Transcriptome and Metabolome Analysis of the Mechanism of Environmental Adaptability in Populus Roots
Abstract
1. Introduction
2. Results
2.1. Environmental Differences Among the Three Sites
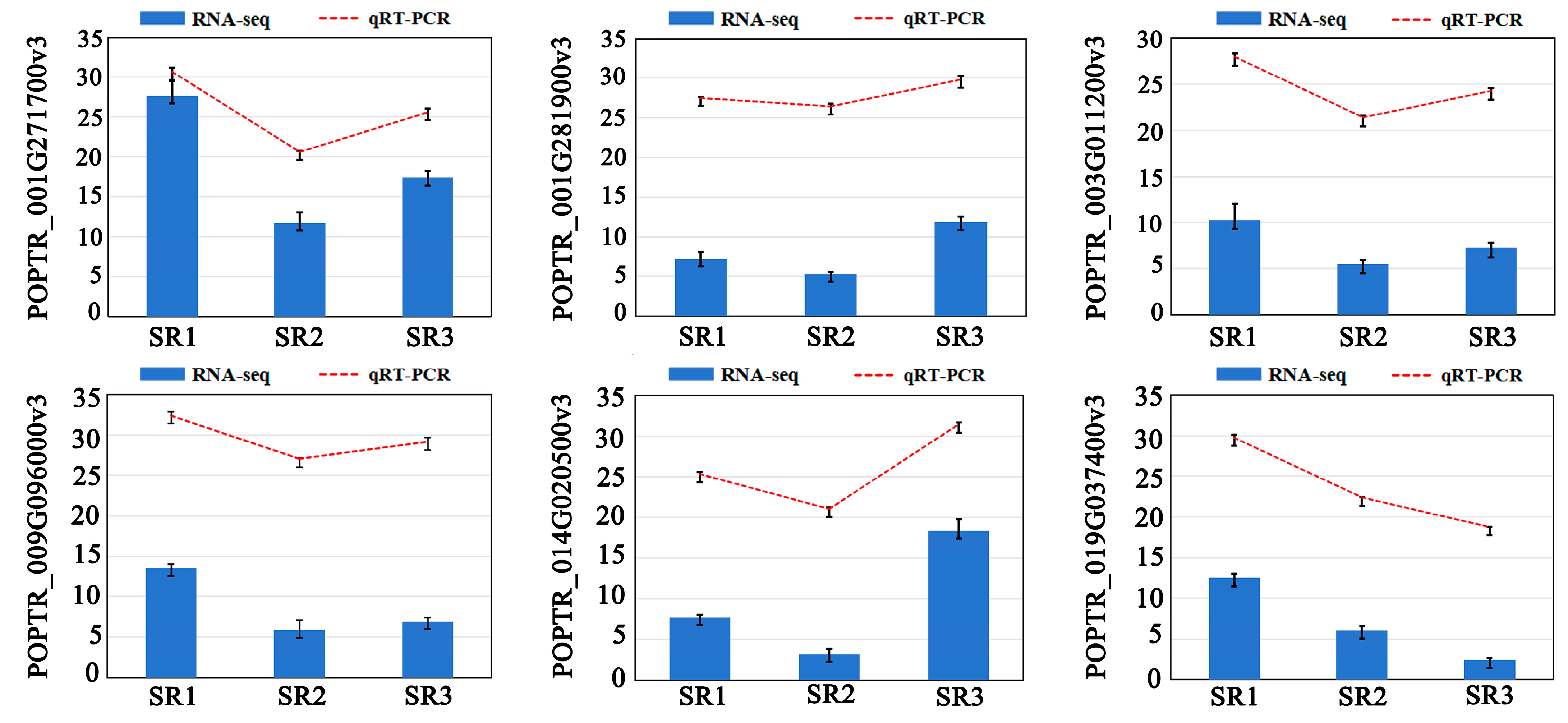
2.2. The Quality Evaluation of Transcriptome and qRT-PCR Verification
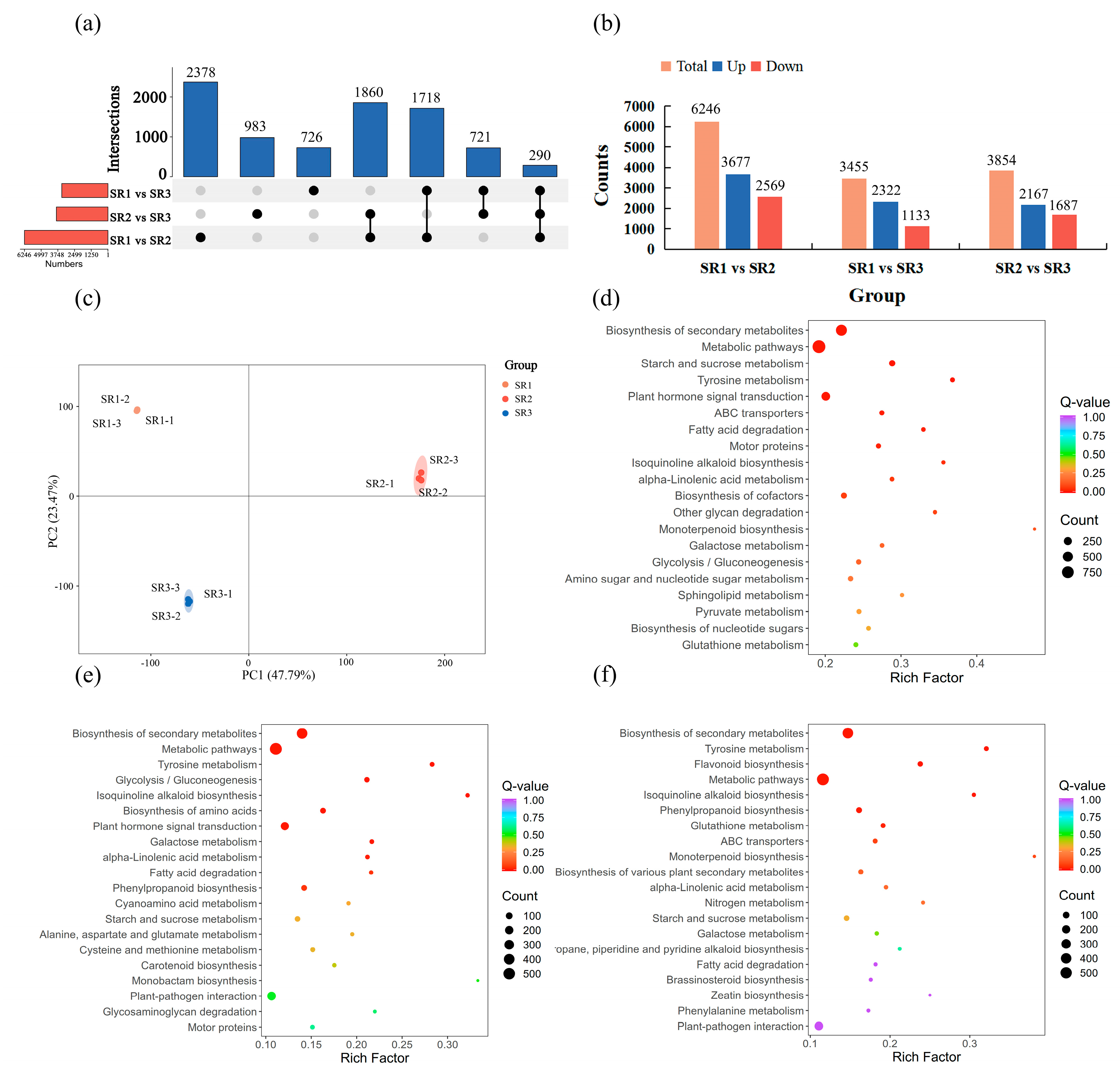
2.3. The Analysis of Differential Expression Genes
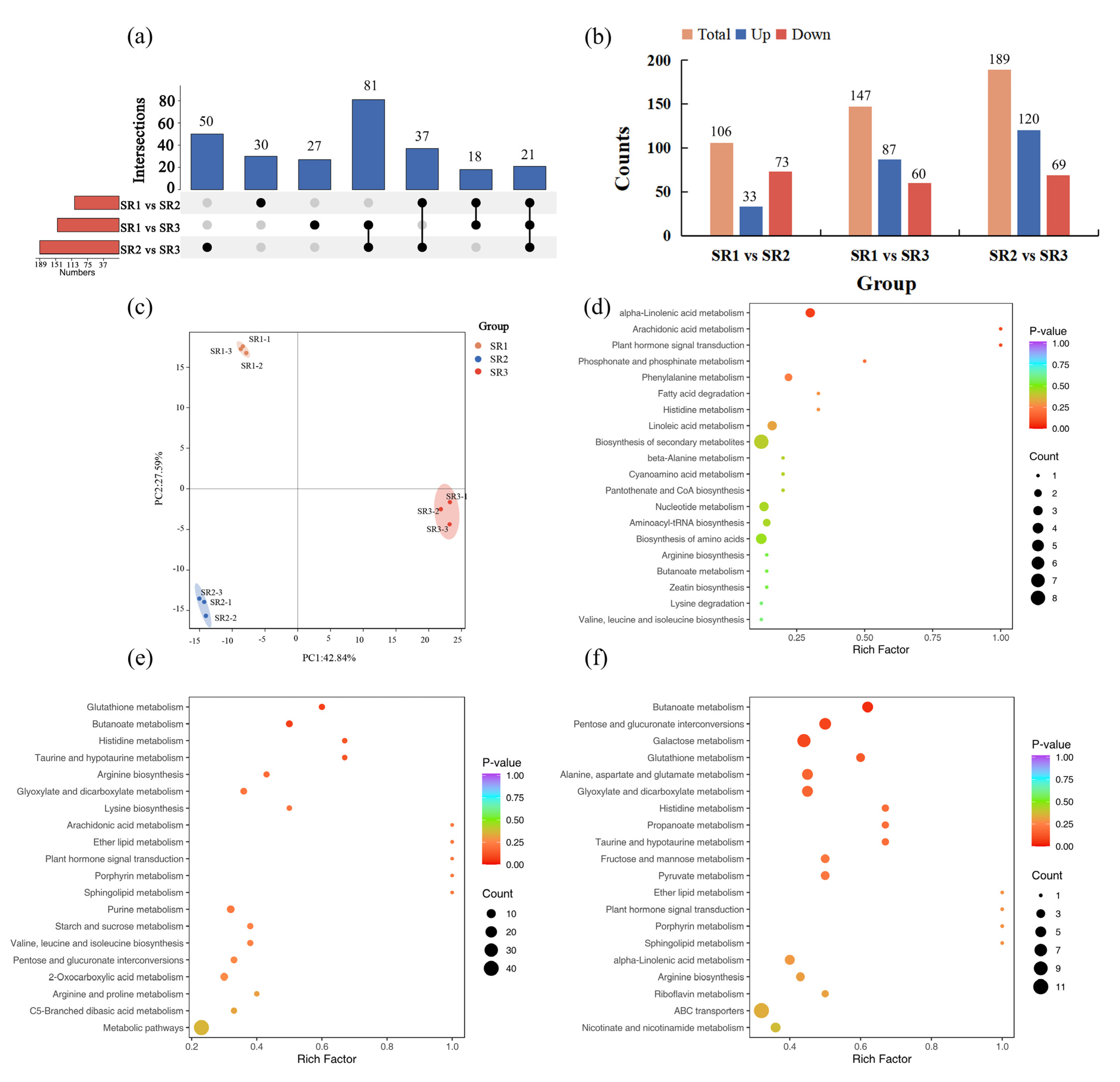
2.4. The Analysis of Differentially Accumulated Metabolites
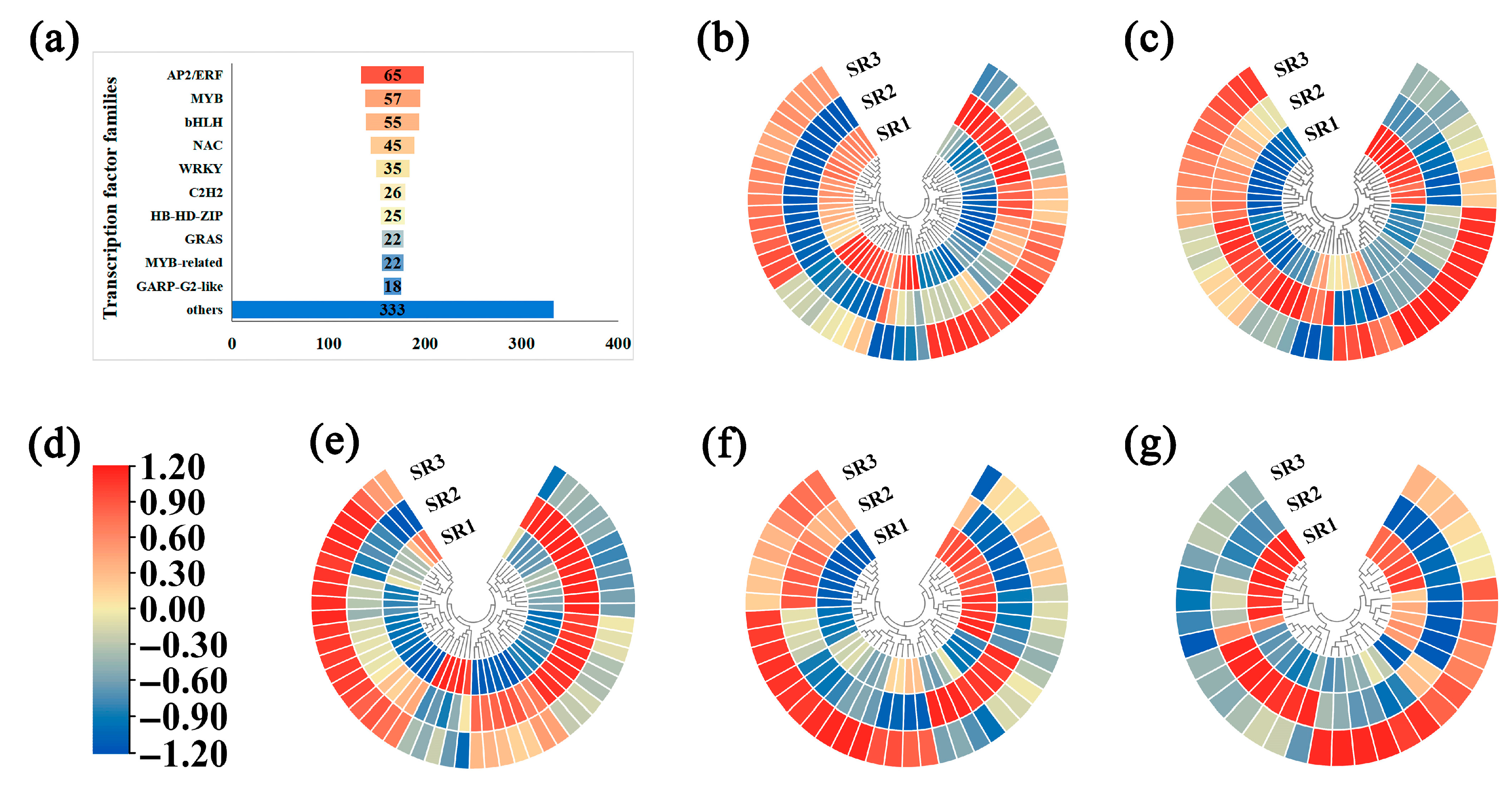
2.5. The Analysis of Transcription Factors
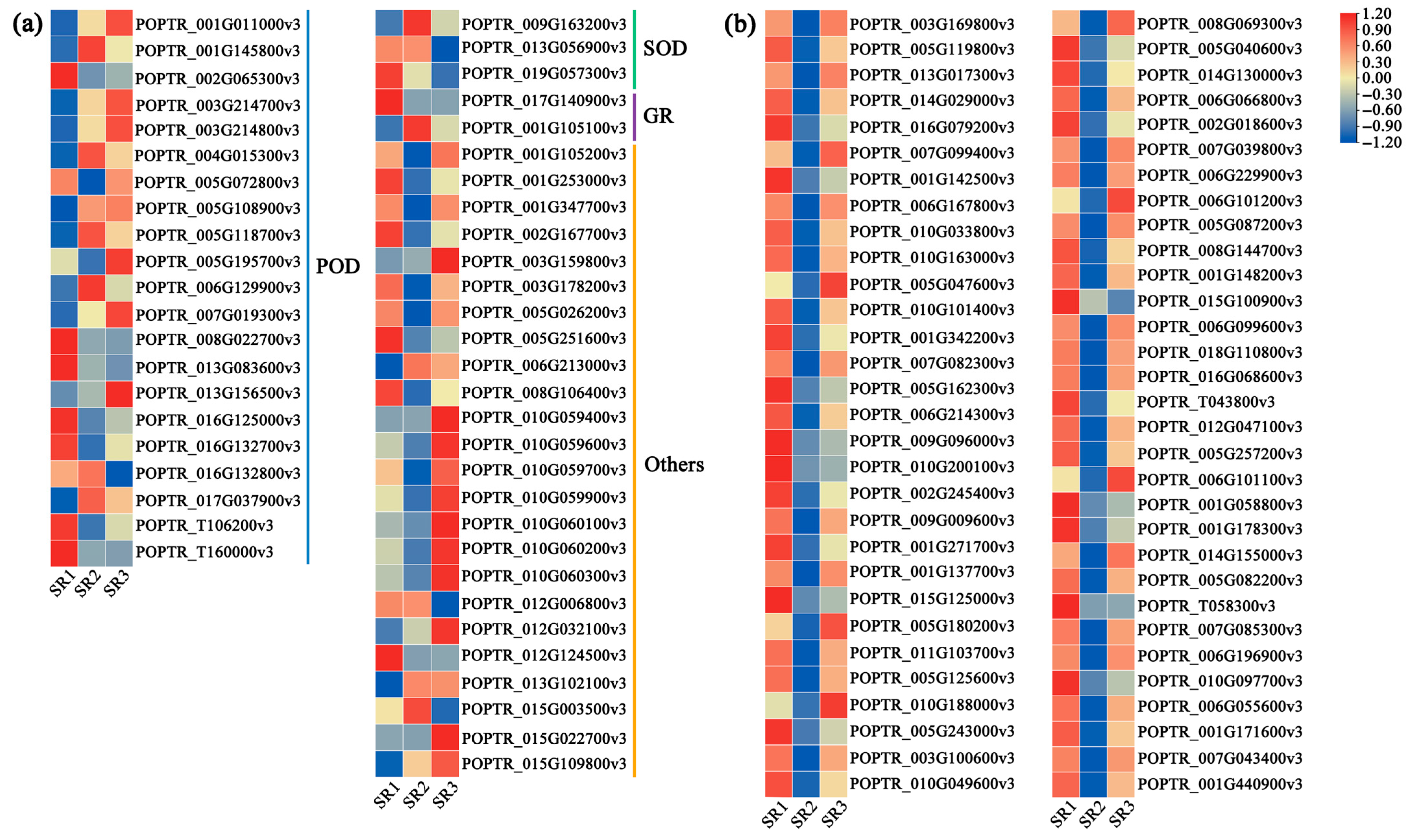
2.6. The Expression Pattern of Genes Related to Antioxidant Enzymes and Stimulus Response
2.7. The Analysis of Genes Involved in Plant Hormone Signal Transduction
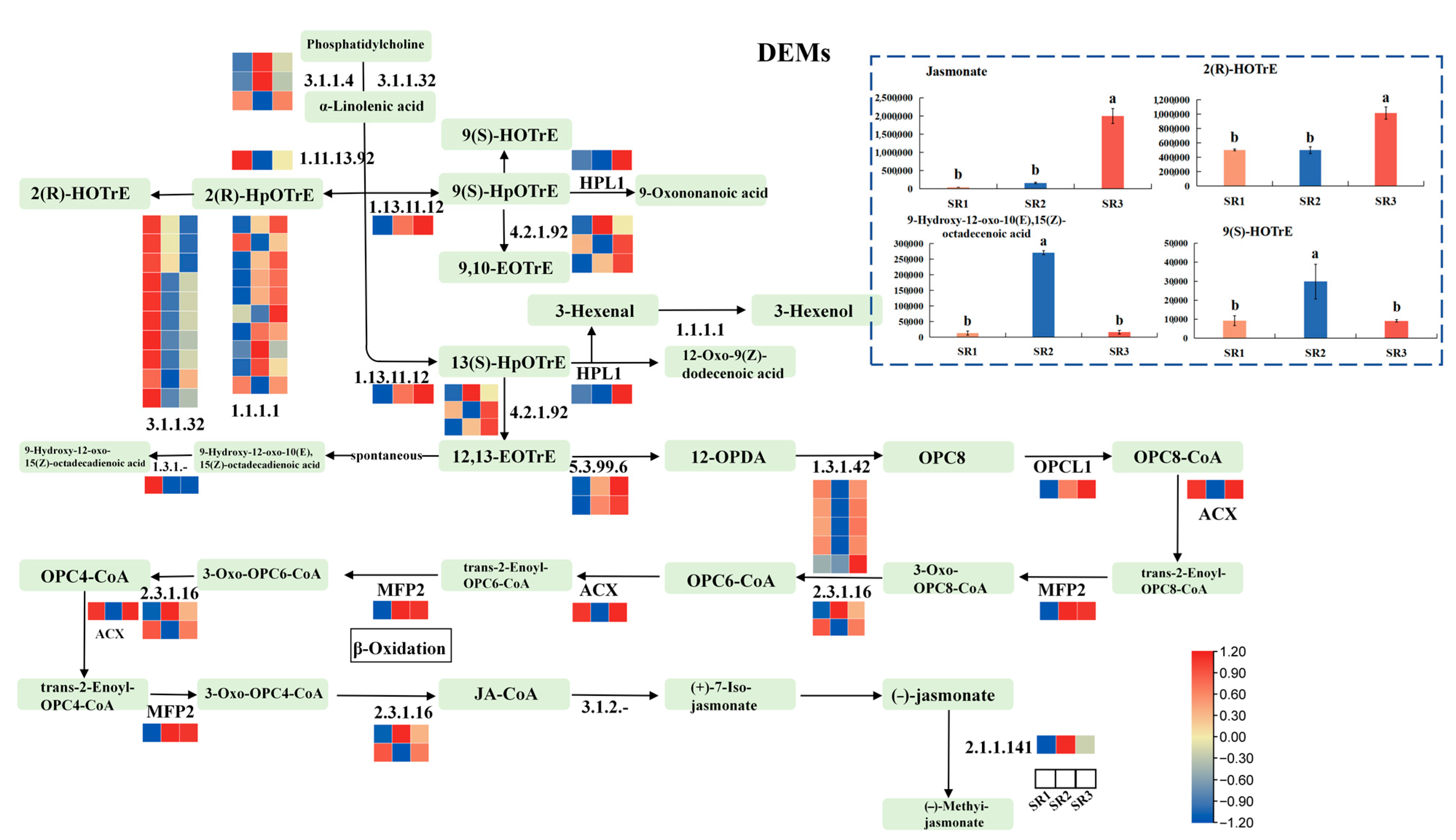
2.8. The Analysis of Genes and Metabolites Involved in Alpha-Linolenic Acid Metabolism
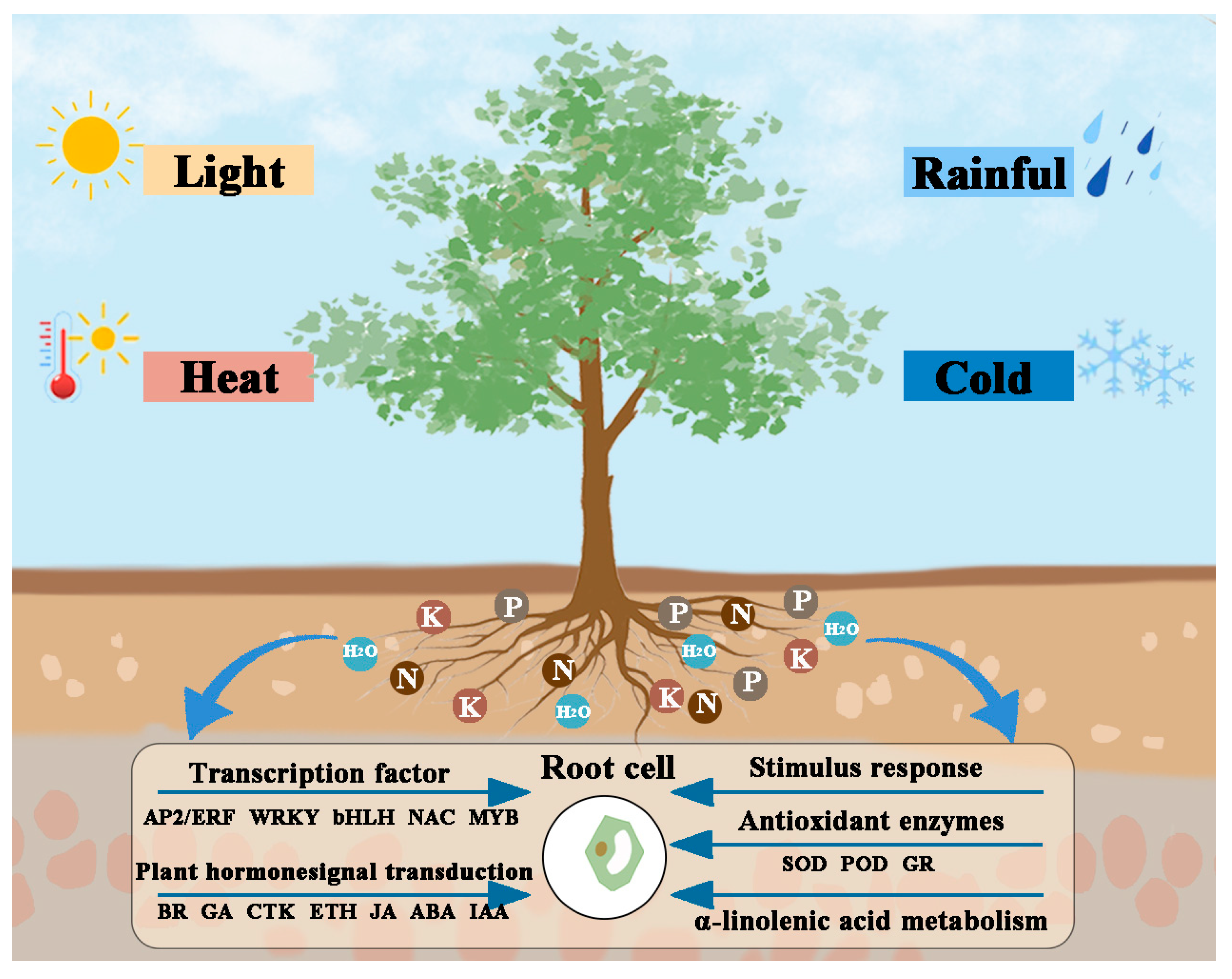
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. The Determination of Soil Element Content
4.3. Total RNA Isolation, Library Construction, and Transcriptome Sequencing
4.4. Transcriptome Analysis
4.5. Sampling Preparation and Metabolite Extraction
4.6. Metabolome Analysis
4.7. qRT-PCR Verification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schreiber, S.G.; Hamann, A.; Hacke, U.G.; Thomas, B.R. Sixteen years of winter stress: An assessment of cold hardiness, growth performance and survival of hybrid poplar clones at a boreal planting site. Plant Cell Environ. 2013, 36, 419–428. [Google Scholar] [CrossRef]
- Sögüt, T.; çalişkan, M.E.; Ertürk, E.; Boydak, E.; Arioğlu, H. Growth, Yield, and Quality of Sweet Potato (İpomoea batatas (l.) Lam.) Cultivars in the Southeastern Anatolian and East Mediterranean Regions of Turkey. Turk. J. Agric. For. 2007, 31, 213–227. [Google Scholar]
- Que, Q.; Kunxi, O.; Chunmei, L.; Buye, L.; Huiyun, S.; Pei, L.; Ruiqi, P.; Huaqing, L.; Chen, X.; Peng, C. Geographic variation in growth and wood traits of Neolamarckia cadamba in China. For. Res. 2022, 2, 12. [Google Scholar] [CrossRef]
- Liu, S.; Liu, H.; Wu, A.; Hou, Y.; An, Y.; Wei, C. Construction of fingerprinting for tea plant (Camellia sinensis) accessions using new genomic SSR markers. Mol. Breed. 2017, 37, 93. [Google Scholar] [CrossRef]
- Berrill, J.; Hara, K.L.O. Estimating site productivity in irregular stand structures by indexing the basal area or volume increment of the dominant species. Can. J. For. Res. 2014, 44, 92–100. [Google Scholar] [CrossRef]
- Jiang, L.; Xu, X.; Cai, Q.; Han, R.; Tigabu, M.; Jiang, T.; Zhao, X. Variations in Growth and Photosynthetic traits of Polyploid Poplar Hybrids and Clones in Northeast China. Genes 2022, 13, 2161. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Pei, X.; Hu, Y.; Chiang, V.L.; Zhao, X. Effects of environment and genotype on growth traits in poplar clones in Northeast China. Euphytica 2021, 217, 169. [Google Scholar] [CrossRef]
- Souch, C.A.; Stephens, W. Growth, productivity and water use in three hybrid poplar clones. Tree Physiol. 1998, 18, 829–835. [Google Scholar] [CrossRef] [PubMed]
- Rönnberg-Wästljung, A.C.; Adler, A.; Karacic, A.; Liepins, K.; Richards, T.J.; Ingvarsson, P.; Weih, M. Phenotypic plasticity in Populus trichocarpa clones across environments in the Nordic-Baltic region. Scand. J. For. Res. 2022, 37, 1–5. [Google Scholar] [CrossRef]
- Rae, A.M.; Pinel, M.P.C.; Bastien, C.; Sabatti, M.; Street, N.R.; Tucker, J.; Dixon, C.; Marron, N.; Dillen, S.Y.; Taylor, G. QTL for yield in bioenergy populus: Identifying G×E interactions from growth at three contrasting sites. Tree Genet. Genomes 2007, 4, 97–112. [Google Scholar] [CrossRef]
- Kuchma, O.; Janz, D.; Leinemann, L.; Polle, A.; Krutovsky, K.; Gailing, O. Hybrid and Environmental Effects on Gene expression in Poplar Clones in pure and Mixed with Black Locust Stands. Forests 2020, 11, 1075. [Google Scholar] [CrossRef]
- Wang, D.; Fan, J.; Jing, P.; Cheng, Y.; Ruan, H. Analyzing the impact of climate and management factors on the productivity and soil carbon sequestration of poplar plantations. Environ. Res. 2016, 144, 88–95. [Google Scholar] [CrossRef]
- Du, C.; Sun, P.; Cheng, X.; Zhang, L.; Wang, L.; Hu, J. QTL mapping of drought-related traits in the hybrids of Populus deltoides ‘Danhong’ × Populus simonii ‘Tongliao1’. BMC Plant Biol. 2022, 22, 238. [Google Scholar] [CrossRef]
- Strømme, C.; Sivadasan, U.; Nissinen, K.; Lavola, A.; Randriamanana, T.; Julkunen-Tiitto, R.; Nybakken, L. Interannual variation in UV-B and temperature effects on bud phenology and growth in Populus tremula. Plant Physiol. Biochem. 2019, 134, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Benomar, L.; Moutaoufik, M.T.; Elferjani, R.; Isabel, N.; Desrochers, A.; El Guellab, A.; Khlifa, R.; Idrissi Hassania, L.A. Thermal acclimation of photosynthetic activity and RuBisCO content in two hybrid poplar clones. PLoS ONE 2019, 14, e206021. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Lian, J.; Liang, J.; Wei, H.; Chen, H.; Hu, W.; Tang, M. Arbuscular mycorrhizal symbiosis modulates nitrogen uptake and assimilation to enhance drought tolerance of Populus cathayana. Plant Physiol. Biochem. 2024, 210, 108648. [Google Scholar] [CrossRef]
- Xia, Z.; He, Y.; Yu, L.; Lv, R.; Korpelainen, H.; Li, C. Sex-specific strategies of phosphorus (p) acquisition in Populus cathayana as affected by soil P availability and distribution. New Phytol. 2020, 225, 782–792. [Google Scholar] [CrossRef]
- Liu, C.L.C.; Kuchma, O.; Krutovsky, K.V. Mixed-species versus monocultures in plantation forestry: Development, benefits, ecosystem services and perspectives for the future. Glob. Ecol. Conserv. 2018, 15, e00419. [Google Scholar] [CrossRef]
- An, Y.; Geng, Y.; Liu, Y.; Han, X.; Huang, L.; Zeng, W.; Zhang, J.; Lu, M. The glutamate receptor gene glr3.3: A bridge of calcium-mediated root development in poplar. Hortic. Plant J. 2024, 10, 1449–1462. [Google Scholar] [CrossRef]
- Sun, X.; Chen, F.; Yuan, L.; Mi, G. The physiological mechanism underlying root elongation in response to nitrogen deficiency in crop plants. Planta 2020, 251, 84. [Google Scholar] [CrossRef] [PubMed]
- An, Y.; Han, X.; Tang, S.; Xia, X.; Yin, W. Poplar GATA transcription factor PdGNC is capable of regulating chloroplast ultrastructure, photosynthesis, and vegetative growth in Arabidopsis under varying nitrogen levels. Plant Cell Tissue Organ Cult. (PCTOC) 2014, 119, 313–327. [Google Scholar] [CrossRef]
- Legué, V.; Rigal, A.; Bhalerao, R.P. Adventitious root formation in tree species: Involvement of transcription factors. Physiol. Plant. 2014, 151, 192–198. [Google Scholar] [CrossRef]
- Harfouche, A.; Meilan, R.; Altman, A. Molecular and physiological responses to abiotic stress in forest trees and their relevance to tree improvement. Tree Physiol. 2014, 34, 1181–1198. [Google Scholar] [CrossRef] [PubMed]
- Cabal, C.; Martínez-García, R.; de Castro Aguilar, A.; Valladares, F.; Pacala, S.W. The exploitative segregation of plant roots. Science 2020, 370, 1197–1199. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Wang, D.; Wang, D.; Li, S.; Chen, Y.; Guo, Y. Phenotypic Plasticity and Local adaptation of Leaf Cuticular Waxes Favor Perennial Alpine Herbs Under Climate Change. Plants 2022, 11, 120. [Google Scholar] [CrossRef]
- Sauvadet, M.; Dickinson, A.K.; Somarriba, E.; Phillips-Mora, W.; Cerda, R.H.; Martin, A.R.; Isaac, M.E. Genotype-environment interactions shape leaf functional traits of cacao in agroforests. Agron. Sustain. Dev. 2021, 41, 31. [Google Scholar] [CrossRef]
- Javaid, M.H.; Khan, A.R.; Salam, A.; Neelam, A.; Azhar, W.; Ulhassan, Z.; Gan, Y. Exploring the Adaptive Responses of Plants to Abiotic Stresses Using Transcriptome Data. Agriculture 2022, 12, 211. [Google Scholar] [CrossRef]
- Zhang, Y.; Lin, S.; Zhou, Y.; Wen, J.; Kang, X.; Han, X.; Liu, C.; Yin, W.; Xia, X. PdNF-YB21 positively regulated root lignin structure in poplar. Ind. Crops Prod. 2021, 168, 113609. [Google Scholar] [CrossRef]
- Yang, M.; Wang, L.; Wang, X.; Li, Y.; Huang, H. Transcriptomic Response to Drought Stress in Populus davidiana Dode. Forests 2023, 14, 1465. [Google Scholar] [CrossRef]
- Li, H.; Shen, J.; Ding, Y.; Li, Y.; Du, J.; Jiang, T.; Kong, X.; Han, R.; Zhang, X.; Zhao, X. Transcriptomic and metabolomic analysis of poplar response to feeding by Hyphantria cunea. BMC Plant Biol. 2024, 24, 920. [Google Scholar] [CrossRef]
- Patel, M.K.; Pandey, S.; Kumar, M.; Haque, M.I.; Pal, S.; Yadav, N.S. Plants Metabolome Study: Emerging Tools and Techniques. Plants 2021, 10, 2409. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Qu, L.; Fan, Z.; Hou, L.; Hu, J.; Wang, L. Dynamic Metabolic Responses of Resistant and Susceptible Poplar Clones Induced by Hyphantria cunea Feeding. Biology 2024, 13, 723. [Google Scholar] [CrossRef]
- Pastierovič, F.; Kalyniukova, A.; Hradecký, J.; Dvořák, O.; Vítámvás, J.; Mogilicherla, K.; Tomášková, I. Biochemical Responses in Populus tremula: Defending Against Sucking and Leaf-Chewing Insect Herbivores. Plants 2024, 13, 1243. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Cao, L.; Chang, R.; Zhang, H.; Yu, J.; Li, C.; Liu, G.; Yan, J.; Xu, Z. Network Analysis of Metabolome and Transcriptome Revealed Regulation of Different Nitrogen Concentrations on Hybrid Poplar Cambium Development. Int. J. Mol. Sci. 2024, 25, 1017. [Google Scholar] [CrossRef]
- Yang, X.; Li, Y.; Yu, R.; Zhang, L.; Yang, Y.; Xiao, D.; Li, A.; Wang, Y. Integrated transcriptomic and metabolomic profiles reveal adaptive responses of three poplar varieties against the bacterial pathogen Lonsdalea populi. Plant Cell Environ. 2023, 46, 306–321. [Google Scholar] [CrossRef]
- Hamanishi, E.T.; Barchet, G.L.; Dauwe, R.; Mansfield, S.D.; Campbell, M.M. Poplar trees reconfigure the transcriptome and metabolome in response to drought in a genotype- and time-of-day-dependent manner. BMC Genom. 2015, 16, 329. [Google Scholar] [CrossRef]
- Wang, K.; Guo, H.; Yin, Y. AP2/ERF transcription factors and their functions in Arabidopsis responses to abiotic stresses. Environ. Exp. Bot. 2024, 222, 105763. [Google Scholar] [CrossRef]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2012, 1819, 86–96. [Google Scholar] [CrossRef]
- Radani, Y.; Li, R.; Korboe, H.M.; Ma, H.; Yang, L. Transcriptional and Post-Translational Regulation of Plant bHLH Transcription Factors during the Response to Environmental Stresses. Plants 2023, 12, 2113. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Zhang, H.; Li, X.; Ai, Q.; Yu, D. bHLH transcription factor bHLH115 regulates iron homeostasis in Arabidopsis thaliana. J. Exp. Bot. 2017, 68, 1743–1755. [Google Scholar] [CrossRef]
- Chen, X.; Wu, X.; Han, C.; Jia, Y.; Wan, X.; Liu, Q.; He, F.; Zhang, F. A WRKY transcription factor, PyWRKY71, increased the activities of antioxidant enzymes and promoted the accumulation of cadmium in poplar. Plant Physiol. Biochem. 2023, 205, 108163. [Google Scholar] [CrossRef]
- Niu, Y.; Li, X.; Xu, C.; Ajab, Z.; Liu, Q.; Majeed, Z.; Guan, Q. Analysis of drought and salt-alkali tolerance in tobacco by overexpressing WRKY39 gene from Populus trichocarpa. Plant Signal. Behav. 2021, 16, 1918885. [Google Scholar] [CrossRef]
- Qi, M.; Wu, R.; Song, Z.; Dong, B.; Chen, T.; Wang, M.; Cao, H.; Du, T.; Wang, S.; Li, N.; et al. Sorbitol Reduces Sensitivity to Alternaria by Promoting Ceramide kinases (CERK) Expression through Transcription Factor Pswrky25 in Populus (Populus simonii Carr.). Genes 2022, 13, 405. [Google Scholar] [CrossRef]
- Yu, X.; Pan, Y.; Dong, Y.; Lu, B.; Zhang, C.; Yang, M.; Zuo, L. Cloning and overexpression of PeWRKY31 from Populus × euramericana enhances salt and biological tolerance in transgenic Nicotiana. BMC Plant Biol. 2021, 21, 80. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Huan, X.; Zhao, K.; Jin, X.; Hu, J.; Du, S.; Han, Y.; Wang, S. PagMYB205 Negatively Affects Poplar Salt Tolerance Through Reactive Oxygen Species Scavenging and Root Vitality Modulation. Int. J. Mol. Sci. 2023, 24, 15437. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, H.; Chen, Y.; Huang, M.; Zhu, S. The Over-Expression of two R2R3-MYB genes, PdMYB2R089 and PdMYB2R151, Increases the Drought-Resistant Capacity of Transgenic Arabidopsis. Int. J. Mol. Sci. 2023, 24, 13466. [Google Scholar] [CrossRef]
- Niu, M.X.; Feng, C.H.; He, F.; Zhang, H.; Bao, Y.; Liu, S.J.; Liu, X.; Su, Y.; Liu, C.; Wang, H.L.; et al. The miR6445-NAC029 module regulates drought tolerance by regulating the expression of glutathione S-transferase U23 and reactive oxygen species scavenging in Populus. New Phytol. 2024, 242, 2043–2058. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zang, W.; Li, X.; Wang, C.; Wang, R.; Jiang, T.; Zhou, B.; Yao, W. Ectopic Expression of PsnNAC090 enhances Salt and Osmotic Tolerance in Transgenic Tobacco. Int. J. Mol. Sci. 2023, 24, 8985. [Google Scholar] [CrossRef]
- Zhang, X.; Cheng, Z.; Fan, G.; Yao, W.; Li, W.; Chen, S.; Jiang, T. Functional analysis of PagNAC045 transcription factor that improves salt and ABA tolerance in transgenic tobacco. BMC Plant Biol. 2022, 22, 261. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Zhang, X.; Duan, H.; Lian, C.; Liu, C.; Yin, W.; Xia, X. Three stress-responsive NAC transcription factors from Populus euphratica differentially regulate salt and drought tolerance in transgenic plants. Physiol. Plant. 2018, 162, 73–97. [Google Scholar] [CrossRef]
- Rajput, V.D.; Harish; Singh, R.K.; Verma, K.K.; Sharma, L.; Quiroz-Figueroa, F.R.; Meena, M.; Gour, V.S.; Minkina, T.; Sushkova, S.; et al. Recent Developments in Enzymatic Antioxidant Defence Mechanism in Plants with Special Reference to Abiotic Stress. Biology 2021, 10, 267. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Song, R.; Cai, K.; Pang, Z.; Qian, C.; Xu, S.; Zhang, Y.; Bai, H.; Zhan, W.; Xiao, R.; et al. Exploring the molecular mechanisms of melatonin-induced tolerance to salt-alkali stress in Populus cathayana × canadansis ‘Xinlin 1’. Ind. Crops Prod. 2024, 215, 118638. [Google Scholar] [CrossRef]
- Gratão, P.L.; Polle, A.; Lea, P.J.; Azevedo, R.A. Making the Life of Heavy Metal-Stressed Plants a Little Easier. Funct. Plant Biol. 2005, 32, 481–494. [Google Scholar] [CrossRef]
- Zhang, P.; Liu, S.; Chen, K. Characterization and Expression Analysis of a Glutathione Reductase Gene from Antarctic Moss pohlia nutans. Plant Mol. Biol. Rep. 2013, 31, 1068–1076. [Google Scholar] [CrossRef]
- Yu, L.; Tang, S.; Guo, C.; Korpelainen, H.; Li, C. Differences in ecophysiological responses of Populus euphratica females and males exposed to salinity and alkali stress. Plant Physiol. Biochem. 2023, 198, 107707. [Google Scholar] [CrossRef]
- Jan, M.; Muhammad, S.; Jin, W.; Zhong, W.; Zhang, S.; Lin, Y.; Zhou, Y.; Liu, J.; Liu, H.; Munir, R.; et al. Modulating root system architecture: Cross-talk between auxin and phytohormones. Front. Plant Sci. 2024, 15, 1343928. [Google Scholar] [CrossRef]
- El Sabagh, A.; Islam, M.S.; Hossain, A.; Iqbal, M.A.; Mubeen, M.; Waleed, M.; Reginato, M.; Battaglia, M.; Ahmed, S.; Rehman, A.; et al. Phytohormones as Growth Regulators During Abiotic Stress Tolerance in Plants. Front. Agron. 2022, 4, 765068. [Google Scholar] [CrossRef]
- Willige, B.C.; Ghosh, S.; Nill, C.; Zourelidou, M.; Dohmann, E.M.N.; Maier, A.; Schwechheimer, C. The DELLA Domain of GA INSENSITIVE Mediates the Interaction with the GA INSENSITIVE DWARF1A Gibberellin Receptor of Arabidopsis. Plant Cell 2007, 19, 1209–1220. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.; Li, Z.; Yuan, H.; Fang, P.; Chen, X.; Li, W. Proper gibberellin localization in vascular tissue is required to regulate adventitious root development in tobacco. J. Exp. Bot. 2013, 64, 3411–3424. [Google Scholar] [CrossRef]
- Björklund, S.; Antti, H.; Uddestrand, I.; Moritz, T.; Sundberg, B. Cross-talk between gibberellin and auxin in development of populus wood: Gibberellin stimulates polar auxin transport and has a common transcriptome with auxin. Plant J. 2007, 52, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xu, J.; Xu, Z.; Xue, H. Brassinosteroids Stimulate Plant Tropisms through Modulation of Polar Auxin Transport in Brassica and Arabidopsis. Plant Cell 2005, 17, 2738–2753. [Google Scholar] [CrossRef]
- Manickam, S.; Rajagopalan, V.R.; Kambale, R.; Rajasekaran, R.; Kanagarajan, S.; Muthurajan, R.; Sveriges, L. Plant Metabolomics: Current Initiatives and Future Prospects. Curr. Issues Mol. Biol. 2023, 45, 8894–8906. [Google Scholar] [CrossRef]
- Nakamura, Y.; Li-Beisson, Y. Lipids: From Chemical Structures, Biosynthesis, and Analyses to Industrial Applications. In Lipids in Plant and Algae Development; Springer International Publishing AG: Cham, Switzerland, 2016; Volume 86, pp. 1–18. [Google Scholar]
- Gasulla, F.; Vom Dorp, K.; Dombrink, I.; Zähringer, U.; Gisch, N.; Dörmann, P.; Bartels, D. The role of lipid metabolism in the acquisition of desiccation tolerance in Craterostigma plantagineum: A comparative approach. Plant J. 2013, 75, 726–741. [Google Scholar] [CrossRef]
- Hou, Q.; Ufer, G.; Bartels, D. Lipid signalling in plant responses to abiotic stress. Plant Cell Environ. 2016, 39, 1029–1048. [Google Scholar] [CrossRef]
- Gaude, N.; Bréhélin, C.; Tischendorf, G.; Kessler, F.; Dörmann, P. Nitrogen deficiency in Arabidopsis affects galactolipid composition and gene expression and results in accumulation of fatty acid phytyl esters. Plant J. Cell Mol. Biol. 2007, 49, 729–739. [Google Scholar] [CrossRef]
- Dal Santo, S.; Zenoni, S.; Sandri, M.; De Lorenzis, G.; Magris, G.; De Paoli, E.; Di Gaspero, G.; Del Fabbro, C.; Morgante, M.; Brancadoro, L.; et al. Grapevine field experiments reveal the contribution of genotype, the influence of environment and the effect of their interaction (G×E) on the berry transcriptome. Plant J. 2018, 93, 1143–1159. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Tran, L.S.P.; Van Nguyen, D.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Yamaguchi Shinozaki, K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007, 51, 617–630. [Google Scholar] [CrossRef]
- Liu, J.; Feng, L.; Gu, X.; Deng, X.; Qiu, Q.; Li, Q.; Zhang, Y.; Wang, M.; Deng, Y.; Wang, E.; et al. An H3K27me3 demethylase-HSFA2 regulatory loop orchestrates transgenerational thermomemory in Arabidopsis. Cell Res. 2019, 29, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Li, Y.; Wang, Y.; Liu, X.; Ma, L.; Zhang, Z.; Mu, C.; Zhang, Y.; Peng, L.; Xie, S.; et al. Initiation and amplification of SnRK2 activation in abscisic acid signaling. Nat. Commun. 2021, 12, 2456. [Google Scholar] [CrossRef]
- Liu, N.; Du, Y.; Warburton, M.L.; Xiao, Y.; Yan, J. Phenotypic plasticity contributes to maize adaptation and heterosis. Mol. Biol. Evol. 2021, 38, 1262–1275. [Google Scholar] [CrossRef] [PubMed]
- Lasky, J.R.; Josephs, E.B.; Morris, G.P. Genotype-environment associations to reveal the molecular basis of environmental adaptation. Plant Cell 2023, 35, 125–138. [Google Scholar] [CrossRef]
- Li, J.; Liu, H.; Wang, J.; Macdonald, C.A.; Singh, P.; Cong, V.T.; Klein, M.; Delgado-Baquerizo, M.; Singh, B.K. Drought-induced plant microbiome and metabolic enrichments improve drought resistance. Cell Host Microbe 2025, 33, 882–900.E7. [Google Scholar] [CrossRef]
- Lang, Z.; Wang, Y.; Tang, K.; Tang, D.; Datsenka, T.; Cheng, J.; Zhang, Y.; Handa, A.K.; Zhu, J. Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc. Natl. Acad. Sci. USA 2017, 114, E4511–E4519. [Google Scholar] [CrossRef]
- Li, Q.; Song, J.; Zhou, Y.; Chen, Y.; Zhang, L.; Pang, Y.; Zhang, B. Full-Length Transcriptomics Reveals Complex Molecular Mechanism of Salt Tolerance in Bromus inermis L. Front. Plant Sci. 2022, 13, 917338. [Google Scholar]
- Hood-Nowotny, R.; Umana, N.H.; Inselbacher, E.; Oswald-Lachouani, P.; Wanek, W. Alternative Methods for Measuring Inorganic, Organic, and Total Dissolved Nitrogen in Soil. Soil Sci. Soc. Am. J. 2010, 74, 1018–1027. [Google Scholar] [CrossRef]
- Tafere, D.A.; Teshager, M.A. Determination of the Levels of Selected Essential Metals in Sycamore (Ficus sycomorus L) Fruit and Seed Using Flame Atomic Absorption Spectrophotometry. J. Chem. 2023, 2023, 3937604. [Google Scholar] [CrossRef]
- Matula, J. Differences in available phosphorus evaluated by soil tests in relation to detection by colorimetric and ICP-AES techniques. Plant Soil Environ. 2010, 56, 297–304. [Google Scholar] [CrossRef]
- Jeannotte, R.; Sommerville, D.; Hamel, C.; Whalen, J. A microplate assay to measure soil microbial biomass phosphorus. Biol. Fertil. Soils 2004, 40, 201–205. [Google Scholar] [CrossRef]
- Kiss, T.; Karácsony, Z.; Gomba-Tóth, A.; Szabadi, K.L.; Spitzmüller, Z.; Hegyi-Kaló, J.; Cels, T.; Otto, M.; Golen, R.; Hegyi, Á.I.; et al. A modified CTAB method for the extraction of high-quality RNA from mono-and dicotyledonous plants rich in secondary metabolites. Plant Methods 2024, 20, 62. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Wu, S.; Da, L.; Xiao, Q.; Pan, Q.; Zhang, J.; Yang, J. ASAP: A platform for gene functional analysis in Angelica sinensis. BMC Genom. 2024, 25, 96. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. Tbtools: An Integrative ToolKit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Zou, S.; Wu, J.; Shahid, M.Q.; He, Y.; Lin, S.; Liu, Z.; Yang, X. Identification of key taste components in loquat using widely targeted metabolomics. Food Chem. 2020, 323, 126822. [Google Scholar] [CrossRef]
- Da Silva, J.M.G.; de Almeida, R.F.; Zeraik, M.L. Comparative metabolite Profiling of Three Savannic Species of Banisteriopsis (Malpighiaceae) via UPLC-MS/MS and Chemometric tools. Chem. Biodivers. 2024, 21, e202400679. [Google Scholar] [CrossRef]
- Blaženović, I.; Kind, T.; Ji, J.; Fiehn, O. Software Tools and Approaches for Compound Identification of LC-MS/MS Data in Metabolomics. Metabolites 2018, 8, 31. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-time Quantitative PCA and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
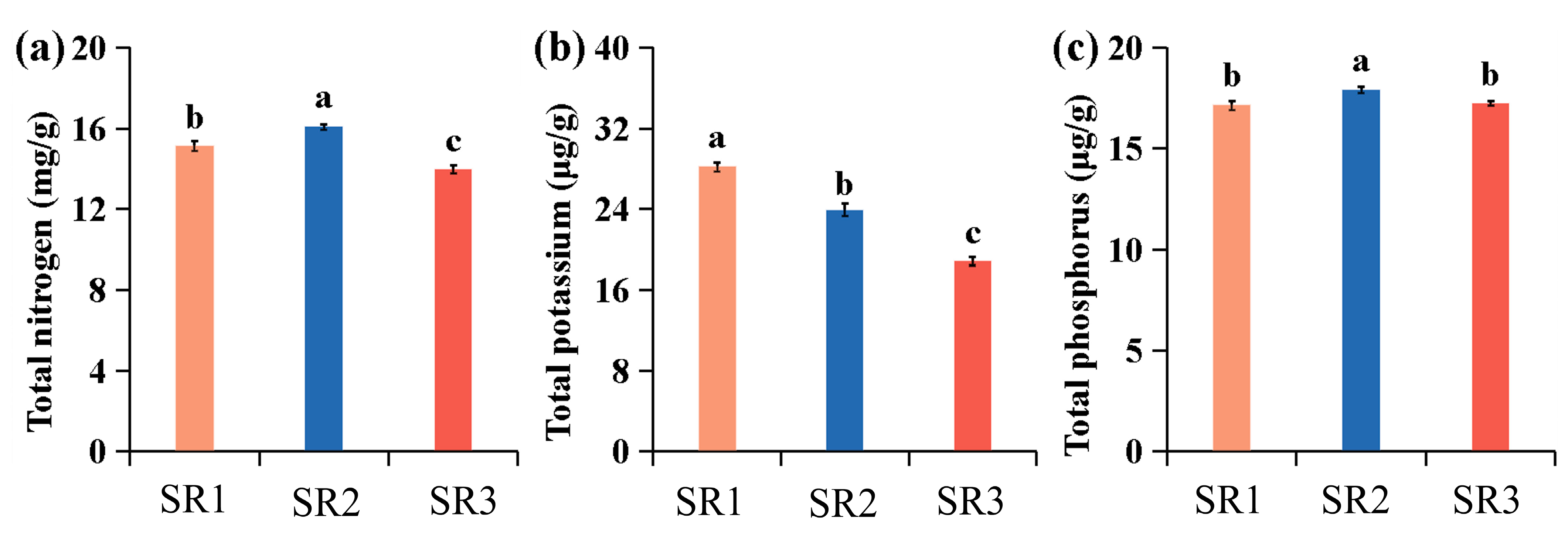

| Site | Longitude | Latitude | Average Temperature | Precipitation | Wind Speed | Relative Humidity | Tree Height | Diameter of Breast Height |
|---|---|---|---|---|---|---|---|---|
| SR1 (Lishu) | 124°54′ | 43°62′ | 6.9 °C | 480 mm | 3.83 m/s | 61.88% | 16.00 m | 15.90 cm |
| SR2 (Xinmin) | 122°56′ | 41°88′ | 8.2 °C | 600 mm | 3.56 m/s | 57.08% | 17.73 m | 19.33 cm |
| SR3 (Cuohai) | 122°85′ | 47 °45′ | 3.4 °C | 465 mm | 3.52 m/s | 56.54% | 13.15 m | 13.32 cm |
| Gene ID | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| POPTR_001G271700v3 | CAGCCAATGAGGCCTGTTGGTG | GACTCATCAACTGATTGCTCGGCA |
| POPTR_001G281900v3 | TCCGTCTAGCAGTGAGAGATCTTCA | GCAATTGAACCGGTCTCTCTGC |
| POPTR_003G011200v3 | TTCCACCGTCGTCGACTCTG | CACGAACAGAAGTCATGCCGA |
| POPTR_009G096000v3 | CCAGTACTCCTTCGGGATCTGA | CCAACTGTCCAGACTCCATCC |
| POPTR_014G020500v3 | TGGAAATTGTAATGGAAGAGGAGGGA | GCCCTGACCTAACATCTACTTCTCTA |
| POPTR_019G037400v3 | ATGGCAAATTACGCACCGGGAA | CGAAGCAGTTTGATCTCACGAAGG |
| PsnActin | CCAGCAACCGCAATACAA | CTTCACCATTCCAGTTCCATT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, P.; Luo, J.; Zhao, Q.; Yu, M.; Pei, X.; Jiang, L.; Han, R.; Zhao, X. Transcriptome and Metabolome Analysis of the Mechanism of Environmental Adaptability in Populus Roots. Plants 2025, 14, 2691. https://doi.org/10.3390/plants14172691
Chen P, Luo J, Zhao Q, Yu M, Pei X, Jiang L, Han R, Zhao X. Transcriptome and Metabolome Analysis of the Mechanism of Environmental Adaptability in Populus Roots. Plants. 2025; 14(17):2691. https://doi.org/10.3390/plants14172691
Chicago/Turabian StyleChen, Panrui, Jiaxin Luo, Qiushuang Zhao, Miao Yu, Xiaona Pei, Luping Jiang, Rui Han, and Xiyang Zhao. 2025. "Transcriptome and Metabolome Analysis of the Mechanism of Environmental Adaptability in Populus Roots" Plants 14, no. 17: 2691. https://doi.org/10.3390/plants14172691
APA StyleChen, P., Luo, J., Zhao, Q., Yu, M., Pei, X., Jiang, L., Han, R., & Zhao, X. (2025). Transcriptome and Metabolome Analysis of the Mechanism of Environmental Adaptability in Populus Roots. Plants, 14(17), 2691. https://doi.org/10.3390/plants14172691






