Genome-Wide Identification of Natural Resistance-Associated Macrophage Protein (NRAMP) and Expression Analysis Under Heavy Metal Stress in Sorghum bicolor L.
Abstract
1. Introduction
2. Results
2.1. Identification of SbNRAMP Gene Family Members and Analysis of Their Physicochemical Properties
2.2. SbNRAMP Gene Members Were Mapped on Seven Chromosomes
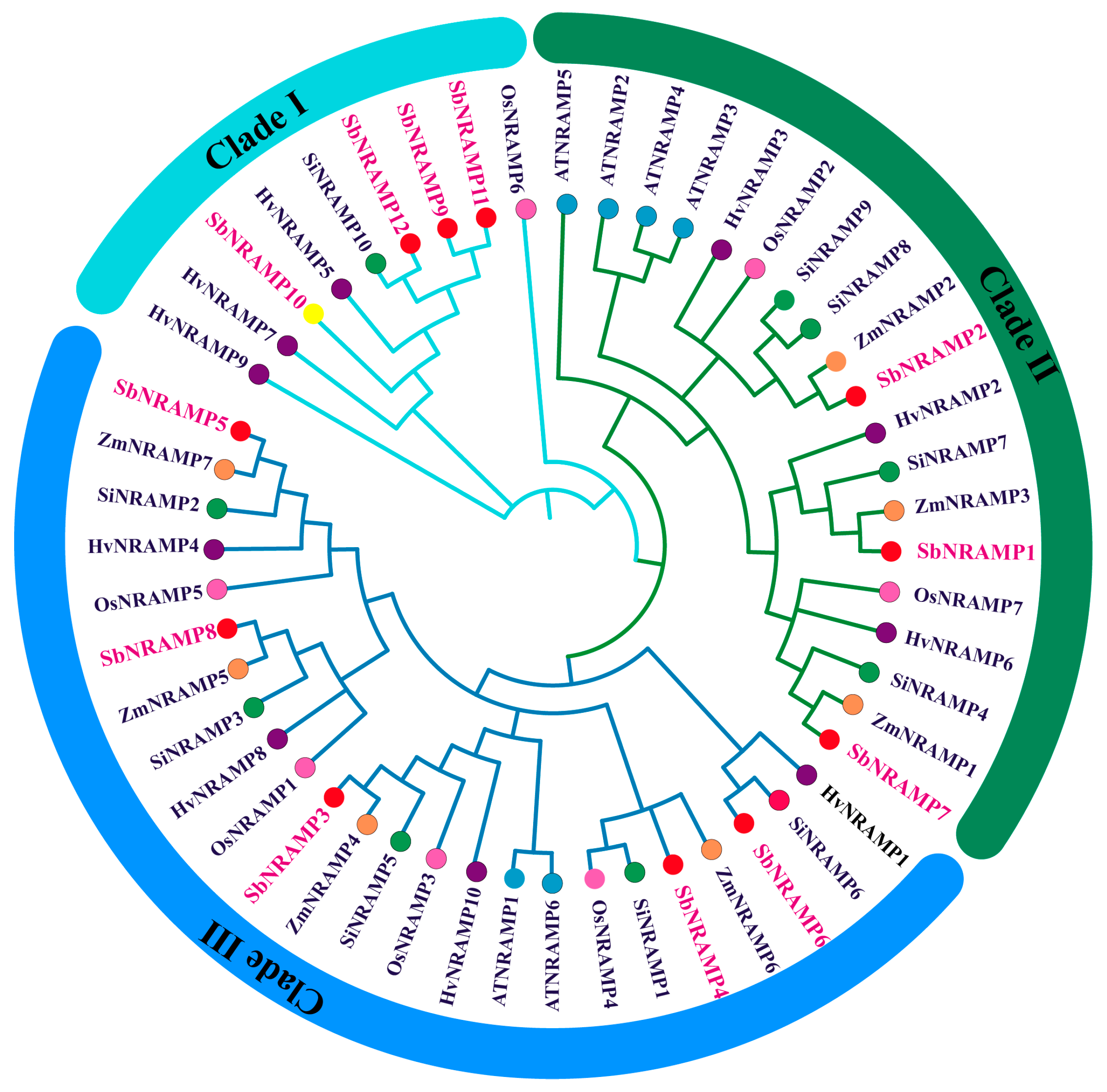
2.3. The Evolutionary Relationships of the Sorghum NRAMP Family in Comparison to Homologous Genes from Various Plant Species
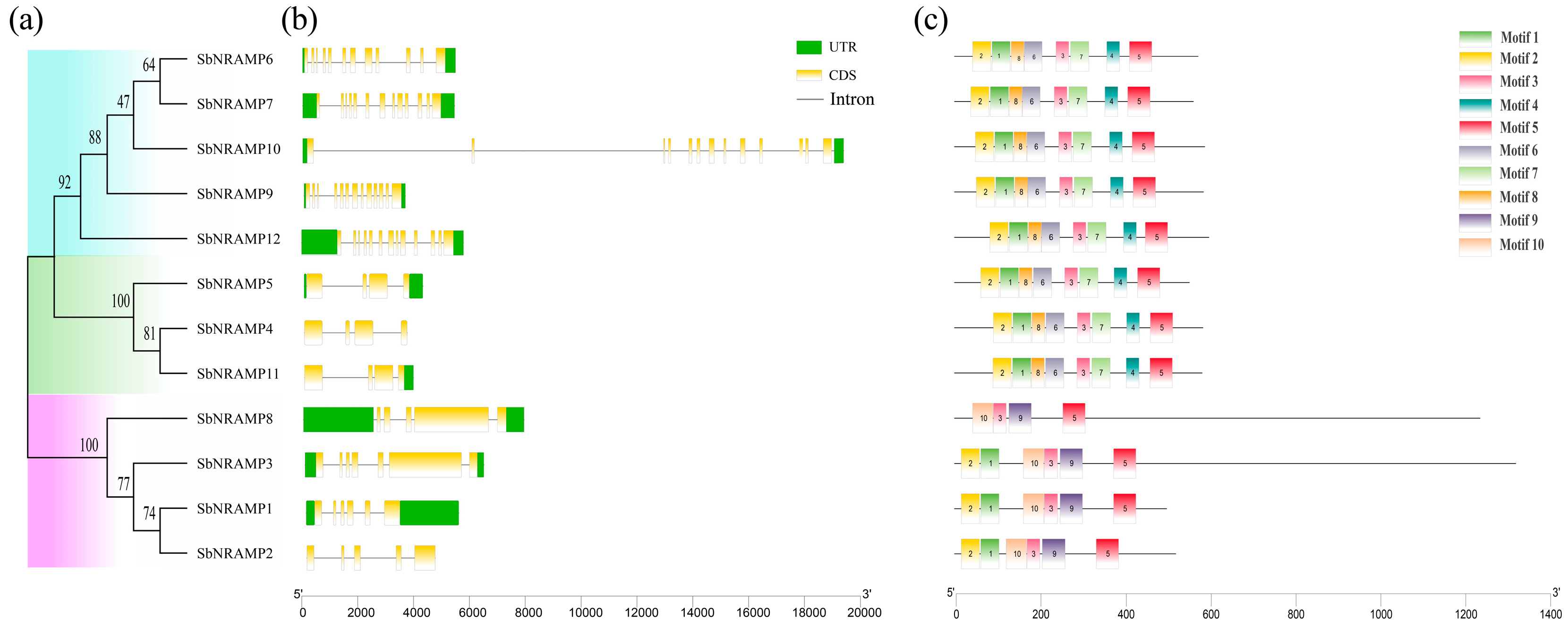
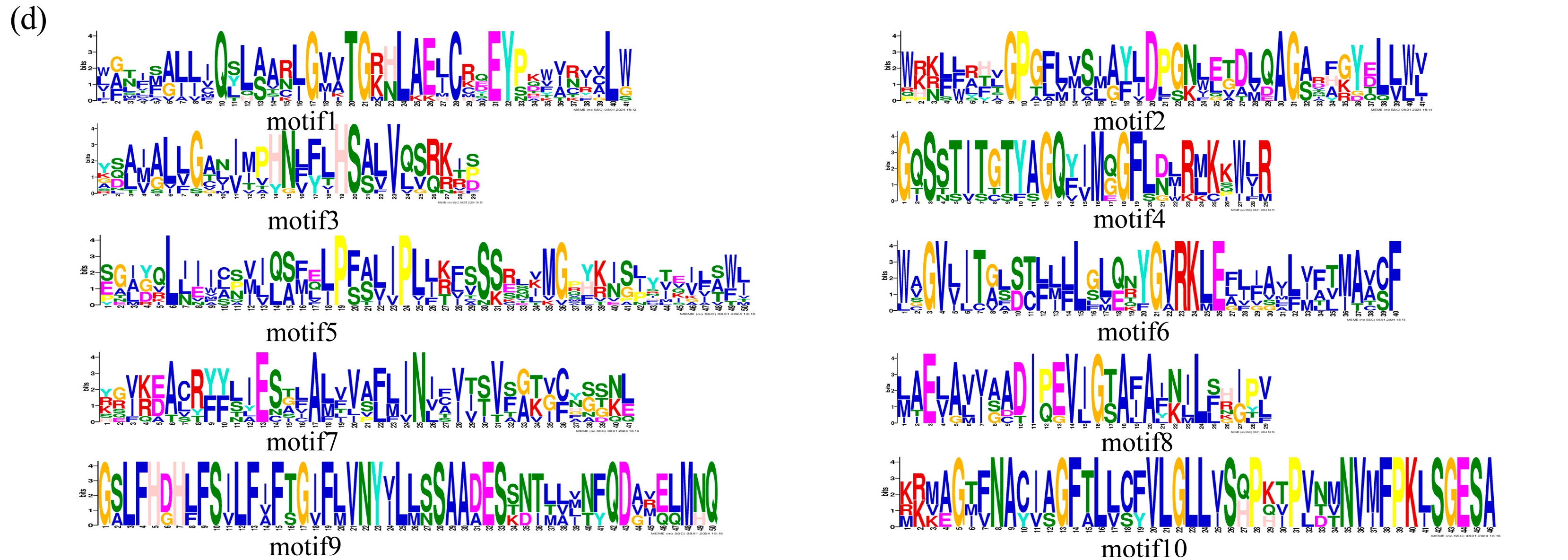
2.4. Comparative Analysis of SbNRAMP Phylogenetic Relationships, Gene Structures, and Conserved Motifs in Sorghum bicolor
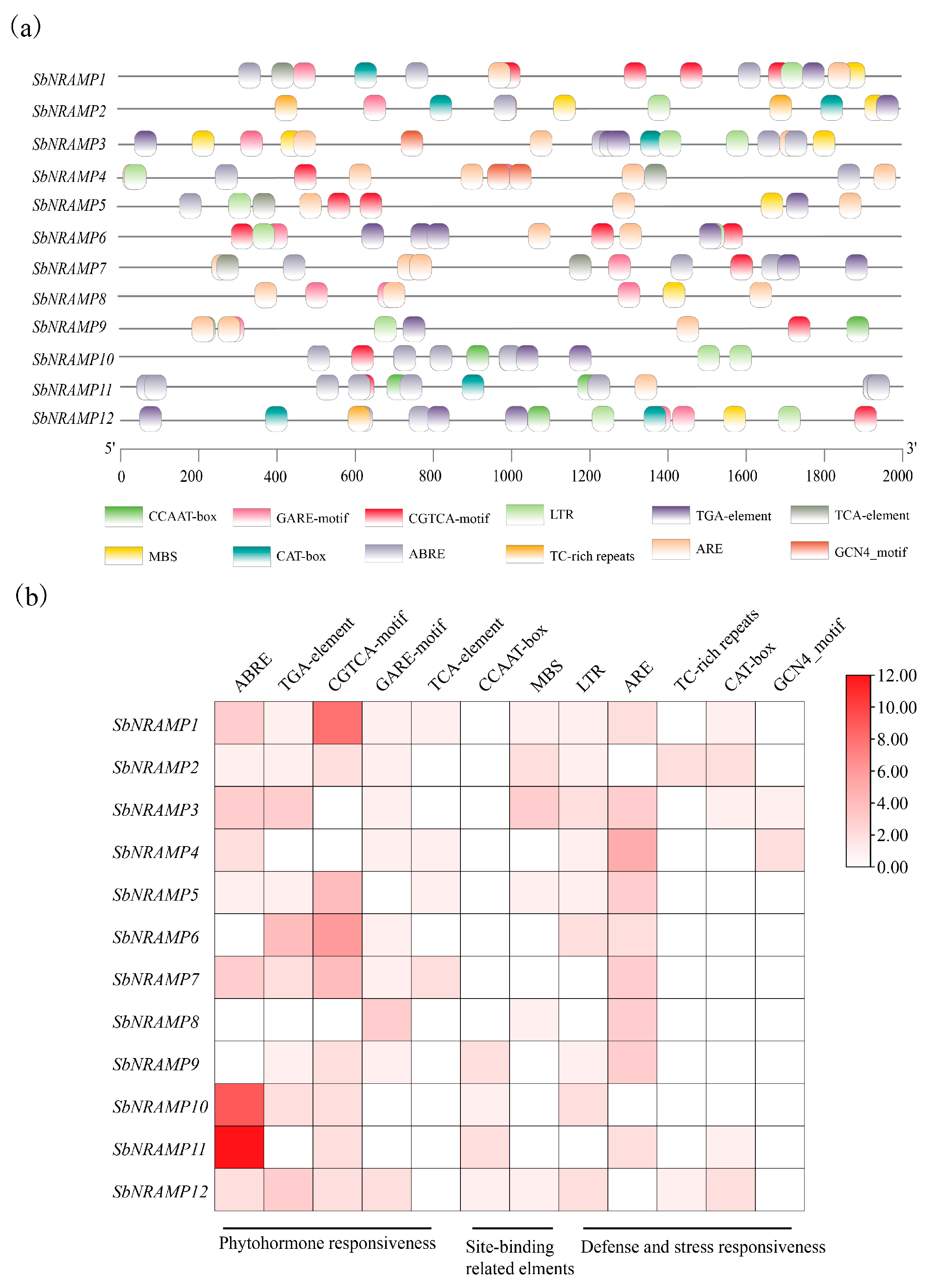
2.5. Analysis of SbNRAMP Cis-Acting Elements
2.6. Intraspecific and Interspecific Collinearity Analysis of SbNRAMP
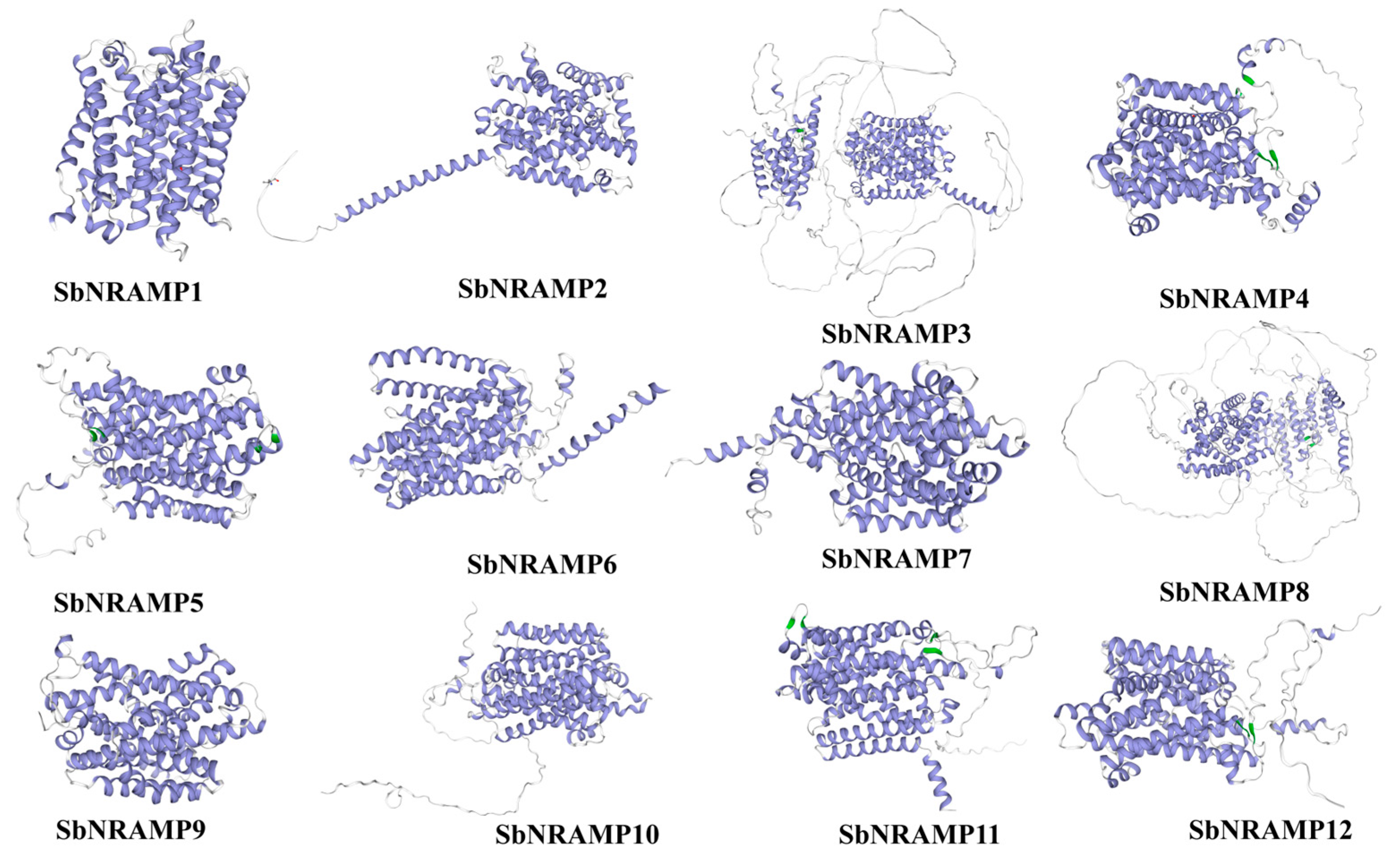
2.7. Subcellular Localization Prediction, Transmembrane Domain Prediction, Protein Structure, and Sequence Analyses of SbNRAMP
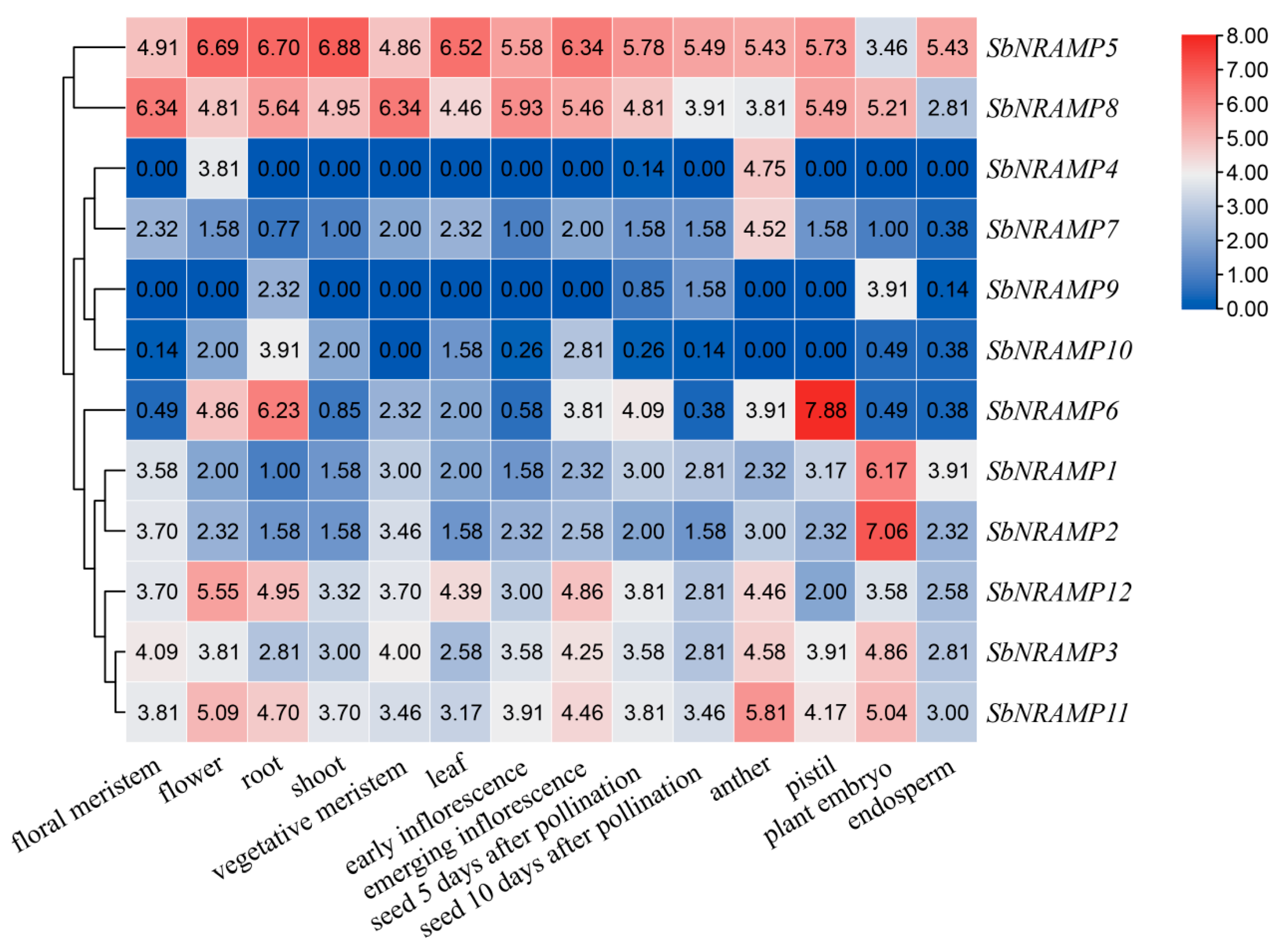
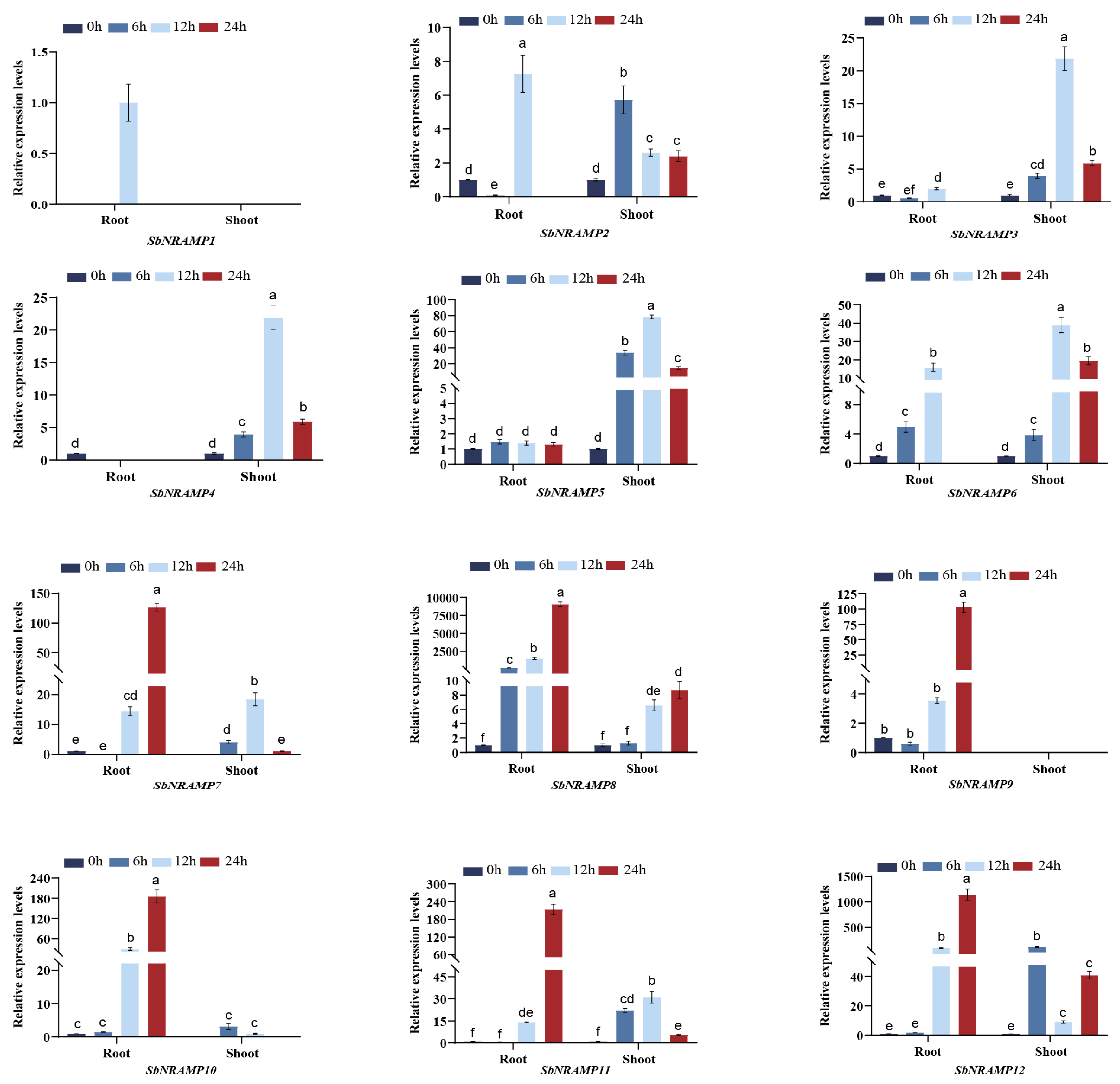
2.8. Tissue Expression Analysis of SbNRAMP
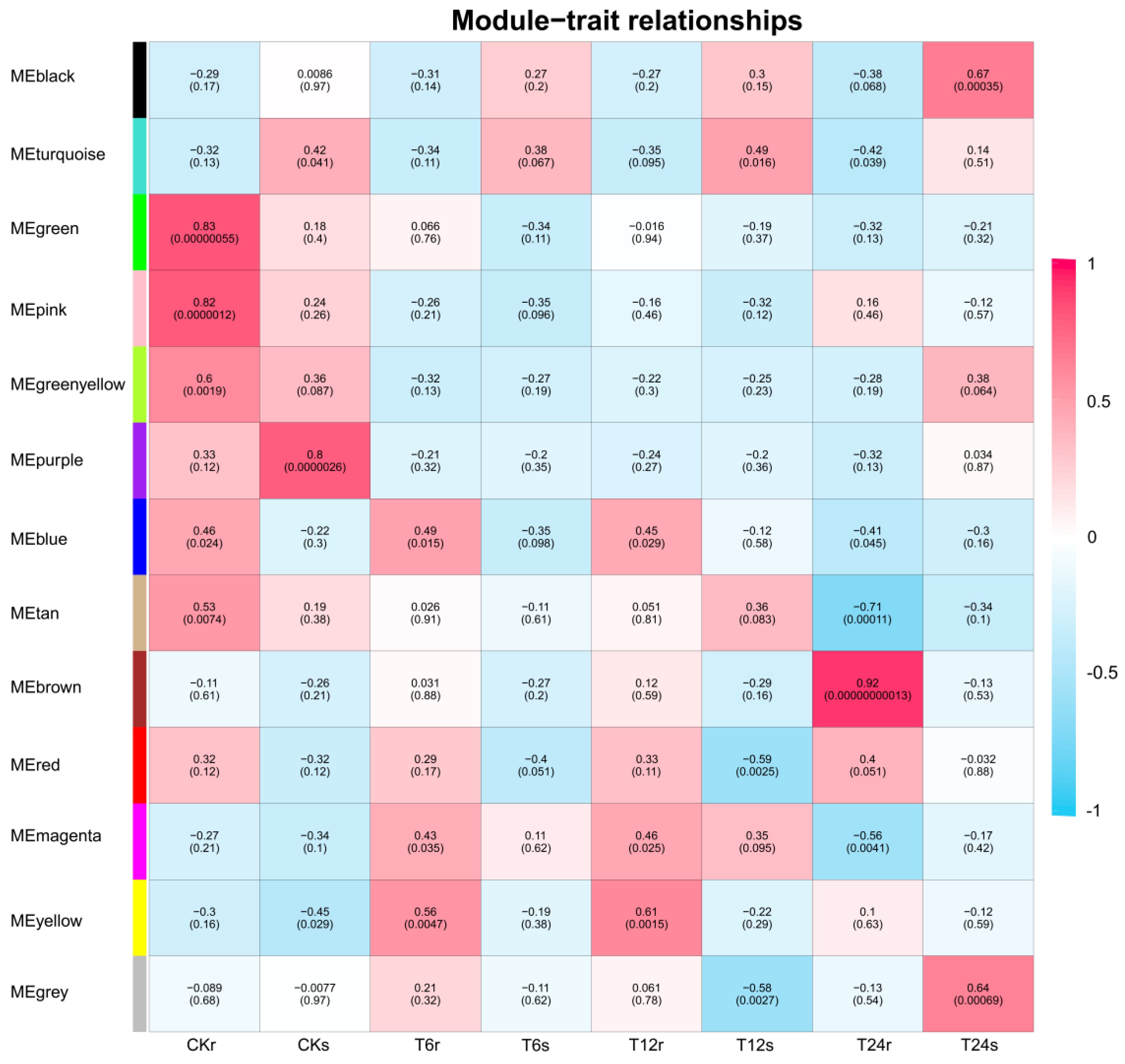
2.9. WGCNA for Identification of Hub Genes
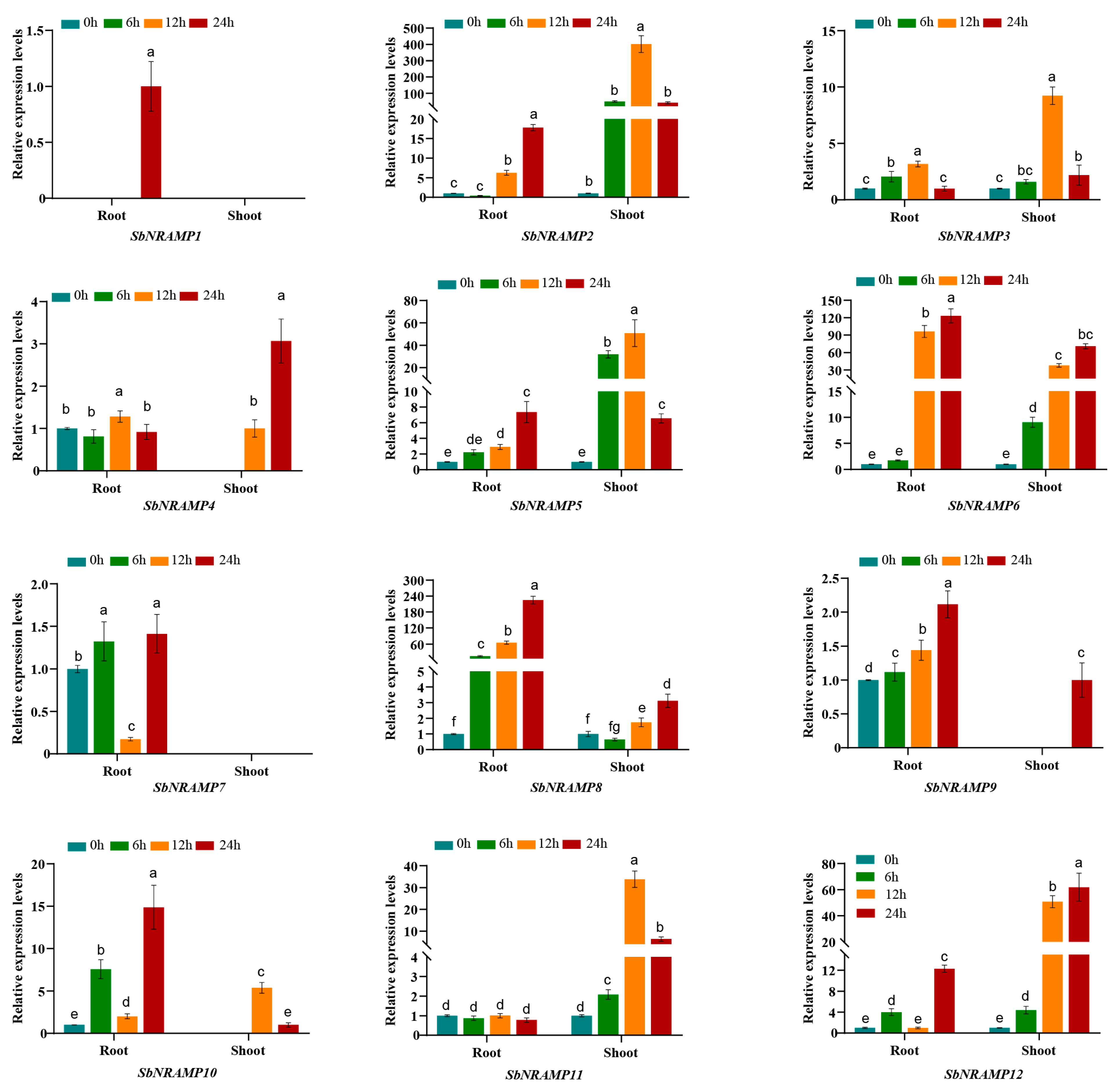
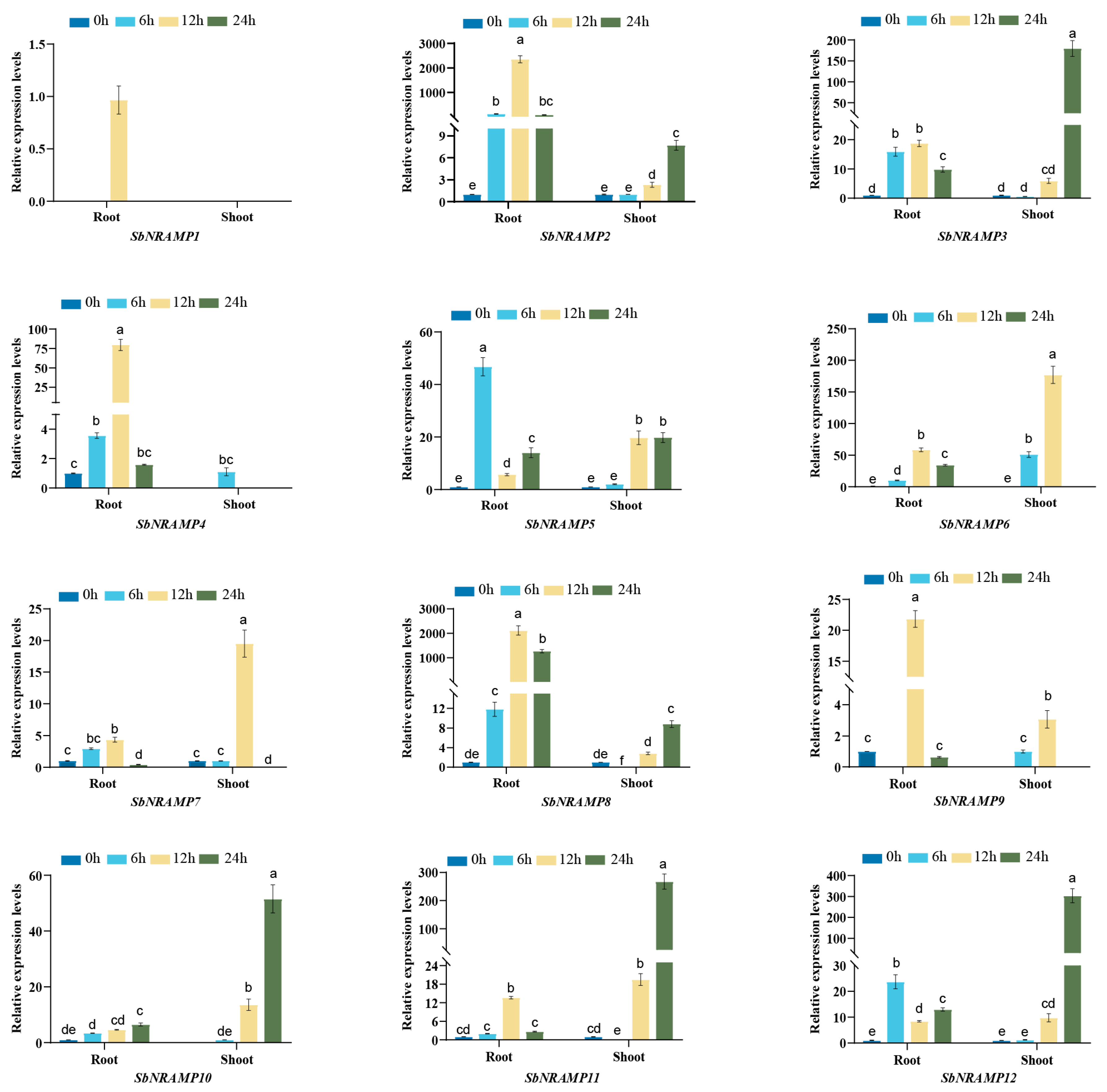
2.10. Cd, Mn, or Zn Induced the Expression of SbNRAMP
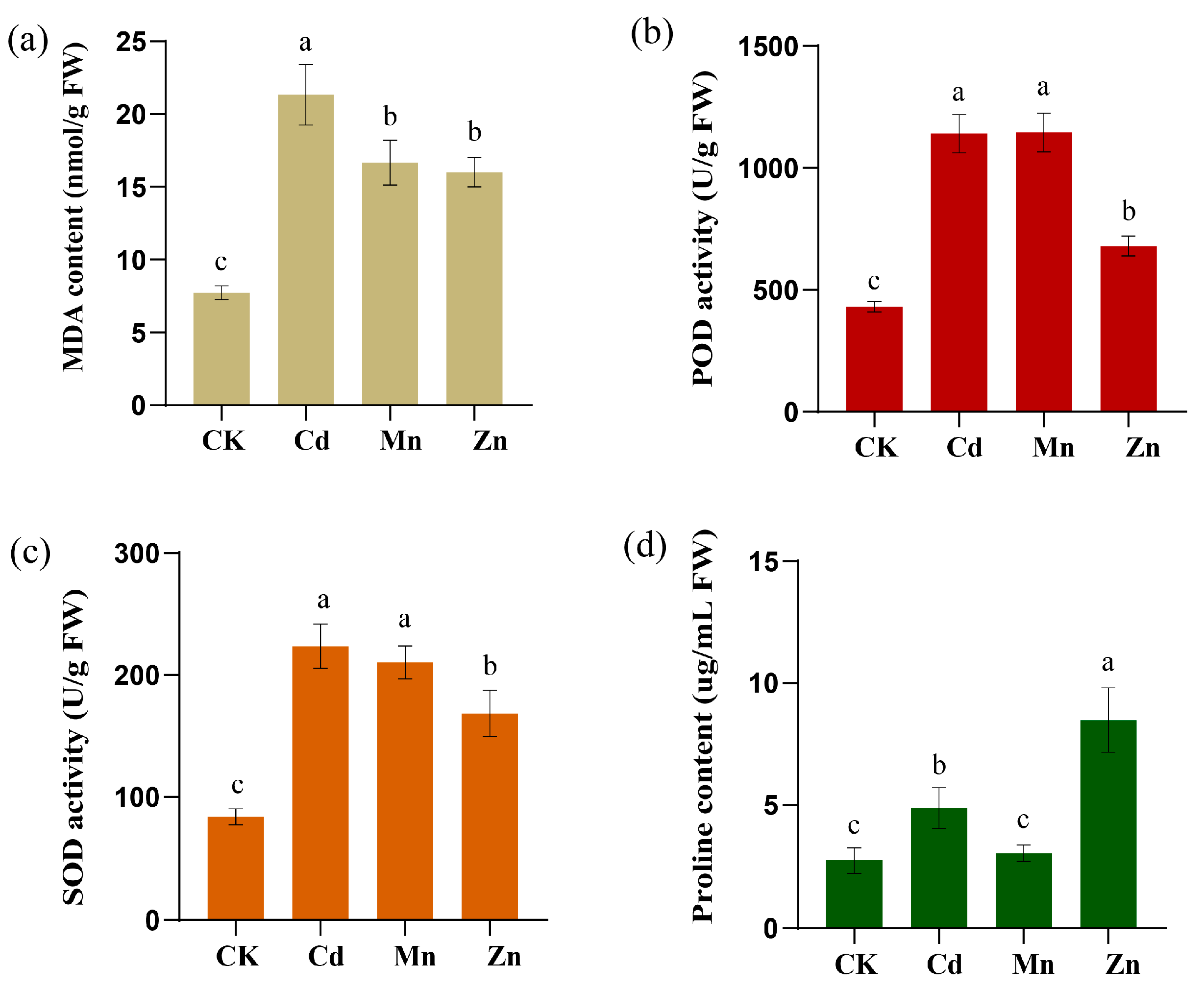
2.11. Effects of Metal Ions Stress on Physiological Indexes of Sorghum Seedlings
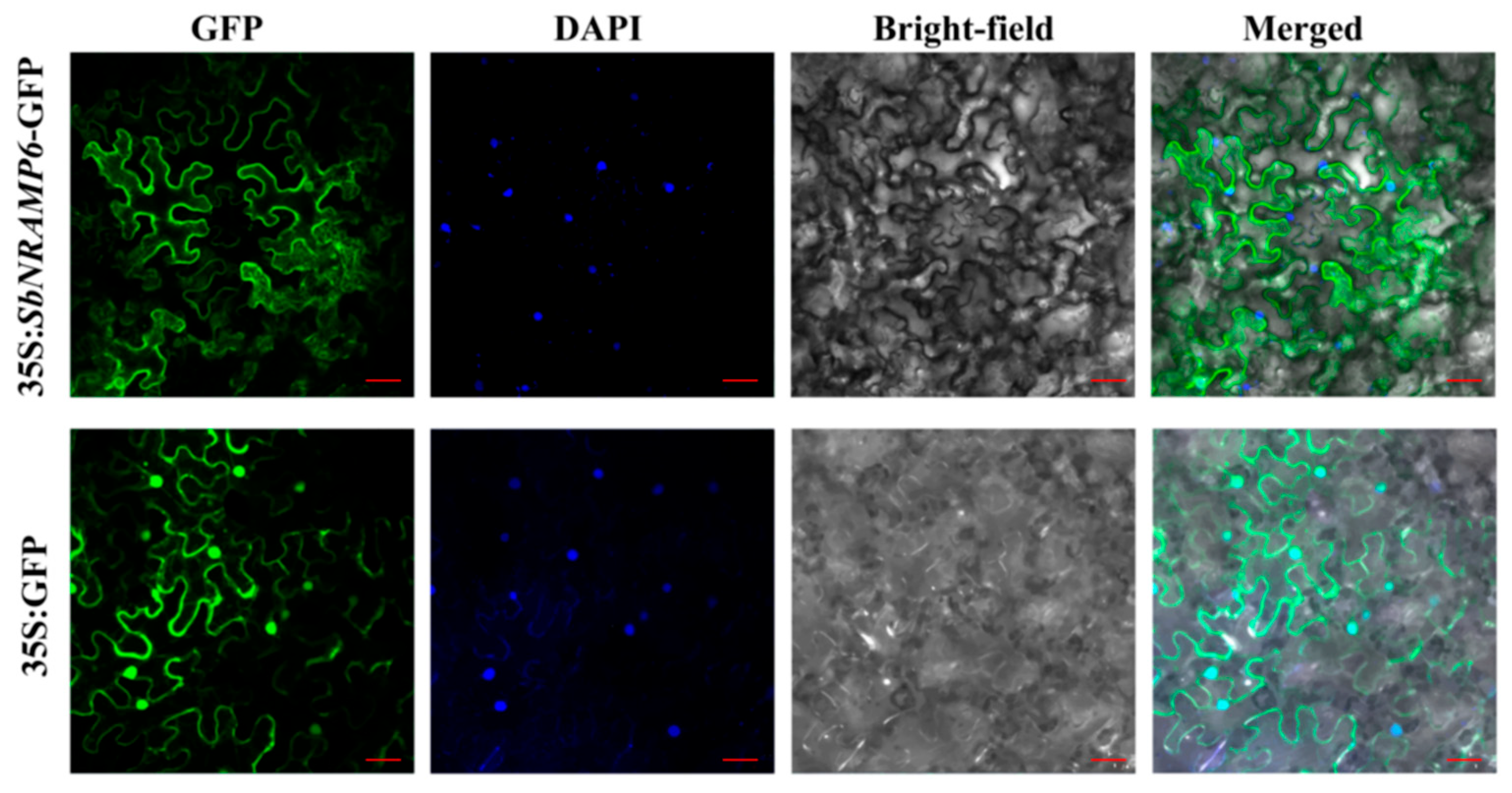
2.12. SbNRAMP6 Was Localized in the Cell Membrane
3. Discussion
4. Materials and Methods
4.1. Material Cultivation and Treatment
4.2. Total RNA Extraction, cDNA Synthesis, and NRAMP Gene Expression Analyses
4.3. Determination of Physiological Indexes
4.4. Subcellular Localization Analysis
4.5. Identification and Physicochemical Properties Analysis of SbNRAMP
4.6. Chromosomal Localization, Protein Structure, and Subcellular Localization Prediction
4.7. Multiple Sequence Alignment and Phylogenetic Analysis
4.8. Conserved Domain Analysis of SbNRAMPs
4.9. Analysis of Cis-Acting Elements of SbNRAMP Promoter
4.10. Replication Events Between SbNRAMPs
4.11. SbNRAMP Expression Profile Mapping
4.12. Weighted Gene Co-Expression Network Analysis (WGCNA)
4.13. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| Gene Name | Primer Name | Primer Sequence (5′–3′) | Tm (°C) | Length (bp) |
|---|---|---|---|---|
| SbNRAMP1 | F | AGCACTCGTGGTCTCAATGG | 60 | 193 |
| R | ACTCCTGGTGGCAAATCTCG | |||
| SbNRAMP2 | F | ACCAGCAAGATAACCGAGGC | 60 | 184 |
| R | TCTTTCAGGAACAGCAGCGT | |||
| SbNRAMP3 | F | GGCTGTTTCGTCAGAATGGC | 59 | 157 |
| R | CAGTGAAGCCTGCCAAACAC | |||
| SbNRAMP4 | F | TCCACCCACCTATGAAGCCGA | 62 | 181 |
| R | CTGAGGCATAGAAGGCACCCT | |||
| SbNRAMP5 | F | GCGCTCGATTGCTTCATCTT | 59 | 175 |
| R | TGATGCAGCCCACAATTCCA | |||
| SbNRAMP6 | F | TCGCAGAGGTGGCTGTAATC | 60 | 119 |
| R | CTTCCGCACCCCGTATCTTT | |||
| SbNRAMP7 | F | GGTGTAGCAATGTTCGTGGC | 60 | 198 |
| R | TTGCCCAGCGTAAGTACCAG | |||
| SbNRAMP8 | F | ATATCTTCTCGGCGCAGTCG | 60 | 208 |
| R | TTGAACAGCCACCCTGATCC | |||
| SbNRAMP9 | F | AATGCAGACAACCTCTCCCC | 59 | 140 |
| R | ATGATGACCTGCCCAGCAAA | |||
| SbNRAMP10 | F | TGTGGAGCAAGCACTTTCCT | 60 | 148 |
| R | TGGCATAAACCCCTCGCAAT | |||
| SbNRAMP11 | F | GCCCTAACACCCAAGCTGTA | 59 | 175 |
| R | CTGCTGTGGACTGCTTTGAC | |||
| SbNRAMP12 | F | TGTCCACCGTCTCCCAAGTA | 60 | 166 |
| R | ATGTCCTCTCGGGGCAAATG |

References
- Kapilan, R.; Vaziri, M.; Zwiazek, J.J. Regulation of Aquaporins in Plants under Stress. Biol. Res. 2018, 51, 4. [Google Scholar] [CrossRef]
- Feil, S.B.; Pii, Y.; Valentinuzzi, F.; Tiziani, R.; Mimmo, T.; Cesco, S. Copper Toxicity Affects Phosphorus Uptake Mechanisms at Molecular and Physiological Levels in Cucumis sativus Plants. Plant Physiol. Biochem. 2020, 157, 138–147. [Google Scholar] [CrossRef]
- Farhat, N.; Elkhouni, A.; Zorrig, W.; Smaoui, A.; Abdelly, C.; Rabhi, M. Effects of Magnesium Deficiency on Photosynthesis and Carbohydrate Partitioning. Acta Physiol. Plant. 2016, 38, 145. [Google Scholar] [CrossRef]
- Gate, T.; Hill, L.; Miller, A.J.; Sanders, D. AtIAR1 Is a Zn Transporter That Regulates Auxin Metabolism in Arabidopsis thaliana. J. Exp. Bot. 2023, 75, 1437–1450. [Google Scholar] [CrossRef]
- Mary, V.; Ramos, M.S.; Gillet, C.; Socha, A.L.; Giraudat, J.; Agorio, A.; Merlot, S.; Clairet, C.; Kim, S.A.; Punshon, T.; et al. Bypassing Iron Storage in Endodermal Vacuoles Rescues the Iron Mobilization Defect in the Natural Resistance Associated-Macrophage Protein 3 Natural Resistance Associated-Macrophage Protein 4 Double Mutant. Plant Physiol. 2015, 169, 748–759. [Google Scholar] [CrossRef]
- Pirooz, P.; Amooaghaie, R.; Bakhtiari, S. Interactive Effect of Silicon and Nitric Oxide Effectively Contracts Copper Toxicity in Salvia officinalis L. Int. J. Phytoremediat. 2023, 25, 1801–1809. [Google Scholar] [CrossRef]
- Niekerk, L.-A.; Carelse, M.F.; Bakare, O.O.; Mavumengwana, V.; Keyster, M.; Gokul, A. The Relationship between Cadmium Toxicity and the Modulation of Epigenetic Traits in Plants. Int. J. Mol. Sci. 2021, 22, 7046. [Google Scholar] [CrossRef]
- Zhu, T.; Li, L.; Duan, Q.; Liu, X.; Chen, M. Progress in Our Understanding of Plant Responses to the Stress of Heavy Metal Cadmium. Plant Signal. Behav. 2021, 16, 1836884. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, T.; Joshi, N.; Fahad, S.; Saud, S.; Ur Rahman, T.; Khan, M.N.R.; Hassan, S. Solar-Powered N2-Fixing Cyanobacteria for Bio-Nitrogen Fertilizer Production and Soil Health Improvement: A Sustainable Farming Approach. In Environment, Climate, Plant and Vegetation Growth; Fahad, S., Saud, S., Nawaz, T., Gu, L., Ahmad, M., Zhou, R., Eds.; Springer Nature: Cham, Switzerland, 2024; pp. 75–113. ISBN 978-3-031-69417-2. [Google Scholar]
- Khalid, S.; Shahid, M.; Niazi, N.K.; Murtaza, B.; Bibi, I.; Dumat, C. A Comparison of Technologies for Remediation of Heavy Metal Contaminated Soils. J. Geochem. Explor. 2017, 182, 247–268. [Google Scholar] [CrossRef]
- Zheng, H.; Dang, Y.; Sui, N. Sorghum: A Multipurpose Crop. J. Agric. Food Chem. 2023, 71, 17570–17583. [Google Scholar] [CrossRef]
- Schmidt, J.J.; Yerka, M.K.; Pedersen, J.F.; Lindquist, J.L. Growth, Fitness, and Overwinter Survival of a Shattercane (Sorghum bicolor ssp. drummondii) × Grain Sorghum (Sorghum bicolor ssp. bicolor) F2 Population. Weed Sci. 2018, 66, 634–641. [Google Scholar] [CrossRef]
- Ma, J.; Liu, Z.; Guo, Z.; Wang, X.; Zou, C.; Zhang, C.; Gai, Z. Identification and Function Analysis of Drought-Responsive miRNAs in Sorghum (Sorghum bicolor). Braz. J. Bot. 2025, 48, 30. [Google Scholar] [CrossRef]
- Li, H.; Xu, J.; Sun, Q.; Wang, X.; Lin, J.; Chen, S.; Ma, H.; Zhong, M. Two Petroleum-Induced Small Heat Shock Proteins of Mirabilis Jalapa Confer Tunicamycin Tolerance in Transgenic Saccharomyces Cerevisiae. Environ. Eng. Sci. 2020, 37, 826–837. [Google Scholar] [CrossRef]
- Smith, N.E.G.; Tooley, E.G.; Maricle, B.R. Inorganic Fertilizer and Salt Tolerance in Sorghum bicolor (L.) Moench ssp. Bicolor. J. Plant Nutr. 2020, 43, 1390–1399. [Google Scholar] [CrossRef]
- Rahman, T.U.; Shah, S.; Hassan, S.; Fahad, S. Food Security Challenges and Adaptation Strategies in China amidst Global Climate Change. J. Umm Al-Qura Univ. Appll. Sci. 2025. [Google Scholar] [CrossRef]
- Qin, L.; Han, P.; Chen, L.; Walk, T.C.; Li, Y.; Hu, X.; Xie, L.; Liao, H.; Liao, X. Genome-Wide Identification and Expression Analysis of NRAMP Family Genes in Soybean (Glycine max L.). Front. Plant Sci. 2017, 8, 1436. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhao, X.; Li, G.; Kumar, S.; Sun, Z.; Li, Y.; Guo, W.; Yang, J.; Hou, H. Genome-Wide Identification of the Nramp Gene Family in Spirodela polyrhiza and Expression Analysis under Cadmium Stress. Int. J. Mol. Sci. 2021, 22, 6414. [Google Scholar] [CrossRef] [PubMed]
- Mani, A.; Sankaranarayanan, K. In Silico Analysis of Natural Resistance-Associated Macrophage Protein (NRAMP) Family of Transporters in Rice. Protein J. 2018, 37, 237–247. [Google Scholar] [CrossRef]
- Yang, Y.; Zheng, J.; Liang, Y.; Wang, X.; Li, K.; Chen, L.; Aduragbemi, A.; Han, Y.; Sun, Z.; Li, H.; et al. Natural Resistance-Associated Macrophage Protein (Nramp) Family in Foxtail Millet (Setaria italica): Characterization, Expression Analysis and Relationship with Metal Content Under Cd Stress. Agronomy 2023, 13, 2000. [Google Scholar] [CrossRef]
- Zhang, X.D.; Meng, J.G.; Zhao, K.X.; Chen, X.; Yang, Z.M. Annotation and Characterization of Cd-Responsive Metal Transporter Genes in Rapeseed (Brassica napus). Biometals 2017, 31, 107–121. [Google Scholar] [CrossRef]
- Mäser, P.; Thomine, S.; Schroeder, J.I.; Ward, J.M.; Hirschi, K.; Sze, H.; Talke, I.N.; Amtmann, A.; Maathuis, F.J.; Sanders, D.; et al. Phylogenetic Relationships within Cation Transporter Families of Arabidopsis. Plant Physiol. 2001, 126, 1646–1667. [Google Scholar] [CrossRef]
- Meng, J.G.; Zhang, X.D.; Tan, S.K.; Zhao, K.X.; Yang, Z.M. Genome-Wide Identification of Cd-Responsive NRAMP Transporter Genes and Analyzing Expression of NRAMP 1 Mediated by miR167 in Brassica napus. Biometals 2017, 30, 917–931. [Google Scholar] [CrossRef]
- Takahashi, R.; Ishimaru, Y.; Nakanishi, H.; Nishizawa, N.K. Role of the Iron Transporter OsNRAMP1 in Cadmium Uptake and Accumulation in Rice. Plant Signal. Behav. 2011, 6, 1813–1816. [Google Scholar] [CrossRef]
- Wang, N.; Qiu, W.; Dai, J.; Guo, X.; Lu, Q.; Wang, T.; Li, S.; Liu, T.; Zuo, Y. AhNRAMP1 Enhances Manganese and Zinc Uptake in Plants. Front. Plant Sci. 2019, 10, 415. [Google Scholar] [CrossRef]
- Yu, Y.; Rong, K.; Sui, X.; Zhang, L.; Zhang, M.; Hu, H.; Jia, J.; Wu, J.; Li, C. Analysis of NRAMP Genes in the Triticeae Reveals That TaNRAMP5 Positively Regulates Cadmium (Cd) Tolerance in Wheat (Triticum aestivum). Plant Physiol. Biochem. 2025, 219, 109321. [Google Scholar] [CrossRef] [PubMed]
- Toyokura, K.; Naito, K.; Makabe, K.; Nampei, M.; Natsume, H.; Fukazawa, J.; Kusaba, M.; Ueda, A. A Chromosome-Level Genome Sequence Reveals Regulation of Salt Stress Response in Mesembryanthemum crystallinum. Physiol. Plant. 2025, 177, e70057. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Zhang, Y.; Zhang, L.; Hu, J.; Zhang, X.; Lu, K.; Dong, H.; Wang, D.; Zhao, F.-J.; Huang, C.-F.; et al. OsNRAMP5 Contributes to Manganese Translocation and Distribution in Rice Shoots. J. Exp. Bot. 2014, 65, 4849–4861. [Google Scholar] [CrossRef]
- Yue, J.; Tan, Y.; Wei, R.; Wang, X.; Mubeen, S.; Chen, C.; Cao, S.; Wang, C.; Chen, P. Genome-Wide Identification of bHLH Transcription Factors in Kenaf (Hibiscus cannabinus L.) and Gene Function Analysis of HcbHLH88. Physiol. Mol. Biol. Plants 2024, 30, 1517–1532. [Google Scholar] [CrossRef]
- Nelson, N. Metal Ion Transporters and Homeostasis. EMBO J. 1999, 18, 4361–4371. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Long, T.; Chen, Z.; Liu, J.; Cui, W.; Leng, H.; Xing, Y.; Rodriguez, L.G.; Gao, Y.; Yao, Y. Genome-Wide Identification of NRAMP Family Genes in Populus trichocarpa and Their Roles in Transport of Heavy Metals. Tree Genet. Genomes 2023, 19, 51. [Google Scholar] [CrossRef]
- Kanwal, F.; Riaz, A.; Ali, S.; Zhang, G. NRAMPs and Manganese: Magic Keys to Reduce Cadmium Toxicity and Accumulation in Plants. Sci. Total Environ. 2024, 921, 171005. [Google Scholar] [CrossRef]
- Belouchi, A.; Kwan, T.; Gros, P. Cloning and Characterization of the OsNramp Family from Oryza sativa, a New Family of Membrane Proteins Possibly Implicated in the Transport of Metal Ions. Plant Mol. Biol. 1997, 33, 1085–1092. [Google Scholar] [CrossRef]
- Zhou, G.; An, Q.; Liu, Z.; Bao, W.; Wan, Y. Systematic Analysis of NRAMP Family Genes in Areca Catechu and Its Response to Zn/Fe Deficiency Stress. Int. J. Mol. Sci. 2023, 24, 7383. [Google Scholar] [CrossRef]
- Li, J.; Duan, Y.; Han, Z.; Shang, X.; Zhang, K.; Zou, Z.; Ma, Y.; Li, F.; Fang, W.; Zhu, X. Genome-Wide Identification and Expression Analysis of the NRAMP Family Genes in Tea Plant (Camellia sinensis). Plants 2021, 10, 1055. [Google Scholar] [CrossRef]
- Liu, Q.; Li, S.; Du, G.; An, X. Genome-Wide Analysis of the Nramp Gene Family in Kenaf (Hibiscus cannabinus): Identification, Expression Analysis, and Response to Cadmium Stress. Plants 2024, 13, 2514. [Google Scholar] [CrossRef]
- Zhang, Y.; Sharan, S.; Rinnan, Å.; Orlien, V. Survey on Methods for Investigating Protein Functionality and Related Molecular Characteristics. Foods 2021, 10, 2848. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Li, J.; Guan, J.; Wang, C.; Zhang, Z.; Shi, G. Genome-Wide Identification and Expression Analysis Reveals Roles of the NRAMP Gene Family in Iron/Cadmium Interactions in Peanut. Int. J. Mol. Sci. 2023, 24, 1713. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, Y.; Zheng, L.; Li, Y.; Zhou, X.; Li, J.; Gu, D.; Xu, E.; Lu, Y.; Chen, X.; et al. The Intracellular Transporter AtNRAMP6 Is Involved in Fe Homeostasis in Arabidopsis. Front. Plant Sci. 2019, 10, 1124. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Pan, T.; Zhang, X.; Fan, L.; Quintero, F.J.; Zhao, H.; Su, X.; Li, X.; Villalta, I.; Mendoza, I.; et al. K Efflux Antiporters 4, 5, and 6 Mediate pH and K Homeostasis in Endomembrane Compartments. Plant Physiol. 2018, 178, 1657–1678. [Google Scholar] [CrossRef]
- Segond, D.; Dellagi, A.; Lanquar, V.; Rigault, M.; Patrit, O.; Thomine, S.; Expert, D. NRAMP Genes Function in Arabidopsis thaliana Resistance to Erwinia chrysanthemi Infection. Plant J. 2009, 58, 195–207. [Google Scholar] [CrossRef]
- Lanquar, V.; Ramos, M.S.; Lelièvre, F.; Barbier-Brygoo, H.; Krieger-Liszkay, A.; Krämer, U.; Thomine, S. Export of Vacuolar Manganese by AtNRAMP3 and AtNRAMP4 Is Required for Optimal Photosynthesis and Growth under Manganese Deficiency. Plant Physiol. 2010, 152, 1986–1999. [Google Scholar] [CrossRef]
- Cailliatte, R.; Schikora, A.; Briat, J.-F.; Mari, S.; Curie, C. High-Affinity Manganese Uptake by the Metal Transporter NRAMP1 Is Essential for Arabidopsis Growth in Low Manganese Conditions. Plant Cell 2010, 22, 904–917. [Google Scholar] [CrossRef]
- Cailliatte, R.; Lapeyre, B.; Briat, J.-F.; Mari, S.; Curie, C. The NRAMP6 Metal Transporter Contributes to Cadmium Toxicity. Biochem. J. 2009, 422, 217–228. [Google Scholar] [CrossRef]
- Yamaji, N.; Sasaki, A.; Xia, J.X.; Yokosho, K.; Ma, J.F. A Node-Based Switch for Preferential Distribution of Manganese in Rice. Nat. Commun. 2013, 4, 2442. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, R.; Ishimaru, Y.; Senoura, T.; Shimo, H.; Ishikawa, S.; Arao, T.; Nakanishi, H.; Nishizawa, N.K. The OsNRAMP1 Iron Transporter Is Involved in Cd Accumulation in Rice. J. Exp. Bot. 2011, 62, 4843–4850. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, A.; Yamaji, N.; Yokosho, K.; Ma, J.F. Nramp5 Is a Major Transporter Responsible for Manganese and Cadmium Uptake in Rice. Plant Cell 2012, 24, 2155–2167. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhang, Y.; Huang, C. Reduction in Cadmium Accumulation in Japonica Rice Grains by CRISPR/Cas9-Mediated Editing of OsNRAMP5. J. Integr. Agric. 2019, 18, 688–697. [Google Scholar] [CrossRef]
- Ishimaru, Y.; Bashir, K.; Nakanishi, H.; Nishizawa, N.K. OsNRAMP5, a Major Player for Constitutive Iron and Manganese Uptake in Rice. Plant Signal. Behav. 2012, 7, 763–766. [Google Scholar] [CrossRef] [PubMed]
- Ueno, D.; Sasaki, A.; Yamaji, N.; Miyaji, T.; Fujii, Y.; Takemoto, Y.; Moriyama, S.; Che, J.; Moriyama, Y.; Iwasaki, K.; et al. A Polarly Localized Transporter for Efficient Manganese Uptake in Rice. Nat. Plants 2015, 1, 15170. [Google Scholar] [CrossRef]
- James, A.B.; Sullivan, S.; Nimmo, H.G. Global Spatial Analysis of Arabidopsis Natural Variants Implicates 5′UTR Splicing of Late Elongated Hypocotyl in Responses to Temperature. Plant Cell Environ. 2018, 41, 1524–1538. [Google Scholar] [CrossRef]
- Zhang, T.; Li, C.; Zhu, J.; Li, Y.; Wang, Z.; Tong, C.-Y.; Xi, Y.; Han, Y.; Koiwa, H.; Peng, X.; et al. Structured 3′ UTRs Destabilize mRNAs in Plants. Genome Biol. 2024, 25, 54. [Google Scholar] [CrossRef]
- Nie, F.; Wang, M.; Liu, L.; Ma, X.; Zhao, J. Genome-Wide Identification and Bioinformatics Analysis of the FK506 Binding Protein Family in Rice. Genes 2024, 15, 902. [Google Scholar] [CrossRef]
- Sharma, V.K.; Kumar, N.; Brahmachari, S.K.; Ramachandran, S. Abundance of Dinucleotide Repeats and Gene Expression Are Inversely Correlated: A Role for Gene Function in Addition to Intron Length. Physiol. Genom. 2007, 31, 96–103. [Google Scholar] [CrossRef][Green Version]
- Zhao, Y.; Xie, Q.; Yang, Q.; Cui, J.; Tan, W.; Zhang, D.; Xiang, J.; Deng, L.; Guo, Y.; Li, M.; et al. Genome-Wide Identification and Evolutionary Analysis of the NRAMP Gene Family in the AC Genomes of Brassica Species. BMC Plant Biol. 2024, 24, 311. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Li, W.; Chen, X. Transcription Factor Is Not Just a Transcription Factor. Trends Plant Sci. 2022, 27, 1087–1089. [Google Scholar] [CrossRef]
- Han, H.; Wang, C.; Yang, X.; Wang, L.; Ye, J.; Xu, F.; Liao, Y.; Zhang, W. Role of bZIP Transcription Factors in the Regulation of Plant Secondary Metabolism. Planta 2023, 258, 13. [Google Scholar] [CrossRef]
- Alejandro, S.; Cailliatte, R.; Alcon, C.; Dirick, L.; Domergue, F.; Correia, D.; Castaings, L.; Briat, J.-F.; Mari, S.; Curie, C. Intracellular Distribution of Manganese by the Trans-Golgi Network Transporter NRAMP2 Is Critical for Photosynthesis and Cellular Redox Homeostasis. Plant Cell 2017, 29, 3068–3084. [Google Scholar] [CrossRef]
- Huang, H.; Yamaji, N.; Huang, S.; Ma, J.F. Uptake and Accumulation of Cobalt Is Mediated by OsNramp5 in Rice. Plant Cell Environ. 2025, 48, 3–14. [Google Scholar] [CrossRef]
- Hao, X.; Mo, Y.; Ji, W.; Yang, X.; Xie, Z.; Huang, D.; Li, D.; Tian, L. The OsNramp4 Aluminum Transporter Is Involved in Cadmium Accumulation in Rice Grains. Reprod. Breed. 2022, 2, 125–132. [Google Scholar] [CrossRef]
- Yuce, M.; Ekinci, M.; Turan, M.; Agar, G.; Aydin, M.; Ilhan, E.; Yildirim, E. Chrysin Mitigates Copper Stress by Regulating Antioxidant Enzymes Activity, Plant Nutrient and Phytohormones Content in Pepper. Sci. Hortic. 2024, 328, 112887. [Google Scholar] [CrossRef]
- de Oliveira, R.L.L.; de Mello Prado, R.; Felisberto, G.; Checchio, M.V.; Gratão, P.L. Silicon Mitigates Manganese Deficiency Stress by Regulating the Physiology and Activity of Antioxidant Enzymes in Sorghum Plants. J. Soil Sci. Plant Nutr. 2019, 19, 524–534. [Google Scholar] [CrossRef]
- Tauqeer, H.M.; Ali, S.; Rizwan, M.; Ali, Q.; Saeed, R.; Iftikhar, U.; Ahmad, R.; Farid, M.; Abbasi, G.H. Phytoremediation of Heavy Metals by Alternanthera bettzickiana: Growth and Physiological Response. Ecotoxicol. Environ. Saf. 2016, 126, 138–146. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, S.; Shan, X.-Q.; Christie, P. Combined Toxicity of Cadmium and Arsenate to Wheat Seedlings and Plant Uptake and Antioxidative Enzyme Responses to Cadmium and Arsenate Co-Contamination. Ecotoxicol. Environ. Saf. 2007, 68, 305–313. [Google Scholar] [CrossRef]
- Ye, L.; Yu, J.; Zhang, X.; Yu, F.; Zeng, T.; Gu, L.; Zhu, B.; Wang, H.; Du, X. Physiological, Transcriptomic and Metabolomic Analyses Reveal That Exogenous Arginine Alleviate the Response of Sorghum bicolor L. to Cadmium Stress. Ind. Crops Prod. 2025, 229, 120970. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Renu, K.; Chakraborty, R.; Haritha, M.; Rajeshwari, K.; Famurewa, A.C.; Madhyastha, H.; Balachandar, V.; George, A.; Abilash, V.G. Molecular Mechanism of Heavy Metals (Lead, Chromium, Arsenic, Mercury, Nickel and Cadmium) Induced Hepatotoxicity—A Review. Chemosphere 2021, 271, 129735. [Google Scholar] [CrossRef]
- DeMell, A.; Mendoza, M.R.; Scholthof, H.B. A Tomato Bushy Stunt Virus-Based Vector for Simultaneous Editing and Sensing to Survey the Host Antiviral RNA Silencing Machinery. PNAS Nexus 2023, 3, pgad436. [Google Scholar] [CrossRef] [PubMed]
- Min, X.; Liu, Z.; Wang, Y.; Liu, W. Comparative Transcriptomic Analysis Provides Insights into the Coordinated Mechanisms of Leaves and Roots Response to Cold Stress in Common Vetch. Ind. Crops Prod. 2020, 158, 112949. [Google Scholar] [CrossRef]
- Wang, H.; Ye, L.; Zhou, L.; Yu, J.; Pang, B.; Zuo, D.; Gu, L.; Zhu, B.; Du, X.; Wang, H. Co-Expression Network Analysis of the Transcriptome Identified Hub Genes and Pathways Responding to Saline–Alkaline Stress in Sorghum bicolor L. Int. J. Mol. Sci. 2023, 24, 16831. [Google Scholar] [CrossRef] [PubMed]



| Gene Name | Protein ID | Aa a | MW b kDa | pI d | Natoms | Instability | Aliphatic Index | GRAVY c | Exons | Introns |
|---|---|---|---|---|---|---|---|---|---|---|
| SbNRAMP1 | A0A1B6QIP3 | 466 | 50.75 | 7.16 | 7281 | 26.38 | 127.34 | 0.83 | 6 | 5 |
| SbNRAMP2 | A0A1Z5S5F1 | 486 | 53.06 | 5.99 | 7533 | 33.4 | 116.98 | 0.622 | 5 | 4 |
| SbNRAMP3 | A0A1B6QIN8 | 1236 | 134.71 | 6.02 | 18,946 | 41.62 | 94.73 | 0.048 | 7 | 6 |
| SbNRAMP4 | A0A1B6QJE9 | 546 | 59.95 | 5.57 | 8509 | 38.01 | 107.91 | 0.441 | 4 | 3 |
| SbNRAMP5 | C5WTQ4 | 516 | 56.35 | 6.17 | 8048 | 34.24 | 114.38 | 0.546 | 4 | 3 |
| SbNRAMP6 | A0A1W0W334 | 535 | 58.38 | 7.07 | 8383 | 38.33 | 121.4 | 0.569 | 12 | 11 |
| SbNRAMP7 | C5 × 2P7 | 525 | 56.37 | 6.74 | 8097 | 38.77 | 127.41 | 0.767 | 13 | 12 |
| SbNRAMP8 | C5XKD6 | 1157 | 125.88 | 6.07 | 17,699 | 47.16 | 91.21 | −0.036 | 5 | 5 |
| SbNRAMP9 | A0A194YMG4 | 547 | 59.39 | 6.03 | 8478 | 32.7 | 114.28 | 0.576 | 13 | 12 |
| SbNRAMP10 | A0A1Z5RJE5 | 559 | 60.15 | 8.52 | 8614 | 43.34 | 118.28 | 0.68 | 13 | 13 |
| SbNRAMP11 | C5YQG7 | 544 | 58.89 | 4.89 | 8376 | 32.8 | 111.23 | 0.469 | 4 | 3 |
| SbNRAMP12 | C5Z7T5 | 550 | 59.49 | 8.45 | 8521 | 32.69 | 118.95 | 0.578 | 13 | 12 |
| Protein Name | TM Domains | Subcellular Localization | Alpha Helix (%) | Extended Strand (%) | Beta Turn (%) | Random Coil (%) |
|---|---|---|---|---|---|---|
| SbNRAMP1 | 11 | Plasma membrane | 53.86 | 16.95 | 2.58 | 26.61 |
| SbNRAMP2 | 10 | Plasma membrane | 55.97 | 12.76 | 2.88 | 28.40 |
| SbNRAMP3 | 10 | Plasma membrane. nucleus | 38.35 | 13.35 | 3.16 | 45.15 |
| SbNRAMP4 | 10 | Plasma membrane | 56.23 | 11.72 | 2.93 | 29.12 |
| SbNRAMP5 | 9 | Plasma membrane | 59.11 | 10.47 | 3.49 | 26.94 |
| SbNRAMP6 | 11 | Plasma membrane | 52.90 | 12.71 | 2.99 | 31.40 |
| SbNRAMP7 | 11 | Plasma membrane | 54.29 | 13.52 | 3.24 | 28.95 |
| SbNRAMP8 | 9 | Plasma membrane. Chloroplast | 35.52 | 11.67 | 2.59 | 50.22 |
| SbNRAMP9 | 10 | Plasma membrane | 57.04 | 12.98 | 3.29 | 26.69 |
| SbNRAMP10 | 10 | Plasma membrane | 58.50 | 11.63 | 3.58 | 26.30 |
| SbNRAMP11 | 10 | Plasma membrane | 58.27 | 12.68 | 2.94 | 26.10 |
| SbNRAMP12 | 12 | Plasma membrane | 53.09% | 13.64% | 2.91 | 30.36 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, X.; Li, X.; Zhu, B.; Gu, L.; Zeng, T.; Yu, F.; Liu, L.; Wang, H.; Du, X. Genome-Wide Identification of Natural Resistance-Associated Macrophage Protein (NRAMP) and Expression Analysis Under Heavy Metal Stress in Sorghum bicolor L. Plants 2025, 14, 2660. https://doi.org/10.3390/plants14172660
Hu X, Li X, Zhu B, Gu L, Zeng T, Yu F, Liu L, Wang H, Du X. Genome-Wide Identification of Natural Resistance-Associated Macrophage Protein (NRAMP) and Expression Analysis Under Heavy Metal Stress in Sorghum bicolor L. Plants. 2025; 14(17):2660. https://doi.org/10.3390/plants14172660
Chicago/Turabian StyleHu, Xiaopan, Xiaoxue Li, Bin Zhu, Lei Gu, Tuo Zeng, Feng Yu, Lang Liu, Hongcheng Wang, and Xuye Du. 2025. "Genome-Wide Identification of Natural Resistance-Associated Macrophage Protein (NRAMP) and Expression Analysis Under Heavy Metal Stress in Sorghum bicolor L." Plants 14, no. 17: 2660. https://doi.org/10.3390/plants14172660
APA StyleHu, X., Li, X., Zhu, B., Gu, L., Zeng, T., Yu, F., Liu, L., Wang, H., & Du, X. (2025). Genome-Wide Identification of Natural Resistance-Associated Macrophage Protein (NRAMP) and Expression Analysis Under Heavy Metal Stress in Sorghum bicolor L. Plants, 14(17), 2660. https://doi.org/10.3390/plants14172660







