal, E.; Espin, G.; Ayala, M.; Rodríguez-Salazar, J.; Pardo-López, L. Evaluation of a Bacterial Group 1 LEA Protein as an Enzyme Protectant from Stress-Induced Inactivation. Appl. Microbiol. Biotechnol. 2022, 106, 5551–5562. [Google Scholar] [CrossRef]
- Futers, T.S.; Vaugha, T.J.; Sharp, P.J.; Cuming, A.C. Molecular Cloning and Chromosomal Location of Genes Encoding the “Early-Methionine-Labelled” (Em) Polypeptide of Triticum aestivum L. Var. Chinese Spring. Theor. Appl. Genet. 1990, 80, 43–48. [Google Scholar] [CrossRef]
- Yu, J.; Lai, Y.; Wu, X.; Wu, G.; Guo, C. Overexpression of OsEm1 Encoding a Group I LEA Protein Confers Enhanced Drought Tolerance in Rice. Biochem. Biophys. Res. Commun. 2016, 478, 703–709. [Google Scholar] [CrossRef]
- Zou, Z.; Fu, X.; Yi, X.; Li, C.; Huang, J.; Zhao, Y. Integrative Analysis Provides Insights into Genes Encoding LEA_5 Domain-Containing Proteins in Tigernut (Cyperus esculentus L.). Plants 2025, 14, 762. [Google Scholar] [CrossRef]
- Shih, M.D.; Huang, L.T.; Wei, F.J.; Wu, M.T.; Hoekstra, F.A.; Hsing, Y.I.C. OsLEA1a, a New Em-like Protein of Cereal Plants. Plant Cell Physiol. 2010, 51, 2132–2144. [Google Scholar] [CrossRef]
- Miyoshi, K.; Kagaya, Y.; Ogawa, Y.; Nagato, Y.; Hattori, T. Temporal and Spatial Expression Pattern of the OSVP1 and OSEM Genes during Seed Development in Rice. Plant Cell Physiol. 2002, 43, 307–313. [Google Scholar] [CrossRef]
- Xiang, D.J.; Man, L.L.; Zhang, C.L.; Peng, L.; Li, Z.G.; Zheng, G.C. A New Em-like Protein from Lactuca sativa, LsEm1, Enhances Drought and Salt Stress Tolerance in Escherichia coli and Rice. Protoplasma 2018, 255, 1089–1106. [Google Scholar] [CrossRef]
- Zhou, C.; Niu, S.; El-Kassaby, Y.A.; Li, W. Genome-Wide Identification of Late Embryogenesis Abundant Protein Family and Their Key Regulatory Network in Pinus Tabuliformis Cold Acclimation. Tree Physiol. 2023, 43, 1964–1985. [Google Scholar] [CrossRef]
- Tang, W.; Page, M. Overexpression of the Arabidopsis AtEm6 Gene Enhances Salt Tolerance in Transgenic Rice Cell Lines. Plant Cell. Tissue Organ Cult. 2013, 114, 339–350. [Google Scholar] [CrossRef]
- Campos, F.; Cuevas-Velazquez, C.; Fares, M.A.; Reyes, J.L.; Covarrubias, A.A. Group 1 LEA Proteins, an Ancestral Plant Protein Group, Are Also Present in Other Eukaryotes, and in the Archeae and Bacteria Domains. Mol. Genet. Genom. 2013, 288, 503–517. [Google Scholar] [CrossRef]
- Artur, M.A.S.; Zhao, T.; Ligterink, W.; Schranz, E.; Hilhorst, H.W.M. Dissecting the Genomic Diversification of Late Embryogenesis Abundant (LEA) Protein Gene Families in Plants. Genome Biol. Evol. 2019, 11, 459–471. [Google Scholar] [CrossRef]
- Meilong, X.; Tong, Q.; Wang, Y.; Wang, Z.; Xu, G.; Elias, G.K.; Li, S.; Liang, Z. Transcriptomic Analysis of the Grapevine Lea Gene Family in Response to Osmotic and Cold Stress Reveals a Key Role for Vamdhn3. Plant Cell Physiol. 2020, 61, 775–786. [Google Scholar] [CrossRef]
- Xu, J.; Hu, P.; Tao, Y.; Song, P.; Gao, H.; Guan, Y. Genome-Wide Identification and Characterization of the Lateral Organ Boundaries Domain (Lbd) Gene Family in Polyploid Wheat and Related Species. PeerJ 2021, 9, e11811. [Google Scholar] [CrossRef]
- Huanhuan, J.; Qiangbin, C.; Tong, W.; Gang, C. Genome-Wide Analysis and Stress-Responsive Expression Profiling of the LEA (Late Embryogenesis Abundant) Gene Family in Wild Peanut. Oil Crop Sci. 2025, 10, 100–108. [Google Scholar] [CrossRef]
- Ashfaq, L.; Iqbal, S.; Rasheed, S.; Munir, H.; Niazi, A.K.; Khan, R.S.A. Genome-Wide Identification and Functional Characterization of LEA4-5 Genes in Response to Drought Stress in Brassica Species. J. Plant Growth Regul. 2025, 44, 4678–4704. [Google Scholar] [CrossRef]
- Liu, H.; Xing, M.; Yang, W.; Mu, X.; Wang, X.; Lu, F.; Wang, Y.; Zhang, L. Genome-Wide Identification of and Functional Insights into the Late Embryogenesis Abundant (LEA) Gene Family in Bread Wheat (Triticum aestivum). Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Xie, D.W.; Wang, X.N.; Fu, L.S.; Sun, J.; Zheng, W.; Li, Z.F. Identification of the Trehalose-6-Phosphate Synthase Gene Family in Winter Wheat and Expression Analysis under Conditions of Freezing Stress. J. Genet. 2015, 94, 55–65. [Google Scholar] [CrossRef]
- Zan, T.; Li, L.; Li, J.; Zhang, L.; Li, X. Genome-Wide Identification and Characterization of Late Embryogenesis Abundant Protein-Encoding Gene Family in Wheat: Evolution and Expression Profiles during Development and Stress. Gene 2020, 736, 144422. [Google Scholar] [CrossRef]
- Ren, J.; Cai, K.; Song, X.; Yue, W.; Liu, L.; Ge, F.; Wang, Q.; Wang, J. Genome-Wide Identification and Expression Profiling of ABA-Stress-Ripening (ASR) Gene Family in Barley (Hordeum vulgare L.). Plants 2025, 14, 970. [Google Scholar] [CrossRef]
- Jeffares, D.C.; Penkett, C.J.; Bähler, J. Rapidly Regulated Genes Are Intron Poor. Trends Genet. 2008, 24, 375–378. [Google Scholar] [CrossRef]
- Lin, Y.; She, M.; Zhao, M.; Yu, H.; Xiao, W.; Zhang, Y.; Li, M.; Chen, Q.; Zhang, Y.; Wang, Y.; et al. Genome-Wide Analysis and Functional Validation Reveal the Role of Late Embryogenesis Abundant Genes in Strawberry (Fragaria × Ananassa) Fruit Ripening. BMC Genom. 2024, 25, 228. [Google Scholar] [CrossRef]
- Jin, X.; Cao, D.; Wang, Z.; Ma, L.; Tian, K.; Liu, Y.; Gong, Z.; Zhu, X.; Jiang, C.; Li, Y. Genome-Wide Identification and Expression Analyses of the LEA Protein Gene Family in Tea Plant Reveal Their Involvement in Seed Development and Abiotic Stress Responses. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef]
- Graether, S.P. Methods for Recombinant Production and Purification of Intrinsically Disordered Proteins. Adv. Protein Mol. Struct. Biol. Methods 2022, 41–48. [Google Scholar] [CrossRef]
- Cuevas-Velazquez, C.L.; Reyes, J.L.; Covarrubias, A.A. Group 4 Late Embryogenesis Abundant Proteins as a Model to Study Intrinsically Disordered Proteins in Plants. Plant Signal. Behav. 2017, 12, e1343777. [Google Scholar] [CrossRef]
- Bremer, A.; Wolff, M.; Thalhammer, A.; Hincha, D.K. Folding of Intrinsically Disordered Plant LEA Proteins Is Driven by Glycerol-Induced Crowding and the Presence of Membranes. FEBS J. 2017, 284, 919–936. [Google Scholar] [CrossRef]
- Thalhammer, A.; Bryant, G.; Sulpice, R.; Hincha, D.K. Disordered Cold Regulated15 Proteins Protect Chloroplast Membranes during Freezing through Binding and Folding, But Do Not Stabilize Chloroplast Enzymes in Vivo. Plant Physiol. 2014, 166, 190–201. [Google Scholar] [CrossRef]
- Huang, R.L.; Xiao, D.; Wang, X.; Zhan, J.; Wang, A.Q.; He, L.F. Genome-Wide Identification, Evolutionary and Expression Analyses of LEA Gene Family in Peanut (Arachis hypogaea L.). BMC Plant Biol. 2022, 22, 155. [Google Scholar] [CrossRef]
- Zaidi, I.; Hanin, M.; Saidi, M.N.; Soltani, N.; Brini, F. The Barley Dehydrin 4 and Stress Tolerance: From Gene to Function. Environ. Exp. Bot. 2024, 224, 105802. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Lai, J.; Zhang, H.; Liu, Y.; Liang, L.; Xie, Q. Ectopic Expression of a LEA Protein Gene TsLEA1 from Thellungiella Salsuginea Confers Salt-Tolerance in Yeast and Arabidopsis. Mol. Biol. Rep. 2012, 39, 4627–4633. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, L.; Jiang, S.; Pan, J.; Cai, G.; Li, D. Group 5 LEA Protein, ZmLEA5C, Enhance Tolerance to Osmotic and Low Temperature Stresses in Transgenic Tobacco and Yeast. Plant Physiol. Biochem. 2014, 84, 22–31. [Google Scholar] [CrossRef]
- Piyatissa, S.; Bandupriya, D. Genome-Wide Identification and Analysis of Late Embryogenesis Abundant (LEA) Genes in Musa Acuminata. Trop. Plant Biol. 2021, 14, 295–312. [Google Scholar] [CrossRef]
- Nagaraju, M.; Anil Kumar, S.; Reddy, P.S.; Kumar, A.; Manohar Rao, D.; Kavi Kishor, P.B. Genome-Scale Identification, Classification, and Tissue Specific Expression Analysis of Late Embryogenesis Abundant (LEA) Genes under Abiotic Stress Conditions in Sorghum bicolor L. PLoS ONE 2019, 14, e0209980. [Google Scholar] [CrossRef]
- Cao, J.; Li, X. Identification and Phylogenetic Analysis of Late Embryogenesis Abundant Proteins Family in Tomato (Solanum lycopersicum). Planta 2015, 241, 757–772. [Google Scholar] [CrossRef]
- Ding, M.; Wang, L.; Zhan, W.; Sun, G.; Jia, X.; Chen, S.; Ding, W.; Yang, J. Genome-Wide Identification and Expression Analysis of Late Embryogenesis Abundant Protein-Encoding Genes in Rye (Secale cereale L.). PLoS ONE 2021, 16, e0249757. [Google Scholar] [CrossRef]
- Lan, T.; Gao, J.; Zeng, Q.Y. Genome-Wide Analysis of the LEA (Late Embryogenesis Abundant) Protein Gene Family in Populus Trichocarpa. Tree Genet. Genomes 2013, 9, 253–264. [Google Scholar] [CrossRef]
- Singh, C.M.; Kumar, M.; Pratap, A.; Tripathi, A.; Singh, S.; Mishra, A.; Kumar, H.; Nair, R.M.; Singh, N.P. Genome-Wide Analysis of Late Embryogenesis Abundant Protein Gene Family in Vigna Species and Expression of VrLEA Encoding Genes in Vigna Glabrescens Reveal Its Role in Heat Tolerance. Front. Plant Sci. 2022, 13, 843107. [Google Scholar] [CrossRef]
- Giménez, M.J.; Pistón, F.; Atienza, S.G. Identification of Suitable Reference Genes for Normalization of QPCR Data in Comparative Transcriptomics Analyses in the Triticeae. Planta 2011, 233, 163–173. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zaïdi, I.; Ebel, C.; Touzri, M.; Herzog, E.; Evrard, J.L.; Schmit, A.C.; Masmoudi, K.; Hanin, M. TMKP1 Is a Novel Wheat Stress Responsive MAP Kinase Phosphatase Localized in the Nucleus. Plant Mol. Biol. 2010, 73, 325–338. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Zaidi, I.; González, A.; Touzri, M.; Alvarez, M.C.; Ramos, J.; Masmoudi, K.; Ariño, J.; Hanin, M. The Wheat MAP Kinase Phosphatase 1 Confers Higher Lithium Tolerance in Yeast. FEMS Yeast Res. 2012, 12, 774–784. [Google Scholar] [CrossRef]



| Name | Number of Amino Acids | Molecular Weight (kDa) | Theoretical pI | Grand Average of Hydropathicity (GRAVY) | Predicted α-Helical Content (%) (SOPMA) | Tendency of Disorder (%) (PONDR) | Predicted Subcellular Localization |
|---|---|---|---|---|---|---|---|
| TtEM1 | 113 | 12.333 | 5.23 | −1.388 | 41.59 | 99.23 | Nucleus |
| TtEM2 | 93 | 10.017 | 5.57 | −1.403 | 33.33 | 98.92 | Nucleus |
| TtEM3 | 94 | 10.002 | 5.29 | −1.328 | 37.23 | 98.94 | Nucleus |
| TtEM4 | 133 | 14.604 | 5.56 | −1.474 | 45.13 | 99.25 | Nucleus |
| TtEM5 | 93 | 9.962 | 5.56 | −1.375 | 36.56 | 98.92 | Nucleus |
| TtEM6 | 93 | 9.948 | 5.25 | −1.363 | 34.41 | 98.92 | Nucleus |
| TtEM7 | 114 | 12.575 | 5.27 | −1.361 | 42.98 | 100 | Nucleus |
| TtEM8 | 124 | 13.694 | 5.17 | −1.308 | 45.16 | 100 | Nucleus |
| Motif | Logo |
|---|---|
| 1 |  |
| 2 | 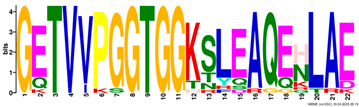 |
| 3 |  |
| 4 | 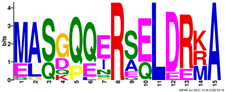 |
| 5 |  |
| 6 |  |
| 7 |  |
| 8 |  |
| 9 | 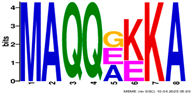 |
| 10 |  |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soltani, N.; Zaidi, I.; Saidi, M.N.; Brini, F. Group 1 LEA Proteins in Durum Wheat: Evolution, Expression, and Roles in Abiotic Stress Tolerance. Plants 2025, 14, 2817. https://doi.org/10.3390/plants14182817
Soltani N, Zaidi I, Saidi MN, Brini F. Group 1 LEA Proteins in Durum Wheat: Evolution, Expression, and Roles in Abiotic Stress Tolerance. Plants. 2025; 14(18):2817. https://doi.org/10.3390/plants14182817
Chicago/Turabian StyleSoltani, Najeh, Ikram Zaidi, Mohamed Najib Saidi, and Faiçal Brini. 2025. "Group 1 LEA Proteins in Durum Wheat: Evolution, Expression, and Roles in Abiotic Stress Tolerance" Plants 14, no. 18: 2817. https://doi.org/10.3390/plants14182817
APA StyleSoltani, N., Zaidi, I., Saidi, M. N., & Brini, F. (2025). Group 1 LEA Proteins in Durum Wheat: Evolution, Expression, and Roles in Abiotic Stress Tolerance. Plants, 14(18), 2817. https://doi.org/10.3390/plants14182817








